Abstract
Given the inherent variability of aquatic systems, predicting the in situ effects of contaminants on such ecosystems still represents a major challenge for ecotoxicology. In this context, transcriptomic tools can help identify and investigate the mechanisms of toxicity beyond the traditional morphometric, physiological and population-level endpoints. In this study, we used the 454 sequencing technology to examine the in situ effects of chronic metal (Cd, Cu) exposure on the yellow perch (Perca flavescens) transcriptome. Total hepatic mRNA from fish sampled along a polymetallic gradient was extracted, reverse transcribed, labeled with unique barcode sequences and sequenced. This approach allowed us to identify correlations between the transcription level of single genes and the hepatic concentrations of individual metals; 71% of the correlations established were negative. Chronic metal exposure was thus associated with a decrease in the transcription levels of numerous genes involved in protein biosynthesis, in the immune system, and in lipid and energy metabolism. Our results suggest that this marked decrease could result from an impairment of bile acid metabolism by Cd and energy restriction but also from the recruitment of several genes involved in epigenetic modifications of histones and DNA that lead to gene silencing.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Human activities have greatly increased the global flux of metals in the Earth’s surficial environment, leading to elevated concentrations notably in aquatic ecosystems. Although considerable progress has been achieved in reducing metal releases to the environment, aquatic organisms living in areas subjected to metal contamination still show evidence of lower condition and overall health (Couture and Kumar 2003; Couture et al. 2008; Levesque et al. 2002; Sherwood et al. 2000). Since it is intrinsically difficult to link observed biological effects to environmental contamination in in situ investigations, past research designed to characterize the effects of contaminants in aquatic environments has historically focused on experimental investigations carried out under short term, acute and simplified exposure conditions (Rasmussen et al. 2008; Schmitt-Jansen et al. 2008). Given that a major aim of ecotoxicology is to measure and predict the effects of contaminants on natural populations, communities and ecosystems, in situ investigations employing suites of biomarkers have received sustained interest over the last 20 years. In the case of metals, such biomarkers include tissue metal concentrations, metallothionein concentrations, in vitro activities of anti-oxidant enzymes, indicators of metabolic capacities and as well as indicators of overall organism health, such as condition index, growth or reproductive endpoints (Zhou et al. 2008). However, metal stress may induce subtle effects at the subcellular level that do not necessarily, or always, translate into changes in these parameters. In addition, these biomarkers may also respond to factors other than environmental contamination, leading to potential ambiguities in their interpretation (Couture et al. 2008; Coyle et al. 2002).
The emergence of the “omics” technologies (proteomics, metabolomics, transcriptomics) now provides ecotoxicologists with promising new tools to investigate the effects of pollutants on organisms (Denslow et al. 2007; Robbens et al. 2007). By allowing the simultaneous measurement of a large number of biological endpoints belonging to several metabolic pathways, these technologies offer a unique research opportunity to reveal and understand new modes and mechanisms of action of toxicants. Among the different technologies, transcriptomic approaches have the potential to detect, characterize and assess the effects of pollutants at the transcriptional level. However, despite the growing interest of ecotoxicologists in gene expression analysis, transcriptomic tools are currently lacking for non-model aquatic species that are perhaps more interesting and more relevant from an ecological and toxicological viewpoint. This has hampered the use of gene expression analysis under field conditions (Denslow et al. 2007).
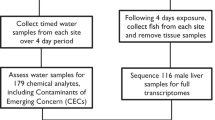
In the present paper, we first present a method that we developed to acquire sequence information by means of next-generation sequencing technology, with the goal of simultaneously identifying genes for which transcription levels were specifically affected by metals in non-model but environmentally relevant natural fish populations of yellow perch (Perca flavescens). Briefly, replicate specimens of yellow perch were collected in the Rouyn-Noranda region (Canada) in four lakes that present a contamination gradient in Cd and Cu concentrations. This region has been subjected to emissions from metal smelters for over 80 years. Total mRNA from the liver of these individuals was extracted, reverse transcribed into complementary DNA (cDNA) and labeled individually with adapters fitted with unique barcode sequences specific to each individual. Then, cDNA samples from each individual fish were pooled in equal amounts and submitted to high-throughput 454 pyrosequencing. In parallel, hepatic Cd and Cu concentrations were determined for each fish. Bioinformatics tools were then used to assemble contigs and identify genes for which transcription levels were significantly affected by metal exposure.
The second goal of this study was indeed to apply this method to obtain quantitative data of gene transcription and carry out correlation analyses between the transcriptional level of a given gene and the hepatic Cd or Cu concentration in each individual fish. Rather than perform comparisons among lake populations, we combined fish from all lakes before carrying out the statistical analyses. Since factors unique to each lake, but unrelated to metal contamination, can also affect the gene expression level, this approach was adopted to strengthen the link between a variation in the transcription level of an individual gene and the contamination level of fish for a given metal.
Materials and methods
Fish sampling
Yellow perch were collected in one clean lake (Opasatica: 48°04′25N, 78°18′10), two moderately metal-contaminated lakes (Adéline: 48°12′17N, 79°10′4; Dufault: 48°18′23′′N, 78°59′58′′O) and one highly metal-contaminated lake (Marlon: 48°15′54′′N, 79°03′53′′O). These lakes are all located within an area corresponding to a 20 km radius. All fish were sampled during April 2009 using a seine net and size-selected to minimize potential allometric bias (length = 6.22 ± 0.06 cm, weight = 2.10 ± 0.06 g, mean ± SE, n = 32, 1 + year old). Eight fish per lake were dissected as soon as possible after their capture (30–60 min) and their livers were immediately stored in liquid nitrogen until needed for analyses. Animal manipulations have been approved by the INRS animal ethics committee.
Liver metal concentrations
Hepatic metal concentrations were determined using inductively coupled plasma mass spectrometry (ICP-MS, Thermo Elemental, Model X-7) according to a method described previously (Pierron et al. 2009). In parallel, certified reference materials from the National Research Council of Canada (TORT2), as well as blanks, were submitted to the same treatment in order to monitor analytical accuracy and recovery.
cDNA preparation for 454 sequencing
Total RNAs were individually extracted from liver of fish using the PureLink™ RNA mini kit (Invitrogen). During this step, samples were submitted to DNAseI treatment, according to the manufacturer’s instructions. Thereafter, the MicroPoly(A)Purist™ Kit was used to remove, at least in part, rRNA and tRNA. Purified mRNAs were then reverse transcribed into cDNA using the SMART™ cDNA Technology kit (Clontech) with the following modification. The primer used for first strand synthesis was a modified oligo-dT primer (5′-AAGCAGTGGTATCAACGCAGAGTTTTCTTTTTTCTTTTTTV–3′). The poly-T stretch is broken by the inclusion of an internal C since long poly(A/T) tails in cDNA may result in sequencing reads of low quality with the Roche GS-FLX DNA Sequencer. Then, cDNA samples were amplified using Advantage 2 Polymerase Mix (Clontech) and SMART™ primer, under the following amplification parameters: 15 min at 37°C followed by 5 min at 94°C and (40 s at 94°C, 1 min at 65°C and 6 min at 72°C) × 17 cycles to not distort the original cDNA profile. At least 2 μg of amplified cDNA were then purified for each individual using the QIAquick PCR Purification Kit (Qiagen). Purified cDNA was fragmented by sonication to produce short, random fragments appropriate for 454 sequencing. To do this, each sample (50 ng μl−1, 50 μl) was submerged in an ice-water cup horn sonicator and sonicated using a Virsonic 475 (Misonix) for at least 1 min (with 30 s bursts followed by 30 s rest periods) with power set at 10%. The effectiveness of the process was checked by gel electrophoresis. When the treatment produced a smear ranging from about 300 to about 800 bp, fragments in this range were selected and purified using the QIAquick Gel extraction Kit (Qiagen). The fragmented cDNA was then polished to obtain blunt-end cDNA molecules and phosphorylated at the 5′-termini by treating simultaneously 100 ng of sample with 3U of T4 DNA polymerase and 3U of T4 polynucleotide kinase in T4 DNA Ligase buffer with 10 mM dNTP and 100 μg ml−1 of BSA (New England Biolabs). The mix was incubated 15 min at 12°C immediately followed by 15 min at 25°C and 20 min at 75°C. To remove enzymes, cDNA fragments were then extracted by phenol/chloroform and precipitated in ethanol. Thereafter, samples were ligated with two unphosphorylated partially double-stranded adaptors (10 μM) containing the standard 454 B primer for the first one and containing the standard 454 A sequence primer with a 10 bp long barcode extension specific of each individual at its 3′ end for the second one (see technical bulletin of Roche No. 005-2009). Adaptors were ligated to fragmented cDNA using 400U of T4 DNA ligase (New England biolabs) during 15 min at 25°C followed by 20 min at 65°C. Following ligation, constructs were purified using the QIAquick Gel extraction Kit (Qiagen) to remove excess adaptors and reagents. Constructs were amplified by PCR using primer A and 5′-biotin-labeled B primer under the following amplification parameters: 94°C for 5 min followed by (94°C for 40 s, 65°C 1 min, 72°C for 3 min) × 17 cycles followed by 72°C for 4 min. Using this protocol, correct constructs containing the A primer on one side and B primer on the other side are preferentially amplified. The final product was again column purified (Qiagen) to remove unincorporated oligonucleotides and quantified using the Quant-iT Picogreen dsDNA Assay Kit (Invitrogen). cDNA from fish of two lakes were pooled in equal amounts to obtain a mix containing more than 5 μg of cDNA. The mixes were visualized on gel electrophoresis and fragments ranging from 350 to 900 bp were purified using the QIAquick Gel extraction Kit (Qiagen) to remove small fragments. The original protocol (Margulies et al. 2005) was then followed from the step of fragment immobilization onto streptavidin beads and mixes were sequenced on two separate half plates on a Roche GS-FLX DNA Sequencer.
Quantitative PCR analyses
For each gene, specific primers were determined (Table S1). First-strand cDNA was synthesized from 500 ng of purified RNA (see above) using the High Capacity RNA-to-cDNA Kit (Applied Biosystems). Real time PCR reactions were performed in duplicate using Sybrgreen in a 7500 Fast Real Time PCR System (Applied Biosystems) following the manufacturer’s instructions. The reaction specificity was determined for each reaction from the dissociation curve of the PCR product and electrophoresis. The relative quantification of each gene expression level was normalized according to the β-actin gene expression as previously described (Pierron et al. 2009). Total RNAs were quantified and 500 ng used for reverse-transcription. During the subsequent qPCR amplifications, the output cycle corresponding to the β-actin gene was investigated. This output was always obtained around the same cycle for control and contaminated individuals, demonstrating the relevance of β-actin as a reference gene under our conditions.
Data treatment
For metal analyses, comparisons among fish groups were performed by analysis of variance (ANOVA), after checking assumptions of normality and homoscedasticity of the error terms. The Least Square Deviation test (LSD) was used to determine whether means between pairs of samples were significantly different from one another. Computations were performed using STATISTICA version 6.1 software (StatSoft, USA). For sequencing data, next-generation sequencing of the prepared cDNA libraries with 454 Life Sciences technology provided us with raw data files in .sff format. Base quality was then extracted from the sff files using PyroBayes beta v0.9 to generate fasta and fasta.qual files containing the sequence and sequence quality information, respectively. The sequences in these files were scanned in order to find and remove the amplification primers and individual tags using an in-house customized Python program (all the code used for the analysis of this article is available upon request). Sequences containing the individual tags were then renamed according to the individual fish from which they originated. Using CLC Genomic Workbench version 3.7 (assembly parameters: de novo assembly, similarity = 0.95, length fraction = 0.5), all the sequences were used in order to create a set of simple contigs, resulting in consensus sequences for 9,996 contigs. The same sequences where then re-assembled using the consensus sequences as a reference set (de novo assembly, similarity = 0.95, length fraction = 0.5).
To annotate these contigs based on similarity with known proteins, they were blasted on both the swissprot and nr protein databases using BLAST 2.2.23+. A suite of Python programs was developed in order to parse these BLAST results and keep only the single most meaningful blast hit for each contig. The meaningfulness of each blast was assessed by using the e-value of the hit and multiplying it by 1 × 10p where p is a penalty based on the presence of certain keywords in the blast hit name. The keywords and associated penalties used are the following: unnamed: 200; unknown: 30; uncharacterized: 30; hypothetical: 20; predicted: 10; similar to: 5; novel protein: 5. This procedure ensured that hits with uninformative content were discarded in favor of more informative and significant hits. Only contigs with an e-value of 1e−30 or lower were kept for further analysis.
Gene transcription was normalized by using RPKM (Reads Per Kilobase per Million mapped reads; Mortazavi et al. 2008). Spearman’s rank correlation coefficient (ρ), as implemented in R’s ‘cor.test’ function (with method = “spearman”) was then used to test for correlation between absolute normalized transcript counts and Cd and Cu contamination levels. In order to increase the power of the analyses, only contigs that were sequenced in at least 20 individuals were analyzed, i.e., 1,582 contigs. The p values were then corrected for multiple testing using the False Discovery Rate (FDR) in the qvalue R package.
Results and discussion
For both Cd and Cu, a significant increase in hepatic metal concentrations was observed along the lake gradient from Opasatica → Adéline → Dufault → Marlon (Fig. 1). Note however that the two metal contamination gradients presented differing intensities: whereas Cd concentrations increased 33-fold from fish sampled in Lake Opasatica compared to those collected in Lake Marlon, this factor only reached 2.9 in the case of Cu concentrations. As in other environmental studies (see Couture et al. 2008 for a review), no significant relationship between metal contamination and fish condition factor was observed (Fig. S1). Fish inhabiting the two moderately metal-contaminated lakes (Ad and Du) displayed a significantly lower condition factor than those inhabiting the clean lake (Op). However, fish inhabiting the highly metal-contaminated lake (Ma) had a Fulton condition factor similar to that of fish from the clean lake (Op).
Concerning sequencing data, more than 778,000 fragments averaging 286 bases in length were sequenced. Individual tags were found in 94.3% of sequenced fragments. After assembly, a total of 9,856 contigs averaging 530 bases were obtained, among which 6,092 could be annotated. For correlation analyses, only contigs sequenced in at least 20 individuals were analyzed; i.e., 1,582 contigs. From these 1,582 contigs, the transcription level of 196 single genes of known function was significantly correlated with hepatic Cd and/or Cu concentrations at P < 0.05 (Table 1 and Tables S2, S3, S4). Examples of such correlations are given in Fig. 2a. Half of these genes were significantly correlated with both metals analyzed. The transcription level of 66 genes was specifically correlated with Cd concentrations and 34 with Cu concentrations. Effects detected in yellow perch were thus mainly associated with Cd, a result consistent with its status as a non-essential metal and with its marked concentration gradient. Interestingly, 71% of these correlations were negative, i.e., the transcription level decreased with increasing metal concentrations. This observation contrasts with laboratory investigations carried out by means of transcriptomic approaches on various fish species experimentally exposed to Cd or Cu and which typically report that the number of up-regulated genes in these fish is either higher or similar to down-regulated genes (Reynders et al. 2006; Santos et al. 2010; Williams et al. 2006). The difference between these laboratory studies and this field investigation presumably reflects the fact that fish in the former are exposed to the metals for short periods at high concentrations, whereas wild fish are chronically exposed. Indeed, whereas short term exposure triggers a common adaptive response that induces genes protecting organisms from toxicity, longer exposure periods bring into play an additional set of genes, the regulation of which allows compensation for the loss of essential pathways specifically caused by the exposure. As the time of exposure increases further, organisms can no longer adapt or compensate and serious adverse effects may occur. In these circumstances, the transcription level of genes involved in growth or reproduction is also affected (Denslow et al. 2007). Moreover, in fish experimentally exposed to increasing Cd or Cu concentrations, Reynerds et al. (2006) and Santos et al. (2010) showed by means of a DNA microarray that the number of up-regulated genes increased with increasing metal concentrations. In contrast to the mostly negative correlations of gene transcription levels with tissue metal concentrations, correlations of transcription levels with fish length and weight were mostly positive (68 and 74% positive correlations, respectively).
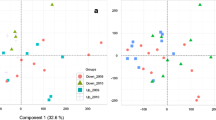
Relationships between the transcription level (a.u.) of complement component C3 (c3), bile salt export pump (bsep), acetyl-coenzyme A synthetase (acs), phosphoenolpyruvate carboxykinase (pepck) and biotinidase (btd) determined from sequencing data and a hepatic metal concentrations (left panel) and b the transcription level of the same genes determined by rt-qPCR (right panel). Spearman coefficient (ρ) and level of significance (P) of correlations are reported on the plots (n = 25–31)
To validate the RNA (cDNA) sequencing data, the transcription level of five genes was measured by quantitative PCR. For all genes tested, expression data obtained by means of 454 sequencing were significantly correlated to those obtained by qPCR (Fig. 2b).
Among genes that showed the most significant correlations with hepatic metal concentrations (Table 1), the three most affected genes encode for rRNA and proteins involved in protein biosynthesis (mitochondrial 16S ribosomal RNA, 60S ribosomal protein L18a, elongation factor 2). The transcription level of these genes was down-regulated in response to chronic metal exposure. The fourth and sixth most strongly down-regulated genes encode for proteins involved in the innate immune system (complement component C3, complement regulatory plasma protein). Finally, among the seven most significant correlations, two genes encoding for enzymes involved in the metabolism of retinol (epidermal retinol dehydrogenase 2) and biliary acids (BAs; bile salt export pump (bsep); Fig. 2a) were also down-regulated. The implications of these variations in gene transcription level with tissue metal concentrations in wild yellow perch sampled along a metal gradient are discussed below.
Retinol and biliary acid metabolism
Retinol, or vitamin A, is a fat-soluble essential nutrient whose dietary absorption requires the presence of BAs (Fig. 3; Noy 2006). BSP is the main protein responsible for the transport of BAs from hepatocytes, i.e., their site of production, to biliary canaliculi (Thomas et al. 2008). In contrast to the negative relationship of bsep transcription level with liver Cd concentration, another gene involved in BA transport, the organic solute transporter alpha (osta), was positively correlated with hepatic Cd concentrations (P = 0.014; Table S3). This enzyme provides an alternative excretion route for BAs into the systemic circulation during abnormal accumulation of BAs into hepatocytes, i.e., cholestasis (Boyer et al. 2006; Thomas et al. 2008). A down-regulation of bsep and an up-regulation of osta are typically observed during chronic cholestasis or after bile duct ligation in mammals (Boyer et al. 2006; Lee et al. 2000). Thus, we hypothesize that chronic exposure to Cd in fish may disturb BA transport, triggering an accumulation of BAs in the hepatocytes. In support of this hypothesis, it has been shown that Cd causes a rapid decrease in bile flow and complete cholestasis in isolated perfused rat livers (Soto et al. 2002). Moreover, cholestasis, by reducing bile flow, triggers malabsorption of dietary retinoids (de Vriese et al. 2001; Weiss et al. 2010). Several studies in rats have shown that dietary retinol associated with chylomicrons is, under normal conditions, taken up by hepatocytes, metabolized into retinaldehyde and retinoic acid by cytosolic alcohol dehydrogenases, retinol dehydrogenases and several aldehyde dehydrogenases before being accumulated, mainly in hepatic stellate cells (Parés et al. 2008). In contrast, in retinol-deficient animals, retinol is directly transferred from hepatocytes to extrahepatic tissues (Blomhoff et al. 1982). Here, whereas the transcription levels of genes encoding for enzymes involved in retinol metabolism were negatively correlated with hepatic metal concentrations, the transcription levels of genes encoding for retinol transport proteins such as transthyretin and sex hormone-binding globulin increased significantly with increasing Cd concentrations (Table S3). These observations support the hypothesis that Cd-contaminated perch could suffer from cholestasis and consequently from retinol depletion. To our knowledge, this is the first study that identifies an impairment in retinol metabolism as a mechanism of Cd toxicity in wild fish. Further studies are required to confirm experimentally this hypothesis. The relationships of retinol metabolism with, and implications for, lipid and energy metabolism are discussed below.
Energy metabolism
In addition to functions listed above, genes encoding for proteins involved in different processes of energy metabolism were down-regulated in response to metal exposure:
-
lipid metabolism (acetyl-coenzyme A synthetase (Fig. 2a), elongation of very long chain fatty acids protein 5, low-density lipoprotein receptor-related protein 1);
-
lipid transport (viglin, apoliporotein B, apolipoprotein A-IV4, long-chain fatty acid transport protein 6);
-
gluconeogenesis (phosphoenolpyruvate carboxykinase [GTP], mitochondrial; Fig. 2a);
-
glycogenesis (UTP-glucose-1-phosphate uridylyltransferase);
-
carbohydrate metabolism and transport (aflatoxin B1 aldehyde reductase member 2, sugar kinase FLJ10986 homolog, solute carrier family 35 member B1).
Such a pattern is consistent with previous studies which showed that yellow perch inhabiting metal-contaminated lakes exhibit lower hepatic triglyceride and glycogen reserves (Levesque et al. 2002). The decrease in the transcription level of genes involved in lipid metabolism and transport is also in agreement with investigations carried out on fish experimentally exposed to Cu (Santos et al. 2010) and with a recent study on Wilson’s disease carried out on mice (Huster et al. 2007). In this disease, the lack of a functional ATP7B (ATPase, Cu2+ transporting, beta polypeptide) leads to Cu accumulation in the liver. Consistent with these studies, genes involved in lipid transport in our study were more consistently down-regulated in response to Cu rather than to Cd contamination (Table 1, Tables S2, S3, S4).
The mechanism by which Cu exerts its toxicity on lipid metabolism is still unclear. In the case of wild yellow perch chronically exposed to metals, down-regulation of genes involved in energy metabolism could also be linked, as evoked earlier, to disturbances in the transport of BAs. Indeed, absorption of dietary lipids requires the presence of BAs. Reduction in bile flow during cholestasis or after bile duct ligation triggers malabsorption of dietary fatty acids (de Vriese et al. 2001). Moreover, it is well established that dietary fatty acids regulate the transcription levels of various genes involved in energy metabolism by activating several families of ligand-activated transcription factors such as the peroxysome proliferator-activated receptor (PPAR; Desvergne and Gerald 2007; Jump et al. 2005; Schoonjans et al. 1995). For example, this could explain why increasing Cd concentrations were associated with a decrease in the transcription level of the gene encoding for acetyl-coenzyme A synthetase, a gene that is known to be up-regulated in response to PPAR activation by dietary fatty acids (Schoonjans et al. 1995). In addition, previous studies have reported that in situ metal exposure impairs aerobic capacities of metal-contaminated yellow perch (Couture and Kumar 2003; Garceau et al. 2010; Pierron et al. 2009). Since lipids are the main fuel of aerobic ATP production, a Cd-induced impairment in lipid metabolism may be involved in the lower aerobic capacities reported in contaminated wild fish.
In the present study, most of the genes encoding for mitochondrial proteins or mitochondrial rRNAs were down-regulated in response to metal exposure (14 genes down-regulated versus 4 genes up-regulated). Interestingly, the up-regulated genes encode for proteins embedded in the inner membrane and for a stress response matrix protein (10 kDa heat shock protein, HSP10). With the exception of hsp10, all genes encoding for proteins located in the mitochondrial matrix were down-regulated. For example, for ATP synthase, genes encoding for membrane subunits (c and f) were up-regulated while genes encoding for subunits located in the matrix (β and γ) were down-regulated (Table 1, Tables S2, S3). As previously described in yellow perch liver (Pierron et al. 2009), such an increase in the transcription levels of genes encoding for membrane subunits could reflect a compensatory mechanism to counteract the toxic effects of Cd and Cu on the mitochondrial inner membrane (Garceau et al. 2010; Pierron et al. 2009). Furthermore, the mitochondrial inner membrane is also known to be a primary target of BA toxicity during cholestasis (Rolo et al. 2000). Such mechanisms of metal toxicity support the observation that yellow perch inhabiting metal-contaminated lakes exhibit a lower growth efficiency (growth increment relative to food consumption rate) compared to fish from clean lakes (Sherwood et al. 2000). Whatever the mechanisms implicated in the down-regulation of genes involved in energy metabolism, our results coupled with previous studies of yellow perch at physiological and biochemical levels strongly support the conclusion that in situ exposure to Cd and Cu leads to a reduction in the energy status of fish, probably at least in part by impairing oxidative phosphorylation and nutrient absorption.
Immune system
Among the most significant correlations established (Table 1), all genes encoding for proteins involved in immunity (n = 6, complement component C3, complement regulatory plasma protein, complement component C3-2, complement component C9, SBCFR-1 protein and C-type lectin domain family 4 member M) were negatively correlated to liver metal concentrations, especially Cd. While Cd was negatively correlated to the transcription level of these six genes, liver Cu concentration was only negatively correlated with three genes. These observations are consistent with the known immunotoxicological effects of Cd and with previous laboratory studies on fish experimentally exposed to Cd (Reynders et al. 2006; Williams et al. 2006). Indeed, these experimental investigations carried out by means of DNA microarrays and suppression subtractive hybridization-PCR also reported a down-regulation of immune-function related genes in response to Cd exposure, notably a down-regulation of complement C3. However, in our case, an alternative hypothesis could be that chronic in situ metal exposure decreases the prevalence of infectious diseases and this, whether directly, by eliminating pathogens or indirectly, by eliminating intermediary hosts that are needed to complete the parasite’s biological cycle. For example, Blanar et al. (2010) have shown that the parasite Discocotyle sagitta is more susceptible to Cu toxicity than is its host fish, Salmo salar, leading to lower infection rates. In highly metal-contaminated lakes of the Rouyn-Noranda region, previous studies have shown that chronic metal pollution leads to the elimination of large metal-sensitive benthic invertebrates, which are possible intermediary hosts of parasites (Rasmussen et al. 2008). In addition, since Scantlebury et al. (2001) have shown that retinoic acid is a potent inducer of complement C3 expression (Fig. 2a), a plasma protein that plays the primary role in the activation of the complement system, disturbances described earlier for genes involved in retinol metabolism and transport could also contribute to the decrease in the transcription level of genes involved in the innate immune system, notably complement C3.
Epigenetics
In contrast to down-regulated genes involved in retinol and energy metabolism, the transcription level of several genes involved in the epigenetic modifications of histones were positively correlated to liver metal concentrations, and these were among the most significant correlations (Table 1). Such genes included VHSV-induced protein-10, poly (ADP-ribose) polymerase (parp) and biotinidase (btd; Fig. 2a). These genes are involved in epigenetic mechanisms that lead to gene silencing through chromatin condensation (Liu et al. 2008). VHSV-induced protein-10 and PARP catalyze the attachment of ADP-ribose units onto target proteins, including histones (Liu et al. 2008). Concerning BTD, several authors have recently proposed an enzymatic mechanism in which BTD acts as biotinyl transferase, leading to histone biotinylation (Peters et al. 2002). Interestingly, glycine N-methyltransferase (gnmt), another gene indirectly involved in histone and DNA modifications but in this case by methylation, was negatively correlated with Cd concentrations (Table 1). GNMT is the key cytosolic enzyme that regulates S-adenosylmethionine (SAM), the common substrate of numerous methyltransferases, including histone and DNA methyltransferases. GNMT disposes of SAM by forming the essentially inactive metabolite sarcosine (Rowling et al. 2002). Previous studies carried out in vitro have shown that long-term low-dose Cd exposure triggers DNA methylation in human embryo lung fibroblast cells (Jiang et al. 2008). Again, such DNA modifications are known to be associated with gene silencing (Liu et al. 2008). These epigenetic phenomena could explain, at least in part, why correlations established between metal concentrations and gene transcription levels were mostly negative (71%).
As epigenetics is still in its infancy, cellular signals and consequent mechanisms activating gene silencing are poorly known. Nevertheless, several studies have shown that gene silencing is stimulated by various stresses, especially by DNA damage (Liu et al. 2008, Pawlak and Deckert 2007). However, in our case, no markers of DNA damage or oxidative stress were increased by metal contamination. On the contrary, the expression of histone H2Ax, a member of the histone H2A family that is known to recruit repair factors after DNA damage, was down-regulated by Cu exposure (Paull et al. 2000, Table 1). In the case of GNMT, previous research carried out on rats reported that retinol up-regulates gnmt transcription levels, leading to DNA hypomethylation (Rowling et al. 2002). The observed decrease in gnmt transcription level with increasing Cd concentrations is thus consistent with an impaired retinol metabolism. For BTD, Peters et al. (2002) proposed that destabilization of the inner mitochondrial membrane following UV irradiation in Jurkat cells triggers a leak of the breakdown product of carboxylases, which are the substrates of BTD, from mitochondria to the cytoplasm, leading to biotinylation of histones. Such a pattern would be consistent with the fact that the mitochondrial inner membrane is a primary target of both metal and BA toxicity (Garceau et al. 2010; Pierron et al. 2009; Rolo et al. 2000).
Protein biosynthesis
In our study, several genes involved in protein biosynthesis (n = 35), including rRNA genes (n = 12), were negatively correlated with metal exposure. In contrast, in fish experimentally exposed to Cu or Cd, the transcription level of genes involved in protein biosynthesis is normally mainly up-regulated (Santos et al. 2010; Williams et al. 2006). This discrepancy between our results and experimental data could be explained, at least in part, by the reduction in the energy status reported for metal-contaminated wild yellow perch. In eukaryotic cells, ribosome production indeed represents the most-energy consuming process, which adapts to changes in intracellular energy status (Murayama et al. 2008). Moreover, recent studies have shown that reduction in energy status leads to deacetylation and methylation of histones, establishing silent chromatin in rDNA loci (Murayama et al. 2008). Epigenetic phenomena impeding gene expression, evoked earlier, could thus represent a response to energy restrictions engendered by long-term metal exposure. During experimental investigations (Santos et al. 2010; Williams et al. 2006), fish (European flounder, stickleback) were exposed to Cd or Cu either by a unique intraperitoneal injection or by waterborne exposure for 1–16 days. This short exposure time could be insufficient to trigger energy depletion in experimental fish. In direct support of this hypothesis, Takiguchi et al. (2003) using rat liver cells showed that whereas prolonged exposure to Cd (10 weeks) triggers DNA hypermethylation, a short term exposure (1 week) to the same conditions induces, on the contrary, DNA hypomethylation. It is therefore likely that the opposite responses of the regulation of genes involved in protein biosynthesis in short-term laboratory exposures compared to chronic exposure in this in situ study are due to the different exposure durations.
Conclusion
Our study illustrates how next-generation sequencing technology can be used to detect, assess and characterize the effects of stressors on the transcriptome. This approach allowed us to reveal new mechanisms of toxicity and action of Cd and Cu in fish populations chronically exposed to in situ metal contamination. Moreover, this approach allowed us to identify genes for which transcription levels are specifically affected by a given metal. Effects detected in yellow perch were mainly associated with Cd, a result consistent with its status as a non-essential metal and with its marked concentration gradient in the system studied. Our results suggest that chronic exposure to Cd and Cu is responsible for disturbances in the innate immune system and in retinol, biliary acid and energy metabolism. Such effects, coupled with effects previously reported at the physiological and biochemical levels, indicate that chronic in situ metal exposure triggers energy restriction. This energy restriction coupled with the recruitment of genes involved in epigenetic modifications of histones and DNA, responsible for chromatin condensation and gene silencing, could explain the numerous negative correlations established between gene transcription levels and hepatic metal concentrations in wild yellow perch. We should add that the possibility of an effect of chronic metal exposure on genetic evolution cannot be discounted (see Hoffmann and Willi 2008 for a review). Although an earlier study carried out on yellow perch in the Rouyn-Noranda region, using neutral microsatellite DNA markers, reported low to moderate differentiation among yellow perch populations inhabiting different lakes with differing metal levels, this study also demonstrated a negative relationship between levels of genetic diversity within populations and the level of metal contamination (Bourret et al., 2008). These previous results suggest that trace metals may have had an evolutionary impact on the genetic makeup of these populations. Thus, in addition to the processes listed above, an effect of structural genomic changes in the expression of genes encoding for proteins involved in resistance and adaptation to environmental stresses cannot be excluded.
References
Blanar C, MacLatchy DL, Kieffer JD, Munkittrick KR (2010) Exposure to a mixture of zinc and copper decreases survival and fecundity of Discocotyle sagittata (Leuckart) parasitizing juvenile Atlantic salmon, Salmo salar L. Bull Environ Contam Toxicol 84:692–697
Blomhoff R, Helgerud P, Rasmussen M, Berg T, Norum KR (1982) In vivo uptake of chylomicron [3H]retinyl ester by rat liver: evidence for retinol transfer from parenchymal to nonparenchymal cells. Proc Natl Acad Sci USA 79:7326–7330
Bourret V, Couture P, Campbell PGC, Bernatchez L (2008) Evolutionary ecotoxicology of wild yellow perch (Perca flavescens) populations chronically exposed to a polymetallic gradient. Aquat Toxicol 86:76–90
Boyer J, Trauner M, Mennone A, Soroka CJ, Cai S-Y, Moustafa T, Zollner G, Lee JY, Ballatori N (2006) Upregulation of a basolateral FXR-dependent bile acid efflux transporter OSTalpha-OSTbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol 290:1124–1130
Couture P, Kumar PR (2003) Impairment of metabolic capacities in copper and cadmium contaminated wild yellow perch (Perca flavescens). Aquat Toxicol 64:107–120
Couture P, Busby P, Gauthier C, Rajotte JW, Pyle GG (2008) Seasonal and regional variations of metal contamination and condition indicators in yellow perch (Perca flavescens) along two polymetallic gradients: I factors influencing tissue metal concentrations. Hum Ecol Risk Assess 14:97–125
Coyle P, Philcox JC, Carey LC, Rofe AM (2002) Metallothionein: the multipurpose protein. Cell Mol Life Sci 59:627–647
de Vriese S, Savelii JL, Poisson JP, Narce M, Kerremans I, Lefebvre R, Dhooge WS, De Greyt W, Christophe AB (2001) Fat absorption and metabolism in bile duct ligated rats. Ann Nutr Metab 45:209–216
Denslow N, Garcia-Reyero NA, Barber DS (2007) Fish ‘n’ chips: the use of microarrays for aquatic toxicology. Mol Biosyst 3:172–177
Desvergne B, Gerald L (2007) RXR: from partnership to leadership in metabolic regulations. Vitam Horm 75:1–32
Garceau N, Pichaud N, Couture P (2010) Inhibition of goldfish mitochondrial metabolism by in vitro exposure to Cd, Cu and Ni. Aquat Toxicol 98:107–112
Hoffmann AA, Willi Y (2008) Detecting genetic responses to environmental change. Nat Rev Genet 9:421–432
Huster D, Purnat TD, Jason L, Burkhead JL, Ralle M, Fiehn O, Stuckert F, Olson NE, Daniel Teupser D, Lutsenko S (2007) High copper selectively alters lipid metabolism and cell cycle machinery in the mouse model of Wilson disease. J Biol Chem 282:8343–8355
Jiang G, Xu L, Song S, Zhu C, Wu Q, Zhang L, Wu L (2008) Effects of long-term low-dose cadmium exposure on genomic DNA methylation in human embryo lung fibroblast cells. Toxicology 244:49–55
Jump D, Botolin D, Wang Y, Xu J, Christian B, Demeure O (2005) Fatty acid regulation of hepatic gene transcription. J Nutr 135:2503–2506
Lee J, Trauner M, Soroka CJ, Stieger B, Meier PJ, Boyer JL (2000) Expression of the bile salt export pump is maintained after chronic cholestasis in the rat. Gastroenterology 118:163–172
Levesque HM, Moon TW, Campbell PGC, Hontela A (2002) Seasonal variation in carbohydrate and lipid metabolism of yellow perch (Perca flavescens) chronically exposed to metals in the field. Aquat Toxicol 60:257–267
Liu L, Li Y, Tollefsbol TO (2008) Gene-environment interactions and epigenetic basis of human diseases. Curr Issues Mol Biol 10:25–36
Margulies M et al (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–380
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, Fukamizu A, Kimura K, Shimizu T, Yanagisawa J (2008) Epigenetic control of rDNA loci in response to intracellular energy status. Cell 133:627–639
Noy N (2006) Vitamin A, biochemical, physiological, & molecular aspects of human nutrition. In: Stipanuk MH (ed) Biochemical, physiological & molecular aspects of human nutrition. Saunders Elsevier, St Louis
Parés X, Farrés J, Kedishvili N, Duester G (2008) Medium- and short-chain dehydrogenase/reductase gene and protein families: medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell Mol Life Sci 65:3936–3949
Paull T, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM (2000) A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 10:886–895
Pawlak S, Deckert J (2007) Histone modifications under environmental stress. Biol Lett 44:65–73
Peters D, Griffin JB, Stanley J, Beck MM, Zempleni J (2002) Exposure to UV light causes increased biotinylation of histones in Jurkat cells. Am J Physiol Cell Physiol 283:878–884
Pierron F, Bourret V, St-Cyr J, Campbell PGC, Bernatchez L, Couture P (2009) Transcriptional responses to environmental metal exposure in wild yellow perch (Perca flavescens) collected in lakes with differing environmental metal concentrations (Cd, Cu, Ni). Ecotoxicology 18:620–631
Rasmussen JB, Gunn JM, Sherwood GD, Iles A, Gagnon A, Campbell PGC, Hontela A (2008) Direct and indirect (foodweb mediated) effects of metal exposure on the growth of yellow perch (Perca flavescens): implications for ecological risk assessment. Hum Ecol Risk Assess 14:317–350
Reynders H, van der Ven K, Moens LN, van Remortel P, De Coen WM, Blust R (2006) Patterns of gene expression in carp liver after exposure to a mixture of waterborne and dietary cadmium using a custom-made microarray. Aquat Toxicol 80:180–193
Robbens J, Van der Ven K, Maras M, Blust R, De Coen W (2007) Ecotoxicological risk assessment using DNA chips and cellular reporters. Trends Biotechnol 25:460–466
Rolo A, Oliveira PJ, Moreno AJM, Palmeira CM (2000) Bile acids affect liver mitochondrial bioenergetics: Possible relevance for cholestasis therapy. Toxicol Sci 57:177–185
Rowling M, McMullen MH, Schalinske KL (2002) Vitamin A and its derivatives induce hepatic glycine N-methyltransferase and hypomethylation of DNA in rats. J Nutr 132:365–369
Santos EM, Ball JS, Williams TD, Wu H, Ortega F, van Aerle R, Katsiadaki I, Falciani F, Viant MR, Chipman JK, Tyler CR (2010) Identifying health impacts of exposure to copper using transcriptomics and metabolomics in a fish model. Environ Sci Technol 44:820–826
Scantlebury T, Sniderman AD, Cianflone K (2001) Regulation by retinoic acid of acylation-stimulating protein and complement C3 in human adipocytes. Biochem J 356:445–452
Schmitt-Jansen M, Veit U, Dudel G, Altenburger R (2008) An ecological perspective in aquatic ecotoxicology: approaches and challenges. Basic Appl Ecol 9:337–345
Schoonjans K, Watanabe M, Suzuki H, Mahfoudi A, Krey G, Wahli W, Grimaldi P, Staels B, Yamamoto T, Auwerx J (1995) Induction of the acyl-coenzyme A synthetase gene by fibrates and fatty acids is mediated by a peroxisome proliferator response element in the C promoter. J Biol Chem 270:19269–19276
Sherwood GD, Rasmussen JB, Rowan DJ, Brodeur J, Hontela A (2000) Bioenergetic costs of heavy metal exposure in yellow perch (Perca flavescens): in situ estimates with a radiotracer (137Cs) technique. Can J Fish Aquat Sci 57:441–450
Soto A, Foy BD, Frazier JM (2002) Effect of cadmium on bromosulfophthalein kinetics in the isolated perfused rat liver system. Toxicol Sci 69:460–469
Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP (2003) Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res 286:355–365
Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K (2008) Targeting bile-acid signaling for metabolic diseases. Nat Rev Drug Discov 7:678–693
Weiss B, Barshack I, Onaca N, Goldberg I, Berkovich Z, Melzer E, Jonas A, Reifen R (2010) Vitamin A deficiency associated with enhanced proliferation of bile duct epithelial cells in the rat. Isr Med Assoc J 12:82–86
Williams TD, Diab AM, George SG, Godfrey RE, Sabine V, Conesa A, Minchin SD, Watts PC, Chipman JK (2006) Development of the GENIPOL European flounder (Platichthys flesus) microarray and determination of temporal transcriptional responses to cadmium at low dose. Environ Sci Technol 40:6479–6488
Zhou Q, Zhang J, Fu J, Shi J, Jiang G (2008) Biomonitoring: an appealing tool for assessment of metal pollution in the aquatic ecosystem. Anal Chim Acta 606:135–150
Acknowledgments
We would like to thank Maxime Bélanger and Charles Gauthier who participated in the field sampling. This work was supported by a Collaborative Research and Development (CRD) grant jointly funded by the Natural Sciences and Engineering Research Council (NSERC) of Canada and by Vale-Inco Ltd. LB and PGCC are supported by the Canada Research Chair program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pierron, F., Normandeau, E., Defo, M.A. et al. Effects of chronic metal exposure on wild fish populations revealed by high-throughput cDNA sequencing. Ecotoxicology 20, 1388–1399 (2011). https://doi.org/10.1007/s10646-011-0696-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-011-0696-z