Abstract
A new metallothionein (MT) gene was cloned from Kandelia candel, a mangrove plant with constitutional tolerance to heavy metals, by rapid amplification of cDNA ends and named KMT, which is composed of two exons and one intron. The full length of KMT cDNA was 728 bp including 121 bp 5′ noncoding domain, 240 bp open reading frame and 384 bp 3′ termination. The coding region of KMT represented a putative 79 amino acid protein with a molecular weight of 7.75 kDa. At each of the amino- and carboxy-terminal of the putative protein, cysteine residues were arranged in Cys–Cys, Cys-X-Cys and Cys-X-X-Cys, indicating that the putative protein was a novel type 2 MT. Sequence and homology analysis showed the KMT protein sequence shared more than 60 % homology with other plant type 2 MT-like protein genes. At amino acid level, the KMT was shown homology with the MT of Quercus suber (83 %), of Ricinus communis (81 %) and of Arabidopsis thaliana (64 %). Function studies using protease-deficient Escherichia coli strain BL21 Star ™(DE3) confirmed the functional nature of this KMT gene in sequestering both essential (Zn) and non-essential metals (Cd and Hg) and the E. coli BL21 with KMT can live in 1,000 μmol/L Zn, 120 μmol/L Hg, and 2,000 μmol/L Cd. The information could provide more details of the causative molecular and biochemical mechanisms (including heavy metal sequestration) of the KMT in K. candel or a scientific basis for marine heavy-metal environment remediation with K. candel. This study also provides a great significance of protecting mangrove species and mangrove ecosystem.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Mangrove ecosystems, possessing great ecological and commercial value, are diverse community in inter-tidal zones of tropical and subtropical coastal rivers, estuaries and bays. Like other wetlands, the mangrove ecosystem has been widely used as sites where effluents are discharged and solid wastes are dumped, including metallic anthropogenic wastes (Peters et al. 1997), and has a large capacity in retaining heavy metals and nutrients (Robertson and Phillips 1995; Tam and Wong 1996). Mangrove ecosystems can act as sinks for heavy metals, which can become pollution sources to plants. Some mangrove plants appear to possess a great tolerance to high levels of heavy metal pollution (Peters et al. 1997; Zhang et al. 2007). As for mangrove plants, previous studies have concentrated on their response to salts tress and water logging, distribution and accumulation of metals, and effect on photosynthesis of heavy metal (Macfarlae and Burchett 2001; Ye et al. 2003). Little information exists on their molecular mechanisms of heavy-metal tolerance. Hence, it is necessary to study the corresponding anti-heavy metal genes including metallothionein (MT) gene.
Metallothioneins (MTs) are low molecular weight proteins with a cysteine (Cys)-rich polypeptide structure and high-affinity for metals, these suggest an important role in the intracellular regulation of these elements (Viarengo 1989). Since MT protein was first isolated from horse kidney in 1957 (Margoshes and Vallee 1957), they have also been found in diverse organisms including fungi, plants, mammals and Cyanobacteria, and shown an extremely heterogeneous composition (Robinson et al. 1993; Yu et al. 1998; Cobbett 2000). The function of MTs is uncertain at present, and considerable evidence has suggested their involvement in metal concentration regulation and detoxification processes. They play important biological functions: (1) trace metal homeostasis, (2) protective role against excess reactive heavy metal ions, (3) free radical scavengers, (4) reservoir of essential metals that can be donated to other metalloproteins and (5) protect the cell against intracellular oxidative damage (Karin 1985). Several studies have shown that MTs may be induced by a variety of stimuli, including elevated concentration of essential and non-essential heavy metals, glucocorticoid hormones, inflammatory agents, and a variety of stress conditions (Winge and Miklossy 1982; Albergoni and Piccinni 1998). According to the location and distribution of the Cys residues in MTs, the deduced MTs proteins from plants are classified into three classes: (i) Class I MTs generally contain two smaller Cys-rich domains (four to eight Cys each) and a large spacer region (30–50 residues) devoid of this amino acids. (ii) Class II MTs are translational monomers in which Cys residues are scattered throughout the entire sequence, such as Ec protein from wheat (Lane et al. 1987). (iii) Class III MTs i.e., phytochelatins, are synthesized enzymatically and consist of peptide chains of variable length (Kotrba et al. 1999; Rauser 1999). Most of plant MTs belong to class I MTs. Based on the distribution of Cys residues, as well as the length of the spacer region, Class I MTs are further sub-divided into four types shown in Table 1. Despite the conformation of the presence of MT genes in various plants, their expression and functions in plants are still unclear, expressing plant MTs in microbial hosts provided important evidence that plant MTs were capable of functioning in metal tolerance. For instance, E. coli transformed with the pea type 1 MT, PsMTa, showed the highest affinity for Cu (Tommey et al. 1991). Arabidopsis MTs have been expressed in MT-deficient strains of yeast and restored the yeast mutant’s heavy metal tolerance (Zhou and Goldsbrough 1994). In addition, some plant’s MTs were found to be involved not only in metal tolerance, but also in Cd detoxification (Cobbett 2000).
Kandelia candel, a species of the mangrove plants, grows in inter-tidal zones of tropical to subtropical coastal rivers, estuaries and bays considered to be major recipients of pollutants with high content of heavy metals, especially (Tam and Wong 1994; Robertson and Phillips 1995; Wong et al. 1995; Tam and Wong 1996). In the last decades, all research for the mangrove have nearly exclusively been concentrated on their physiological and ecological characteristics, and K. candel with high concentration of accumulated metals in tissues have been found (Peters et al. 1997; Macfarlae and Burchett 2001; Zhang et al. 2007). In the case of heavy metal tolerance of K. candel, its underlying mechanisms were still unclear including their corresponding gene. However, at present little information is about these genes including MTs, correlating with heavy metal tolerance, and there is no report on analysis of MTs gene in all mangrove species. In this paper, we aim to resolve some questions as follow: whether there is MTs gene in K. candel; if there is, how structure is the MTs gene; and study on its biological function to different heavy metal.
Materials and methods
Plant materials and metal ion treatment
Viviparous seeds of K. candel were collected from the Neilingding Island-Futian Nature Reserve of Guangdong, and planted in sands at 30/25 °C with a 12 h light/12 h dark cycle. The seedlings were irrigated every 5 days with 2 L liquid fertilizer (half strength Hoagland’s solution contained 10 ‰ NaCl). When two leaves had been grown, the young plants were irrigated with liquid fertilizer (as described above) containing ZnSO4 500 mg/L. After 48 h, the shoots were collected and frozen in liquid nitrogen.
RNA extraction and purification
Total RNA was extracted from the shoots following the procedures of the Concert™ Plant RNA Reagent Kit (Invitrogen). Subsequently, a DNAse I treatment was performed according to the manufacturer’s protocol of TaKaRa (China): using 1U DNase I per 50 μg of total RNA for 30 min at 37 °C. RNA integrity was examined by electrophoresis on a regular 1.0 % agarose gel and stored at −70 °C.
First strand cDNA synthesis
cDNA synthesis was carried out using the 1st Strand cDNA Synthesis Kit (Invitrogen). Ten microliters contained 5 μg of total RNA, 1 μl dNTPs (10 mmol/L), 0.5 μg oligo(dT)12–18 primer, and DEPC treated H2O, and incubated at 65 °C for 5 min, then placed on ice for at least one minuter. Addition of 2 μL 10× RT buffer, 1 U RNase inhibitor, and 40 U SuperScript™ II reverse transcriptase. The first synthesis proceeded at 45 °C for 1 h followed by heat inactivation for 15 min at 70 °C.
Cloning of partial K. candel cDNA
First-strand cDNA was used as a template for reverse transcription polymerase chain reaction (RT-PCR) amplification with degenerate primers designed according to the conservative region in other plants, MT-like proteins (Zhang et al. 2004). The forward primer and the reverse primer were 5′-ATG WSI TGY GGI GGI AAY TG-3′ and 5′-RCA IKT RCA IGG RTY RCA IKT RCA-3′ (W = A/T; S = G/C; Y = C/T; R = A/G; K = G/T; S = A, T, C, G), respectively. PCRs were performed in a total reaction volume of 25 μL containing 10 × PCR buffer, 10 mmol/L dNTPs, 25 mmol/L MgCl2, 1 μ mol/L of each primer and 1.5 U Taq DNA polymerase (Invitrogen). PCR conditions were as follows: an initial denaturation at 94 °C for 4 min, followed by 35 cycles with 94 °C for 40 s, 60 °C for 30 s, 72 °C for 1.5 min and a final extension step of 10 min at 72 °C. PCR products were separated on a 1.5 % agarose gel. Bands were excised and purified with high pure PCR purification kit (TianGen, China), and were then subcloned into pUCm-T Vector (BBI, Canada) and sequenced. The resulting sequence information was used to design gene-specific primers for 3′- and 5′- Rapid amplification of cDNA ends (RACE).
3′-RACE
cDNA synthesis was performed with the kit of 3′RACE System (3′-Full RACE Cort Set, TaKaRa, China). RNA was reversely transcribed using the oligo (dT) anchor primer provided by the kit. 3′-RACE gene specific primer GSP1 (5′-GCC GAG AAG ACC ACT ACC GAG-3′) was designed according to the sequence obtained in step 2.4. PCR was carried out in a total volume of 25 μL containing 5 μL cDNA mixture, 10 pmol of each primer GSP1 and 3 sites Adaptor Primer provided by the kit too, and 1 U TaKaRa Taq polymerase. The PCR reaction was performed under the following conditions: cDNA was denatured at 94 °C for 4 min followed by 35 cycles of amplification (94 °C for 30 s, 55 °C for 30 s, 72 °C for 2 min) and by 10 min at 72 °C. The PCR positive clone was sequenced using M13+/− primers (TaKaRa, China).
5′-RACE
Based on the sequence obtained in step 2.5, the specific primers GSP2 (5′-GCA AGA CTC CCA ACA CAC ACA TAT AGA C-3′), GSP3 (5′-CTT CCC TGT CAC TTT CCC CTC ATT-3′), GSP4 (5′-CTT CCG CAC TTG CAG CCT CCG TTC T-3′) were designed to obtain the 5′ untranslated region of K. candel KMT mRNAs using the GeneRacer™ RACE kit (Invitrogen). First strand cDNA was prepared with primer GSP2 as described above. Terminal deoxynucleotidyl transferase (TdT) was used to add homopolymeric dC-tails to 3′ ends of the cDNA according to the manufacturer’s instructions. The dC-tail cDNA was amplified by PCR using a homopolymer containing anchor primer supplied with the kit and the specific primer GSP3. Then, a second, nested PCR was performed in order to increase PCR specificity, using an abridged amplification primer, supplied with the kit too, and the specific primer GSP4. PCR conditions were adjusted according to the instruction of the kit. PCR products were separated on a 1.5 % agarose gel. Bands were purified from the gel, cloned, colonies screened for appropriately sized inserts and sequenced as described above.
Genomic DNA cloning and analysis
The total genomic DNA was isolated from K. candel. The KMT genomic DNA was amplified by PCR with the following primers: the forward primer (5′- CAC TGA CAT GGA CTG AAG GAG T-3′) and the reverse primer (5′-TGA GCC TAA TTG AAA CTC CT-3′) designed according to the sequences of full-length cDNA (5′- and 3′- untranslated regions, respectively). The PCR was carried out in a programmable heating chamber (Biometra, Germany) for 35 cycles (94 °C for 40 s, 58 °C for 30 s, 72 °C for 1 min) followed by 10 min at 72 °C in 25 μL. The PCR product was purified and cloned into pUCm-T Vector (BBI, Canada) followed by sequencing.
Recombinant DNA manipulations
The plasmid pET100-TOPO, for the expression of KMT in E. coli, was constructed as follow. A 240 bp cDNA fragment containing the full open reading frame of KMT was amplified with the primers (the forward: 5′- CAC CAT GTC TTG CTG TGG TGG AAA CTG T -3′, the reverse: 5′- TCA TTT GCA CGT GCA AGG GTC GCA -3′) by RT-PCR from the RNA of K. candel. According to the manufacturer’s instructions (Champion. pET Directional TOPO® Expression Kits, Invitrogen), the PCR product gel-purified was ligated with the plasmid and the resultant product was used to transform E. coli TOP10 competent cells. Then the transformation mixture was plated on LB agar containing ampicillin at 100 μg/mL. And after the positive clones selected and confirmed were used to transform the protease-deficient E. coli strain BL21 Star ™ (DE3). The E. coli strain BL21 Star ™(DE3) containing the KMT cDNA coding region was named EBKMT, otherwise designated EB.
Sequence analyses
Once the full-length cDNA sequence and genomic DNA sequence of KMT had been determined, the nucleotide sequences were submitted to Genbank. The homology search of the nucleotide sequence obtained, throughout the National Centre for Biotechnology Information (NCBI) database by the basic local alignment search tool (BLAST). A BLAST search calculates an expectation value (E value), which expresses the significance of a math by the number of different alignments with scores equivalent to or better than the current alignment that are expected to occur in a database search by chance. An E value of 1e−5 is considered as a threshold value for a match (Schäffer et al. 1999). Pairwise alignments were performed using BLAST and Clustal W program for generating multiple alignments and dendrogram with the neighbor joining (NJ) method (Saitou and Nei 1987).
Heavy metal tolerance of the recombinant with KMT
The EBKMT and EB was cultured in LB medium at 37 °C by the end of OD600 = 1, then 200 μL of each culture were added 1.0 mmol/L isopropyl β-D-thiogalactoside for induction of protein synthesis, and streaked on plates containing various concentrations of ZnSO4, CuSO4, PbCl2, HgCl2, or CdCl2 for 2 days at 30 °C, and photographed.
Results
Full-length of KMT cDNA sequence
Using degenerate primers (2.4 steps) against well-conserved MT amino acid motifs, PCR was performed with K. candel cDNA as template. PCR product of appropriate size was subcloned and sequenced. Sequence analysis of the PCR fragment revealed that it most likely encoded MT-like homologue. According to this cDNA sequence, specific primer (GSP1) was designed for 3′-RACE. A 531 bp fragment was obtained in which a 3′-untranslated region (UTR) of 384 bp was found downstream from the stop codon (TGA). Three specific primers (GSP2, GSP3, GSP4) designed according to the sequence of 3′-RACE was used for 5′-RACE. A 244 bp fragment was obtained in which a 5′-UTR of 121 bp was found upstream of the start codon (ATG). Upon sequencing, a 728 bp full-length cDNA sequence of K. candel MT-like gene was obtained by assembling the 531 bp of 3′-RACE and the 244 bp of 5′-RACE. The cDNA contained 240 bp open reading frame (ORF) from 121 to 361 bp of the sequence (Fig. 1). 3′-UTR possessed a canonical polyadenylation signal site (AATAAA) and a poly (A) tail, which agrees to the Kozak’s rule of ribosome-binding site consensus sequence (Kozak 1987). The sequence is available in the NCBI Genbank database under accession number DQ414691.
The full-length cDNA sequence and deduced primary structure of MT of K. candel. The first row shows the 121 bp long 5′ UTR. The following ORF starting with a methionine coding ATG (underlined/bold) codes for a protein of 79 residues. The one letter code for amino acids of the deduced primary structure is indicated below the corresponding triplets. The 3′ UTR comprises 367 bp including a polyadenylation signal and a poly(A)-tail (both are bold and double underlined). Arrow indicates the position of intron
Putative amino acids sequence
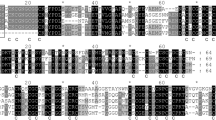
Once the correct reading frame and the initiation and stop translation signals were established, the nucleotide sequence was translated to amino acids, rendering a protein with a theoretical molecular weight of 7.75 kDa, as calculated using the “ProParam” tool, the deduced primary structure yields a protein of 79 amino acids. The amino-terminal and carboxy-terminal domain contained eight and six Cys residues respectively, separated by a central cys free spacer. All the Cys residues were located in both terminal domains with the arrangement of Cys–Cys, Cys-X-Cys and Cys-X-X-Cys, which is a typical of plant class I type 2 MTs (Table 1). The predicted peptide was compared in NCBI and exhibited high homology with type 2 MT of other plants. Sequence and homology analysis showed that the KMT protein sequence shared more than 65 % homology with other plant type 2 MT-like protein genes. The alignment of KMT deduced amino acid sequence with other plant type 2 MT-like proteins was shown in Fig. 2 (Clustal W). We found the KMT was the highest homology (83 %) with the MT clone from Quercus suber (EMBL/GenBank accession number CAC39481) and homology (81 %) with the MT of R. communis (P30564) followed by 77 % with Glycine max (BAD18377), 73 % with Typha latifolia (AAK2802), and 70–65 % with Posidonia oceanica (CAB96155), Avicennia marina (AAK11269), Arabidopsis thaliana (NP_187550), Xerophyta humilis (AAT45000) and with Eichhornia crassipes (CAB53392).
Comparison of the putative amino acid sequence of KMT from K. candel with other plant MT-like proteins. The GenBank accession numbers of the plant proteins are as follows: Q. suber (CAC39481), R. communis (P30564), G. max (BAD18377), A. thaliana (NP-187550), P. oceanica (CAB96155), T. latifolia (AAK2802), X. humilis (AAT45000), A. marina (AAK11269), E. crassipes (CAB53392)
Sequence of KMT gene
A DNA fragment of ~792 bp corresponding to the KMT gene was amplified from K. candel genomic DNA by using PCR (Fig. 3a). The size of the amplification obtained was greater (around 100 bp) than that of the span of primers. This suggests that the amplified fragment contained intron sequences. The PCR product was inserted into pUCm-T Vector. A number of positive clones were obtained and one of them was selected at random for sequencing. Sequence analysis showed that KMT gene coincided with the coding part of the KMT cDNA reported earlier, disrupted by a single intron of 106 bp (Fig. 3b). The 5′ and 3′ intron boundaries were assigned on the basis of the cDNA sequence. The intron boundaries were canonical, begin with GT and end with AG (Fig. 3c). The sequence of the intron showed AT content of 63 %. The exon–intron boundary sequences were 5′-G/GTACA (5′ splice donor) and TGCAG/G-3′ (3′ splice donor). Internal intron similarities are not observed which suggested that they maybe playing a role in the splicing process.
Gene structure and exon/intron boundaries of K. candel. a 1.5 % agarose gel showing an amplified genomic DNA fragment of ~800 bp (lane 1). Lane M is lambda genomic DNA (100 bp + 1.5 kb DNA ladder, BBI, Canada) as marker; b Scheme of the K. candel MT-gene. Exon I comprise the 5′ UTR and the first 22 amino acids encoding part of the polypeptide. Exon II comprise the C-terminal coding part additionally to the 3′ UTR including the polyadenylation signal. Exons and intron are shown to scale; c Exon/intron boundaries of the gene encoding K. candel MT. Conserved 3′ splice sites as well as the 5′ splice sites are marked in black. The intron sequences are indicated with the total intron length (number of base pairs). Five nucleotides of the neighboring exons are shown in addition
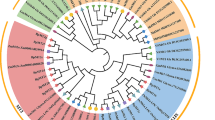
Phylogenetic analysis
To investigate the evolutionary relationships between KMT and other MT-like proteins, a phylogenetic tree was constructed based on the deduced amino acid sequence of predicted KMT and MT-like proteins from other plant species (Table 1). The result showed that KMT shared a cluster with MT-like proteins from G. max, T. latifolia and R. communis, indicating that K. candel has a closer relationship with G. max than with the other two species (Fig. 4). A. thaliana and X. humilis was grouped a cluster, which is the sister clade of the cluster with K. candel. T. latifolia is a marish plant, X. humilis is a xerophile. It may be indicated that KMT might have some unique function, i.e. be in favor of K. candel survived in adversity environment (heavy metals, salt or others stress).
Phylogenetic tree of the K. candel –KMT and other plant MTs-like. All notations are the same as those in Fig. 2
Function of KMT to heavy metal
The pET vector containing the whole ORF of KMT was used for expression of the KMT protein in E. coli BL21 Star ™(DE3) to check whether the KMT gene of K. candel offered the recombinant heavy metal tolerant ability. A vector carrying no KMT gene (EB) was also transformed into the recombinant to serve as negative control. The BL21 transformants were streaked on plates with different concentration of Zn2+ (ZnSO4), Cu2+ (CuSO4), Pb2+ (PbCl2), Hg2+ (HgCl2) and Cd2+ (CdCl2). As shown in Fig. 5, cells transformed with KMT could survive on metal enriched mediums. Furthermore, the expression of KMT gene enabled the cells to be resistant at very high concentrations of heavy metals, as high as 1,000 μmol/L Zn, 120 μmol/L Hg, and 2,000 μmol/L Cd. For Cu and Pb, the tolerances of EBKMT or EB have no difference.
Discussions
Phyto-remediation, due to its cost effectiveness, is a promising technology that uses plants to clean up pollutants (Garbisu and Alkorta 2001). K. candel is studied as bio-accumulator and used as a tool for remediation of heavy metal contaminated environment and for wastewater treatment (Peters et al. 1997; Zhang et al. 2007). However, the molecular mechanism of its heavy metal tolerance is basically unknown. The present study successfully described and characterized of the first MTs gene from this mangrove plant, K. candel. In our experiment, a MTs gene, designated as KMT, had been isolated with a deduced amino acid sequence showing strong homology to deduced type 2 MTs of other plant species (Fig. 4), especially within the amino- and carboxy-terminal domains with less conservation between the central spacer region (43 aa without Cys) linking the two domains. The existence of this central spacer with amino acids is common in most of plant MTs-like proteins, such as 39 aa in Mimulus guttatus (de Miranda et al. 1990), 45 aa in Vicia faba (Foley and Singh 1994) and 41 aa in Typha latifolia (Zhang et al. 2004). The intron with KMT gene was not consistent with other plant MTs-like gene so far, which maybe have unique function to enhance the tolerance of K. candel exposure to heavy metal.
Now, many studies on the function of MTs in higher plants were based on expressing plant MTs in microbial hosts [PsMTa in E. coli (Tommey et al. 1991)], in yeast host [tyMT or mcMT1 in S. cerevisiae mutant (Ma et al. 2003; Zhang et al. 2004)] or transforming animal MTs in plants, such as human MT in tobacco and Brassica napus L. (Misra and Gedamu 1989; Pan et al. 1994). Plant MTs did not only play a role in metal metabolism, but also in detoxification of excess heavy metals (Kotrba et al. 1999; Rauser 1999; Mejare and Bulow 2001). In this paper, the recombinant of E. coli BL21 study (Fig. 5) showed that the KMT gene not only restored the heavy metal tolerance for it, but also offered it with heavy metal tolerant ability, although, no direct evidence for its role in higher plants, which provided further evidence to support the potential of binding heavy metal ions. This was consisted with the results of some previous studies i.e. the correlation between MT gene expression and copper tolerance has been observed in Arabidopsis and Silene ecotypes (Murphy and Taiz 1995; Van Hoof et al. 2001), the ability of some plant MTs to functionally bind copper in both yeast and E. coli cells has been demonstrated (Zhou and Goldsbrough 1994; Foley et al. 1997; Ma et al. 2003), and Zn and Cd also induce some plant MT gene response (Ma et al. 2003; Brkljacic et al. 2004) yielding the corresponding metal-MT aggregates.
The results of Fig. 5 also demonstrated the multi-tolerance ability of the KMT gene to various heavy metals, and this gene was not only limited to the essential metals, such as Zn, but also functional to non-essential metals, for instance, Hg and Cd. This might be one of the mechanisms that help K. candel survive in heavy metal contaminated areas (Wong et al. 1995; Peters et al. 1997). And it appears that the cloned KMT gene is not metal-specific in Zn-induction. Not only copper but also cadmium and Zinc induce MT2a in Arabidopsis (Zhou and Goldsbrough 1995), and Cd, Zn, Fe and Al all elevated the mRNA levels of MT in rice suspension-cultured cells (Hsieh et al. 1995) and in wheat roots (Snowden et al. 1995). Some studies showed that metal exposure did not have any effects on MT-like gene expression (Lane et al. 1987; Choi et al. 1996; Foley et al. 1997). In our experiment, KMT gene of K. candel is induced by Zn, and could be presumed to induce through Cd and Hg from the results of expression in E. coli.
In conclusion, we have isolated and cloned a novel MT-like gene from K. candel. Sequence analysis showed that the gene contains two exons and an intron, and encodes 79 amino acid residues. The alignments with other plant proteins indicate that the polypeptide is belong to type 2 Class I of MTs-like protein in plants. We also expressed the recombinant with KMT in E. coli and demonstrated that the recombinant of KMT was endowed with the tolerance of heavy metal, especially Zn, Cd and Hg. More efforts, however, should be taken to study the distribution and expression of KMT protein, and heavy metal accumulation within the plants. The information could provide more details of the causative molecular and biochemical mechanisms (including heavy metal sequestration) of the KMT in K. candel or shed more information on the diversified nature of similar MT genes in terms of their induction and physiological role to heavy metal tolerance. This study provides a scientific basis for marine heavy-metal environment remediation with K. candel, and is of great significance in protecting mangrove species and mangrove ecosystem.
References
Albergoni V, Piccinni E (1998) Copper and zinc metallothioneins. In: Rainsford KD, Milanino R, Sorenson JRJ, Velo GP (eds) Copper and zinc in inflammation and degenerative dieases. Kluwer Academic, London, pp 61–78
Brkljacic JM, Samardzic JT, Timotijevic GS, Maksimovic VR (2004) Expression analysis of buckwheat (Fagopyrum esculentum Moench) metallothionein-like gene (MT3) under different stress and physiological conditions. J Plant Physiol 161:741–746
Choi D, Kim HM, Yun HK, Park JA, Kim WT, Bok SH (1996) Molecular cloning of a metallothionein-like gene from Nicotiana glutinosa L. and its induction by wounding and tobacco mosaic virus infection. Plant Physiol 112:353–359
Cobbett CS (2000) Phytochelatins and their roles in heavy metal detoxification. Plant Physiol 123:825–832
De Miranda JR, Thomas MA, Thurman DA, Tomsett AB (1990) Metallothionein genes from the flowering plant Mimulus guttatus. FEBS Lett 260:277–280
Foley RC, Singh KB (1994) Isolation of a Vicia faba metallothionein-like gene, expression in foliar trichomes. Plant Mol Biol 26:435–444
Foley RC, Liang ZM, Singh KB (1997) Analysis of type 1 metallothionein cDNAs in Vicia faba. Plant Mol Biol 33:583–591
Garbisu C, Alkorta I (2001) Phytoextraction: a cost-effective plant-based technology for the removal of metals from the environment. Bioresour Technol 77:229–236
Hsieh HM, Liu WK, Huang PC (1995) A novel stress-inducible metallothionein-like gene from rice. Plant Mol Bio 28:381–389
Karin M (1985) Metallothioneins: proteins in search of function. Cell 41:9–10
Kotrba P, Tomas M, Tomas R (1999) Heavy metal-binding peptides and proteins in plant, a review. Collect Czech Chem Commun 64:1057–1086
Kozak MC (1987) At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J Mol Biol 196:947–950
Lane BG, Kajioka R, Kennedy TD (1987) The wheat germ Ec protein is a zinc containing metallothionein. Biochem Cell Biol 65:1001–1005
Ma M, Lau PS, Jia YT, Tsang WK, Lam SKS, Tam NFY, Wong YS (2003) The isolation and characterization of Type 1 metallothionein (MT) cDNA from a heavy-metal-tolerant plant, Festuca rubra cv, Merlin. Plant Sci 164:51–60
Macfarlae GR, Burchett MD (2001) Photosynthetic pigments and peroxidase activity as indicators of heavy metal stress in the grey mangrove, Avicennia marina (Forsk.) Vierh. Mar Pollut Bull 42(2):233–240
Margoshes M, Vallee BL (1957) A cadmium protein from equine kidney cortex. J Am Chem Soc 79:4813–4814
Mejare M, Bulow L (2001) Metal-binding proteins and peptides in bioremediation and phyto-remediation of heavy metals. Trends Biotechnol 19:67–73
Misra S, Gedamu L (1989) Heavy metal tolerant transgenic Brassica napus L. and Nicotiana tabacum L. plants. Theor Appl Genet 78:161–168
Murphy A, Taiz L (1995) Comparison of metallothionein gene expression and nonprotein thiols in ten Arabidopsis ecotypes (Correlation with copper tolerance). Plant Physiol 109:945–954
Pan A, Yang M, Tie F, Li L, Chen Z, Ru B (1994) Expression of mouse metallothionein-I gene confers cadmium resistance in transgenic tobacco plants. Plant Mol Biol 24:341–351
Peters EC, Gassman NJ, Firman JC, Richmond RH, Power EA (1997) Ecotoxicology of tropical marine ecosystems. Environ Toxicol Chem 16:12–40
Rauser WE (1999) Structure and function of metal chelators produced by plants. Cell Biochem Biophys 31:19–48
Robertson AI, Phillips MJ (1995) Mangroves as filter of shrimp pond effluent: prediction and biogeochemical research needs. Hydrobiologia 295:311–321
Robinson NJ, Tommey AM, Kuske C, Jackson PJ (1993) Plant metallothioneins. Biochem J 295:1–10
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Schäffer AA, Wolf YL, Ponting CP, Koonin EV, Aravind L, Altschul SF (1999) IMPALA: matching a protein sequence against a collection of PSI-BLAST-constructed position-specific score matrices. Bioinformatics 15:1000–1011
Snowden KC, Richards KD, Gardner RC (1995) Aluminum-induced genes. Plant Physiol 107:341–348
Tam NFY, Wong YS (1994) Nutrient and heavy metals and plant mineral nutrients. Acta Soc Bot Pol 64(3):265–271
Tam NFY, Wong YS (1996) Retention and distribution of heavy metals in mangroves soils receiving wastewater. Environ Pollut 94:283–291
Tommey AM, Shi WP, Lindsay J, Urwin PE, Robinson NJ (1991) Expression of the pea gene PsMTA in E. coli metal-binding properties of the expressed protein. FEBS Lett 292:48–52
Van Hoof NA, Hassinen VH, Hakvoort HW, Ballintijn KF, Schat H, Verkleij JA, Ernst WH, Karenlampi SO, Tervahauta AI (2001) Enhanced copper tolerance in Silene vulgaris (Moench) Garcke populations from copper mines is associated with increased transcript levels of a 2b type metallothionein gen. Plant Physiol 126:1519–1526
Viarengo A (1989) Heavy metals in marine invertebrates: mechanisms of regulation and toxicity at cellular level. Crit Rev Aquat Sci 1:295–317
Winge DR, Miklossy KA (1982) Domain nature of metallothionein. J Biol Chem 257:3471–3476
Wong YS, Lan CY, Chen GZ, Li SH, Chen XR, Liu ZP, Tam NFY (1995) Effect of wastewater discharge on nutrient contamination of mangrove soils and plant. Hydrobiologia 295:243–254
Ye Y, Tam NFY, Wong MH (2003) Growth and physiological responses of two mangrove species (Bruguiera gymnorrhiza and Kandelia candel) to waterlogging. Environ Exp Bot 49:209–221
Yu LH, Umeda M, Liu JY, Zhao NM, Uchimiya H (1998) A novel MT gene of rice plants is strongly expressed in the node portion of the stem. Gene 206:29–35
Zhang YW, Tan NFY, Wong YS (2004) Cloning and characterization of type 2 metallothionein-like gene from a wetland plant. Typha latifolia 167:869–877
Zhang FQ, Wang YS, Lou ZP, Dong JD (2007) Effect of heavy metal stress on antioxidative enzymes and lipid per oxidation in leaves and roots of two mangrove plant seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere 67:44–50
Zhou J, Goldsbrough PB (1994) Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell 6:875–884
Zhou J, Goldsbrough PB (1995) Structure, organization and expression of the metallothionein gene family in Arabidopsis. Mol Gen Genet 248:318–328
Acknowledgments
This research was supported by the National Natural Science Foundation of China (No. 41176101 and No. 41076070), the projects of knowledge innovation program of Chinese Academy of Sciences (No. KSCX2-SW-132, No. KSCX2-YW-Z-1024, No. KZCX2-YW-Q07-02 and KSCX2-EW-G-12C) and the key projects in the National Science & Technology Pillar Program in the Eleventh Five-year Plan Period (No. 2009BADB2B0606 and No. 2012BAC07B0402).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, FQ., Wang, YS., Sun, CC. et al. A novel metallothionein gene from a mangrove plant Kandelia candel . Ecotoxicology 21, 1633–1641 (2012). https://doi.org/10.1007/s10646-012-0952-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-012-0952-x