Abstract
Background
Exosomes are potential tools for disease control by regulating intercellular communication through carrying proteins and RNAs between cells or remote organs. Exosome activities have aroused wide concerns in cancer biology and malignancy control.
Aims
This study was performed to explore the roles of mesenchymal stem cell (MSC)-derived exosomes in colorectal cancer (CRC) progression.
Methods
MSC-exosomal microRNAs (miRNAs) in CRC tissues were analyzed, and aberrantly expressed miRNAs in CRC tissues were obtained from the data available on the GEO database. Altered expression of miR-3940-5p was introduced to identify its role in CRC invasion and metastasis in both cell and animal models. The binding relationship between miR-3940-5p and Integrin alpha6 (ITGA6) was predicted on TargetScan and validated through a luciferase assay. The effects of ITGA6 on CRC were figured out.
Results
MSC-derived exosomes carried miR-3940-5p into CRC cells. Up-regulation of miR-3940-5p inhibited epithelial–mesenchymal transition (EMT) and invasion of CRC cells, and suppressed the tumor metastasis and growth in vivo. miR-3940-5p was found to directly bind to ITGA6. Overexpression of ITGA6 promoted CRC cell invasion and EMT and tumor progression through upregulating the transforming growth factor-beta1 (TGF-β1) signaling. A TGF-β1-specific antagonist, Disitertide, blocked the functions of ITGA6 both in vivo and in vitro.
Conclusion
MSC-exosomal miR-3940-5p inhibits invasion and EMT of CRC cells as well as growth and metastasis of tumors through targeting ITGA6 and the following TGF-β1 inactivation. This study may provide novel insights into exosome-based treatment for CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer diagnosis and treatment has been a major challenge in recent society. In China, the cancer mortality rate has not seen a significant decline, which has exceeded the world average by 17% [1]. Colorectal cancer (CRC) ranks the 5th most common cancer among all cancer types, and most cases were diagnosed at an old age though patients are also increasingly becoming younger [2]. Although the treating strategies have seen a significant improvement, the overall 5-year survival rate of CRC patients in China remains unfavorable at 31% mainly due to the late diagnosis, recurrence, and metastasis of tumor cells [3]. Metastasis is the most lethal feature of malignant tumors that is responsible for approximately 90% of tumor-related mortality including in CRC-related death [4, 5]. Identifying novel interventions or molecules for metastasis control is of great importance to improve the outcome of CRC patients.
Recently, the ubiquitous mesenchymal stem cells (MSCs) in many tissues have been widely employed in clinical studies because of their unique advantages [6]. They can migrate to the tumor surrounding stroma and fulfill complex functions on tumor growth and progression with the involvement of extracellular vesicles (Evs), mainly exosomes [6, 7]. Exosomes are 30–100 nm Evs abundantly existed in numerous body fluids that can mediate cell-to-cell communication via carrying proteins and RNAs between cells or remote organs [8]. The MSC-derived exosomes (MSC-exos) can exert key functions in cancer progression by carrying microRNAs (miRNAs) and involving a wide array of molecules [9, 10]. The cargoes of exosomes were suggested to participate in several cancer-related processes including metastasis, epithelial–mesenchymal transition (EMT) and drug resistance of CRC [11, 12]. In light of these findings, this study aimed to identify the roles of MSC-exos in CRC progression using human umbilical cord mesenchymal stem cells (hUC-MSCs). Our study identified miR-3940-5p as an MSC-exosomal miRNA that was poorly expressed in CRC tissues. Though it is a relatively seldom studied one, this miRNA has been identified as a tumor suppressor in non-small cell lung cancer [13]. More specifically, miR-3940-5p was reported to be poorly expressed in CRC patient serum and closely linked to prognosis of patients [14]. However, its exact role in CRC metastasis remains unknown yet. In addition, our study confirmed integrin α6 (ITGA6) as a target of miR-3940-5p. ITGA6 has been suggested to promote cancer progression and metastasis in several malignancies such as metastatic breast cancer [15], and CRC [16]. This is potentially due to its function in cancer stem cell maintenance and regulation in tumorigenic microenvironment [17]. High expression of ITGA6 was suggested to be correlated with worse global survival in CRC patients [18]. In addition, the transforming growth factor-beta1 (TGF-β1), reported as an oncogenic signaling that triggers cancer cell migration and invasion [19, 20], was found to be influenced by ITGA6 in this paper. Thereby, we speculated that MSC-exos inhibit CRC metastasis and progression by carrying miR-3940-5p with the involvement of ITGA6 downregulation and TGF-β1 inactivation. Then, gain- and loss-of functions of these molecules were performed in in vitro and in vivo experiments to validate this hypothesis and to figure out the mechanism of action.
Materials and Methods
Bioinformatics Analysis
The miRNAs in MSC-exos and in CRC tissues were searched on EVmiRNA DataBase (http://bioinfo.life.hust.edu.cn/EVmiRNA/#!/). Differentially expressed miRNAs between CRC tissues and normal colon tissues were analyzed using the Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) available on national center for biotechnology information (NCBI). The potential target messenger RNAs (mRNAs) of exosomal miRNA were predicted on TargetScan (http://www.targetscan.org/), and the expression of candidate mRNAs in CRC and tumor tissues was analyzed on http://gepia.cancer-pku.cn/.
Cell Culture and Transfection
hUC-MSCs, CRC cell lines HT-29 and DLD-1, and normal colon cell line FHC were all acquired from American Type Culture Collection (ATCC, USA). The hUC-MSCs were cultured in serum-free medium (Gibco Company, Grand Island, NY, USA), while CRC and normal colon cell lines were in Roswell Park Memorial Institute (RPMI)-1640 supplemented with 10% fetal bovine serum (FBS) in a constant incubator (37 °C, 5% CO2). miR-3940-5p mimic/inhibitor, small interfering RNA (siRNA) targeting ITGA6, pcDNA ITGA6, and the corresponding control vectors were purchased from Gene Pharma Co., Ltd. (Shanghai, China). A TGF-β1-specific antagonist, Disitertide, was acquired from Med Chem Express (Monmouth Junction, NJ, USA). The above vectors were co-cultured with cells for 48 h (h) and then cultivated in 3 μg/mL puromycin-supplemented medium for 10–14 days (d) to screen out the cells harboring stable transfection. Then, the transfection efficiency was determined based on the RNA expression measured by reverse transcription quantitative polymerase chain reaction (RT-qPCR).
RT-qPCR
Total RNA from all cells was extracted using the TRIzol Reagent (Invitrogen, Inc., Carlsbad, CA, USA) and then reversely transcribed into complementary DNA (cDNA) using a PrimeScript™RT Master Mix (TaKaRa Holdings Inc., Kyoto, Japan). RT-qPCR was performed using SYBR Premix Ex Taq™II (TaKaRA) on a StepOnePlus real-time PCR System (Life Technologies, Foster, CA, USA). The expression ratio between genes in experimental group and control group was determined by the 2−∆∆Ct method. U6 small nuclear RNA (snRNA) was served as the internal reference for miRNA and glyceraldehyde-3-phosphate dehydrogenase for mRNAs. Three independent experiments were performed to minimize errors. Primer sequences are presented in Table 1.
Separation and Purification of Exosomes
The acquired hUC-MSCs were cultured in serum-free medium. When the cell confluence reached 90%, the cells were washed in phosphate-buffered saline (PBS) and then cultured in Dulbecco’s modified Eagle’s medium (System Biosciences, PA, USA) supplemented with 10% exosome-depleted FBS for 72 h. Then, the medium was collected and centrifuged at 300g at 4 °C for 20 min, and then at 2000g at 4 °C for 20 min (min) to remove the cells, dead cells and debris. The supernatant was centrifuged at 100,000g at 4 °C for 70 min to precipitate the exosomes. The precipitates were washed twice in PBS, resuspended in 100 μL PBS and preserved at − 80 °C.
Transmission Electron Microscopy (TEM) Observation
The separated particles were fixed in 2% glutaraldehyde at 4 °C overnight. After PBS washes, the particles were fixed in 1% OsO4 for 1 h, dehydrated in ethanol, embedded in epoxy, and cut into sections. The sections were further treated with saturated sodium periodate, 0.1 N hydrochloric acid, and then observed under a TEM (JEM-1010; JEOL, Tokyo, Japan) 10 min later.
Nanoparticle Tracking Analysis (NTA)
The particle size and distribution were further determined using an NTA analyzer (Malvern, UK). The machine was prewarmed, and the sample inlet and sample pool were washed using ultra-pure water. When the setting temperature was reached, 10–20 μL extracted particles was diluted to 1 mL in PBS, and then injected into the sample poor using a clean 1 mL injector. The size and distribution of particles were measured according to the path of particle and Brownian motion principle [21].
Western Blot Analysis
Total proteins from each group of cells were extracted using the cell lysis buffer (P0013, Beyotime Biotechnology Co., Ltd., Shanghai, China). The protein level in supernatant was detected using a bicinchoninic acid kit (P0009, Beyotime). Next, the protein was run on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Bio-Rad, Hercules, CA, USA) and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). Then, the membranes were blocked in 5% bovine serum albumin (BSA)-supplemented tris-buffered saline tween for 1 h to block non-specific binding. After that, the membranes were co-cultured with the 5% BSA-supplemented primary antibodies at 4 °C overnight, and then with secondary antibody at 37 °C for 2 h. Afterward, the protein bands were visualized via enhanced chemiluminescence reagent (WBKLS0100, Millipore) and imaged. Three independent experiments were performed. The antibodies are shown in Table 2.
Transwell Assay
Cell invasion was determined using the Transwell plates. In brief, each apical chamber was pre-coated with 50 μL diluted Matrigel (BD Biosciences, USA) and then loaded with cells and serum-free medium. Each basolateral chamber was loaded with RPMI-1640-10% FBS. The chambers were incubated at 37 °C with 5% CO2 for 16 h. Then, the cells on the upper polycarbonate membrane were removed, and the remaining invaded cells were stained using crystal violet and observed under a microscope with 5 random fields selected.
Animal Experiments
A total of 36 athymic mice (BALB/c, 5 weeks old, 20 ± 2 g) purchased from Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China) were used for in vivo tumor growth and metastasis experiments. As for the tumor metastasis, 24 mice were allocated into 8 groups. For the first batch of metastasis experiments, four groups of mice were injected with 200 μL suspension (5 × 106 cells) of HT-29 cells pre-transfected with exosomes (exo-NC, exo-miR-3940-5p, exosome-inhibitor) or PBS, respectively. For the second batch, four groups of mice were injected with HT-29 cell suspension pre-transfected with pcDNA alone, pcDNA ITGA6, pcDNA ITGA6 + NC or pcDNA ITGA6 + Disitertide, respectively. All transfection was administrated through the caudal vein. After transfection, the mice were euthanized by intravenous injection of Bartiturate (100 mg/kg). Next, the lung tissues were collected and the number of metastatic nodules in lung tissues was calculated by hematoxylin and eosin (HE) staining. As for the tumor growth experiments, 12 mice were allocated into 4 groups. Each mouse was injected with HT-29 cells pre-transfected with PBS, exo-miR-3940-5p, exo-miR-3940-5p + pcDNA, or exo-miR-3940-5p + pcDNA ITGA6 and then euthanized in a same manner to collect the tumor tissues. Then, the tumor weight and volume were measured. The tumor volume (V) was calculated based on the width (W) and length (L) as the following formula: V = (W2 × L)/2.
Dual Luciferase Reporter Gene Assay
The wild type (WT) and mutant type (MT) recombinant vectors (ITGA6-WT and ITGA6-MT) based on the putative binding sequences between miR-3940-5p and ITGA6 were constructed. Well-constructed vectors were co-transfected with either miR-3960-5p mimic/inhibitor or the corresponding negative control into the HT-29 and DLD-1 cells. The cells were harvested 24 h later and incubated with the passive lysis buffer at room temperature for 10 min. The relative luciferase activity was determined using a dual luciferase assay kit (Promega, Madisom, WI, USA) as per the kit’s protocols.
Statistical Analysis
SPSS 22.0 (IBM Corp. Armonk, NY, USA) was used for data analysis. Data were in normal distribution according to Kolmogorov–Smirnov method and exhibited as mean ± standard deviation (mean ± SD). Differences between every two groups were measured using the t test, while differences among multiple groups were analyzed using one-way or two-way analysis of variance (ANOVA). Tukey’s multiple comparisons test was used for the pairwise comparison after ANOVA. p was obtained by two-tailed test and p < 0.05 was regarded to show a statistically significant difference.
Results
Bioinformatics Analysis of Differentially Expressed MSC-exosomal miRNAs Between CRC and Normal Colon Tissues
The potential of MSC-exos in cancer treatment has been reported, but the functions of MSC-exos in CRC remain largely unknown. First, we obtained a total of 108 miRNAs in MSC-exos and 535 in colon tissues according to the data available on EVmiRNA DataBase, with 79 of them were found intersected (Fig. 1a). Next, the data on a GEO microarray GSE53339 based on 6 pairs of CRC and normal colon tissues were analyzed, and miR-3940-5p was found significantly highly expressed in normal tissues (Fig. 1b). According to the ID check by the GEO microarray attachment GPL18058, miR-3940-5p was found as one of the above 79 intersections (Fig. 1c). In addition, data on EVmiRNA DataBase suggested that miR-3940-5p is one of the lowest expressed exosomal miRNAs. Therefore, we inferred that it exerts potential functions in CRC inhibition.
Differentially expressed MSC-exosomal miRNAs between CRC and normal colon tissues measured by bioinformatics analysis. a intersected miRNAs found in MSC-exos and in colon tissues; b heatmap for part differentially expressed miRNAs between CRC and normal colon tissues according to the data on GEO microarray GSE53339; C, miR-3940-5p is highly expressed in normal colon tissues
MSC-exosomal miR-3940-5p Is Poorly Expressed in CRC Cells
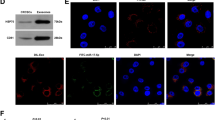
Next, overexpression or knockdown of miR-3940-5p was introduced in hUC-MSCs by miR-3940-5p mimic/inhibitor, and RT-qPCR found that the miR-3940-5p expression was successfully increased or decreased (Fig. 2a). After centrifugation and the removal of dead cells and debris, the particles were collected. TEM observation found that there was a large number of hUC-MSCs-derived vesicles (Fig. 2b). Then, the NTA identified the diameter of the vesicles was 50–150 nm (Fig. 2c). In addition, the western blot analysis confirmed existences of exosome-specific protein biomarkers CD63, CD81 and TSG101 but not β-actin in the vesicles (Fig. 2d). These results by implication meant the vesicles were MSC-exos. In MSCs transfected with NC, it was found the miR-3940-5p expression in the precipitate resuspension was notably higher than that in the supernatant following MSC centrifugation at 10,000g (Fig. 2e). The exosomes from hUC-MSCs pre-transfected with miR-3940-5p mimic/inhibitor or NC were correspondingly defined as exo-miR-3940-5p, exo-inhibitor and exo-NC. Next, miR-3940-5p expression in each group of MSC-exos was determined. Accordingly, we found the highest expression of miR-3940-5p in exo-miR-3940-5p while the lowest in exo-inhibitor (Fig. 2f). In addition, RT-qPCR also found that miR-3940-5p was poorly expressed in HT-29 and DLD-1 cell lines as compared to that in FHC (Fig. 2g).
MSC-exosomal miR-3940-5p is poorly expressed in CRC cells. a miR-3940-5p mimic/inhibitor or the corresponding NC vector was transfected into hUC-MSCs, and then the miR-3940-5p expression was determined by RT-qPCR (one-way ANOVA, **p < 0.01); hUC-MSC-derived vesicles observed under TEM; c size and distribution of particles determined by NTA; d exosome-specific proteins determined by western blot analysis; e miR-3940-5p expression in the purified precipitate and in supernatant following 10,000g centrifugation determined by RT-qPCR (one-way ANOVA, ****p < 0.0001); f miR-3940-5p expression after exo-miR-3940-5p and exo-inhibitor co-culture determined by RT-qPCR (one-way ANOVA, **p < 0.01); g miR-3940-5p expression in HT-29, DLD-1 and FHC cells determined by RT-qPCR (one-way ANOVA, ***p < 0.001). All experiments were repeated 3 times and the average value was collected
Artificial Up-Regulation of miR-3940-5p in MSC-exos Inhibits Metastasis of CRC
Each group of exosomes (exo-miR-3940-5p, exo-inhibitor, exo-NC) or PBS were transfected into HT-29 and DLD-1 cells, and then the levels of EMT-related proteins were determined by western blot analysis. It was found that cells transfected with exo-miR-3940-5p presented the highest level of E-cadherin and the lowest levels of vimentin and N-cadherin, while cells transfected with PBS presented totally reverse trends (Fig. 3a). In addition, the Transwell assay results identified that the cells transfected with exo-miR-3940-5p presented a decline in invasion ability (Fig. 3b). In addition, HT-29 cells with stable transfection of exosomes or PBS were implanted into nude mice. According to HE staining, it was found that the number of metastatic nodules in lung tissues was smallest in mice implanted with HT-29 cells transfected with exo-miR-3940-5p, while mice implanted with cells transfected with PBS showed a largest number of metastatic nodules in lung tissues (Fig. 3c). These findings suggested that up-regulation of miR-3940-5p in MSC-exos inhibits EMT and invasion of CRC cells as well as the metastasis of CRC in vivo.
Up-regulation of miR-3940-5p in MSC-exos inhibits metastasis of CRC. HT-29 and DLD-1 cells were treated with exosomes (exo-miR-3940-5p, exo-inhibitor, exo-NC) or PBS alone. a Protein levels of EMT-related biomarkers E-cadherin, N-cadherin and vimentin in cells determined by western blot analysis (one-way ANOVA, **p < 0.01); b number of invaded cells determined using Transwell assays (one-way ANOVA, *p < 0.05); c exo-treated HT-29 cells were implanted into nude mice through the caudal veins, and the metastatic nodules in 21 d determined by HE staining (n = 3, one-way ANOVA, **p < 0.01). All experiments were repeated 3 times and the average value was collected
miR-3940-5p Targets ITGA6
Next, we identified ITGA6 as a candidate target mRNA of miR-3940-5p on TargetScan (Fig. 4a). RT-qPCR was performed to determine mRNA expression of ITGA6 in CRC cells after exosome transfection. The results showed that exo-miR-3940-5p transfection inhibited mRNA expression of ITGA6, while the exo-inhibitor transfection, accordingly, increased the mRNA expression of ITGA6 (Fig. 4b). In addition, co-transfection of ITGA6-WT vector and miR-3940-5p mimic led to an increase in luciferase activity in cells, indicating miR-3940-5p directly binds to ITGA6 mRNA (Fig. 4c).
miR-3940-5p targets ITGA6. a Binding sites between miR-3940-5p and ITGA6 predicted on TargetScan (http://www.targetscan.org/); b mRNA expression of ITGA6 in CRC cells determined by RT-qPCR; c binding relationship between miR-3940-5p and ITGA6 validated by a dual luciferase reporter gene assay (two-way ANOVA, **p < 0.01). All experiments were repeated 3 times and the average value was collected
High Expression of ITGA6 Promotes CRC Metastasis Through Mediating the TGF-β1 Signaling Pathway
Following the findings above, we further searched ITGA6 expression in 275 CRC tumor tissues and in 349 normal colon tissues on a bio-information website Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/). The results suggested a high expression profile of ITGA6 in CRC tissues (Fig. 5a). In addition, RT-qPCR was performed to determine ITGA6 mRNA expression in HT-29, DLD-1 and FHC cells. It was found that ITGA6 expression was notably higher in CRC cell lines than that in normal colon cells (Fig. 5b). It has been that the TGF-β1 signaling pathway is closely correlated with EMT and CRC progression [22]. Thereafter, we explored the potential correlation between ITGA6 and this signaling. siRNA targeting ITGA6, pcDNA ITGA6 and the corresponding NC vectors were introduced into CRC cells. The RT-qPCR results suggested the transfection was successfully performed (Fig. 5c). In addition, a TGF-β1-specific antagonist, Disitertide, was administrated into cells as well. Then, it was found that siRNA ITGA6 while pcDNA ITGA6 increased TGF-β1 expression. Accordingly, Disitertide presented a significant inhibiting role in TGF-β1 expression (Fig. 5d). To further identify the involvement of TGF-β1 in ITGA6-mediated events, CRC cells were further transfected with pcDNA ITGA6 alone or co-transfected with pcDNA ITGA6 + Disitertide. Then, we found pcDNA ITGA6 transfection led to an increase in vimentin and N-cadherin production and a decline in E-cadherin production, while further administration of Disitertide considerably reversed these changes (Fig. 5e). Likewise, Transwell assays suggested that pcDNA ITGA6 increased the invasion ability of CRC cells, while Disitertide treatment counteracted this effect (Fig. 5f). To further identify their roles in CRC metastasis, HT-29 cells transfected with pcDNA ITGA6 alone or co-transfected with pcDNA ITGA6 + Disitertide were implanted into nude mice, and then the metastatic nodules in lung tissues in mice were observed on day 21. It was found that pcDNA ITGA6-transfected cells led to an increase in metastatic nodules in lung tissues in mice, but further administration of Disitertide led to a decline in metastasis (Fig. 5g).
High expression of ITGA6 promotes CRC metastasis through mediating the TGF-β1 signaling pathway. a ITGA6 expression in 275 CRC tumor tissues and in 349 normal colon tissues according to the data available on GEPIA (http://gepia.cancer-pku.cn/); b ITGA6 expression in CRC and normal colon cells determined by RT-qPCR; c siRNA ITGA6, pcDNA ITGA6 and the corresponding NC vector were transfected into CRC tissues (one-way ANOVA, **p < 0.01); d expression of TGF-β1 after ITGA6 interference or Disitertide administration determined by western blot analysis (one-way ANOVA, * p < 0.05); e pcDNA ITGA6 or pcDNA ITGA6 + Disitertide was transfected into CRC cells, and then the protein levels of EMT-related biomarkers were determined by western blot analysis (one-way ANOVA, *p < 0.05); f number of invaded cells determined by Transwell cells (one-way ANOVA, *p < 0.05); g HT-29 cells transfected with pcDNA alone, pcDNA ITGA6, pcDNA ITGA6 + NC, or pcDNA ITGA6 + Disitertide were implanted into nude mice, and then the metastatic nodules in lung in mice were observed by HE staining on day 21 (n = 3, one way ANOVA, *p < 0.05). All experiments were repeated 3 times and the average value was collected
MSC-exosomal miR-3940-5p Targets ITGA6 to Mediate TGF-β1 Signaling Pathway to Inhibit CRC Tumor Growth in Mice
HT-29 cells were transfected with exo-miR-3940-5p, PBS, exo-miR-3940-5p + pcDNA ITGA6 or exo-miR-3940-5p + pcDNA (NC for ITGA6), and then the protein expression of TGF-β1 was determined by western blot analysis. It was shown that the TGF-β1 expression was highest in the PBS group. TGF-β1 expression was inhibited by exo-miR-3940-5p inhibited and then reversed by pcDNA ITGA6 (Fig. 6a). Next, the cells were implanted into nude mice to induce xenograft tumors. The tumor weight and size were determined 21 d later (Fig. 6b). It was found that the PBS-transfected cells led to enlarged tumor size and weight. Exo-miR-3940-5p inhibited tumor growth, which was further counteracted by pcDNA ITGA6.
MSC-exosomal miR-3940-5p targets ITGA6 to mediate TGF-β1 signaling pathway to inhibit CRC tumor growth in mice. a TGF-β1 expression following exo-miR-3940-5p, PBS or pcDNA ITGA6 transfection determined by western blot analysis (one-way ANOVA, **p < 0.01); B, HT-29 cells co-cultured with exo-3940-5p or exo-3940-5p + pcDNA ITGA6 were implanted into the mice through subcutaneous injection, and then the volume and weight of xenograft tumors in nude mice were determined on the 21 d (n = 3, one-way ANOVA, *p < 0.05)
Discussion
Metastasis remains a main contributor to high mortality rate in CRC patients and a major challenge in the nowadays cancer treatment. MSC-based therapies have aroused concerns for their potential regulation in a wide range of conditions including cancer, and the MSC-exos have presented both tumor-promoting or tumor-suppressing roles through the different cargos [23]. Here in this paper, our study identified that MSC-exos carry miR-3940-5p into CRC cells to inhibit invasive and metastatic potential of cells, during which the inhibition of ITGA6 and TGF-β1 defect are possibly involved.
Initially, the integrated online analysis and miRNA microarray data on GEO suggested that miR-3940-5p might be an influential MSC-exosomal miRNA in CRC. We then identified miR-3940-5p was poorly expressed in CRC cell lines as compared to that in normal colon cells. Next, miR-3940-5p mimic/inhibitor, the corresponding NC, and PBS were transfected into hUC-MSCs and the corresponding exosomes were collected. CD9, CD63, CD81 and TSG101 are well-known enriched proteins in exosomes that were wide used as biomarkers of exosomes [24, 25], which were found in the MSC-derived particles in the current study. In addition, miR-3940-5p expression was found higher in the exo-NC than that in the supernatant, indicating that miR-3940-5p was carried by MSC-exos. Artificial overexpression of miR-3940-5p in MSC-exo led to declines in cell invasion and vimentin and N-cadherin expression but an increase in E-cadherin expression. The changes of these factors explained that miR-3940-5p inhibited the EMT of CRC cells, since E-cadherin is a well-known epithelial marker, while N-cadherin and vimentin are mesenchymal markers [26]. miR-3940-5p is a relatively seldom studied miRNA. It has been noted to inhibit proliferation of gingival MSCs which lead to gingival over-growth [27]. Besides, its suppressive role in tumor progression has been observed in non-small cell lung cancer [28]. Importantly, miR-3940-5p has been observed to be poorly expressed in the serum in CRC patients, thus serving as a potential prognostic marker for CRC [14]. Here, our study identified that MSC-exos carry miR-3940-5p into CRC cells and tissues, and overexpression of miR-3940-5p also presented suppressive effects on CRC metastasis and progression. The animal results presented similar trends since overexpression miR-3940-5p in MSC-exos led to declines in tumor metastasis and growth.
The findings prompted us to explore the involving downstream molecules. The main role of miRNAs in human body is gene regulation by binding to the target mRNAs through the matched sequences [29]. Then, our study identified ITGA6 as a candidate mRNA of miR-3940-5p in CRC. The binding relationship was further validated through a luciferase assay. ITGA6 attracted our attention since it has been noted to promote the progression in several human malignancies. For instance, ITGA6 was suggested to cooperate with RPSA to trigger the invasion and metastasis of pancreatic cancer [30]. Overexpression of ITGA6 and Thy1 was said to promote the metastatic potential of human gallbladder carcinoma [31]. Likewise, suppression in ITGA6 by its upstream miRNAs has been witnessed to lead to inhibited growth, invasion and migration of cancer cells [32, 33]. Thereby, we speculated that the similar trends would be seen by miR-3940-5p in CRC. We found much higher ITGA6 expression in CRC cell lines than that in normal colon cells. Overexpression of ITGA6 led to a significant increase in vimentin and N-cadherin expression while a decrease in E-cadherin expression in CRC cells. Up-regulation of ITGA6 also promoted tumor growth and metastasis in nude mice. It was noteworthy that ITGA6 positively regulated TGF-β1 expression. Aberrant expression of TGF-β1 may lead to fibrosis and dysregulated immune responses, which are also major characteristics of cancers [34]. The TGF-β1 has been noted to play crucial promoting roles in cell migration, invasion and EMT in several human cancer types [20, 35], leaving inhibiting this pathway a potential target in cancer therapy [36]. There is no exception for CRC that inhibition of TGF-β1 is a promising method for metastasis control [37]. In addition, the result that Disitertide partially blocked the roles of ITGA6 overexpression in cell EMT and tumor progression in vivo further validated the involvement of ITGA6-mediated events.
Conclusion
To sum up, this study evidenced that MSC-exo carry miR-3940-5p into CRC cells, leading to further ITGA6 downregulation and TGF-β1 signaling deficit, and consequently the reductions in invasive and metastatic potentials of CRC cells and tumors (Fig. 7). These findings may provide novel insights into MSC-exo-based treatment for CRC therapy. We hope more studies will be conducted in the field to validate our findings and to develop more new thoughts into CRC treatment.
Abbreviations
- ANOVA:
-
Analysis of variance
- BSA:
-
Bovine serum albumin
- CRC:
-
Colorectal cancer
- EMT:
-
Epithelial–mesenchymal transition
- Evs:
-
Extracellular vesicles
- FBS:
-
Fetal bovine serum
- GEO:
-
Gene Expression Omnibus
- HE staining:
-
Hematoxylin and eosin
- hUC-MSCs:
-
Human umbilical cord mesenchymal stem cells
- ITGA6:
-
Integrin α6
- miRNAs:
-
microRNAs
- NTA:
-
Nanoparticle tracking analysis
- MSC:
-
Mesenchymal stem cell
- MSC-exos:
-
MSC-derived exosomes
- MT:
-
Mutant type
- PBS:
-
Phosphate-buffered saline
- RPMI:
-
Roswell Park Memorial Institute
- RT-qPCR:
-
Reverse transcription quantitative polymerase chain reaction
- siRNA:
-
Small interfering RNA
- TEM:
-
Transmission electron microscopy
- TGF-β1:
-
Transforming growth factor-beta 1
- WT:
-
Wild type
References
Ding D, Han S, Zhang H, He Y, Li Y. Predictive biomarkers of colorectal cancer. Comput Biol Chem. 2019;83:107106.
Sun M, Liu J, Hu H, et al. A novel panel of stool-based DNA biomarkers for early screening of colorectal neoplasms in a Chinese population. J Cancer Res Clin Oncol. 2019;145:2423–2432.
Zhang W, Sun Z, Su L, et al. miRNA-185 serves as a prognostic factor and suppresses migration and invasion through Wnt1 in colon cancer. Eur J Pharmacol. 2018;825:75–84.
Hong Y, Rao Y. Current status of nanoscale drug delivery systems for colorectal cancer liver metastasis. Biomed Pharmacother. 2019;114:108764.
Paauwe M, Schoonderwoerd MJA, Helderman R, et al. Endoglin expression on cancer-associated fibroblasts regulates invasion and stimulates colorectal cancer metastasis. Clin Cancer Res. 2018;24:6331–6344.
Zhang X, Tu H, Yang Y, Fang L, Wu Q, Li J. Mesenchymal stem cell-derived extracellular vesicles: roles in tumor growth, progression, and drug resistance. Stem Cells Int. 2017;2017:1758139.
Altaner C, Altanerova U. Mesenchymal stem cell exosome-mediated prodrug gene therapy for cancer. Methods Mol Biol. 2019;1895:75–85.
Zhou J, Tan X, Tan Y, Li Q, Ma J, Wang G. Mesenchymal stem cell derived exosomes in cancer progression, metastasis and drug delivery: a comprehensive review. J Cancer. 2018;9:3129–3137.
Reza A, Choi YJ, Yasuda H, Kim JH. Human adipose mesenchymal stem cell-derived exosomal-miRNAs are critical factors for inducing anti-proliferation signalling to A2780 and SKOV-3 ovarian cancer cells. Sci Rep. 2016;6:38498.
Xu H, Zhao G, Zhang Y, et al. Mesenchymal stem cell-derived exosomal microRNA-133b suppresses glioma progression via Wnt/beta-catenin signaling pathway by targeting EZH2. Stem Cell Res Ther. 2019;10:381.
Cheshomi H, Matin MM. Exosomes and their importance in metastasis, diagnosis, and therapy of colorectal cancer. J Cell Biochem. 2018. https://doi.org/10.1002/jcb.27582.
Kletukhina S, Neustroeva O, James V, Rizvanov A, Gomzikova M. Role of mesenchymal stem cell-derived extracellular vesicles in epithelial–mesenchymal transition. Int J Mol Sci. 2019;20:4813.
Sun Y, Su B, Zhang P, et al. Expression of miR-150 and miR-3940-5p is reduced in non-small cell lung carcinoma and correlates with clinicopathological features. Oncol Rep. 2013;29:704–712.
Matboli M, Shafei AE, Ali MA, et al. Role of extracellular LncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal cancer. Arch Physiol Biochem. 2019. https://doi.org/10.1080/13813455.2019.1650070.
Brooks DL, Schwab LP, Krutilina R, et al. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26.
Guo L, Fu J, Sun S, et al. MicroRNA-143-3p inhibits colorectal cancer metastases by targeting ITGA6 and ASAP3. Cancer Sci. 2019;110:805–816.
Bigoni-Ordonez GD, Czarnowski D, Parsons T, Madlambayan GJ, Villa-Diaz LG. Integrin alpha6 (CD49f), The Microenvironment and Cancer Stem Cells. Curr Stem Cell Res Ther. 2019;14:428–436.
Linhares MM, Affonso RJ Jr, Viana Lde S, et al. Genetic and immunohistochemical expression of integrins ITGAV, ITGA6, and ITGA3 as prognostic factor for colorectal cancer: models for global and disease-free survival. PLoS ONE. 2015;10:e0144333.
Cui M, Chang Y, Du W, et al. Upregulation of lncRNA-ATB by transforming growth factor beta1 (TGF-beta1) promotes migration and invasion of papillary thyroid carcinoma cells. Med Sci Monit. 2018;24:5152–5158.
Magnussen SN, Hadler-Olsen E, Costea DE, et al. Cleavage of the urokinase receptor (uPAR) on oral cancer cells: regulation by transforming growth factor-beta1 (TGF-beta1) and potential effects on migration and invasion. BMC Cancer. 2017;17:350.
Yan W, Wu X, Zhou W, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20:597–609.
Dai G, Sun B, Gong T, Pan Z, Meng Q, Ju W. Ginsenoside Rb2 inhibits epithelial–mesenchymal transition of colorectal cancer cells by suppressing TGF-beta/Smad signaling. Phytomedicine. 2019;56:126–135.
de Araujo Farias V, O’Valle F, Serrano-Saenz S, et al. Exosomes derived from mesenchymal stem cells enhance radiotherapy-induced cell death in tumor and metastatic tumor foci. Mol Cancer. 2018;17:122.
Hu Y, Zhang Y, Ni CY, et al. Human umbilical cord mesenchymal stromal cells-derived extracellular vesicles exert potent bone protective effects by CLEC11A-mediated regulation of bone metabolism. Theranostics. 2020;10:2293–2308.
Wu K, Xing F, Wu SY, Watabe K. Extracellular vesicles as emerging targets in cancer: recent development from bench to bedside. Biochim Biophys Acta Rev Cancer. 2017;1868:538–563.
Paolillo M, Schinelli S. Extracellular matrix alterations in metastatic processes. Int J Mol Sci. 2019;20:4947.
Han X, Yang H, Cao Y, et al. The miR-3940-5p inhibits cell proliferation of gingival mesenchymal stem cells. Oral Dis. 2019;25:1363–1373.
Ren K, Li Y, Lu H, Li Z, Han X. miR-3940-5p functions as a tumor suppressor in non-small cell lung cancer cells by targeting cyclin D1 and ubiquitin specific peptidase-28. Transl Oncol. 2017;10:80–89.
Condrat CE, Thompson DC, Barbu MG, et al. miRNAs as biomarkers in disease: latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9:276.
Wu Y, Tan X, Liu P, et al. ITGA6 and RPSA synergistically promote pancreatic cancer invasion and metastasis via PI3K and MAPK signaling pathways. Exp Cell Res. 2019;379:30–47.
Zhang DH, Yang ZL, Zhou EX, et al. Overexpression of Thy1 and ITGA6 is associated with invasion, metastasis and poor prognosis in human gallbladder carcinoma. Oncol Lett. 2016;12:5136–5144.
Chen W, Zhuang X, Qi R, Qiao T. MiR-302a-5p suppresses cell proliferation and invasion in non-small cell lung carcinoma by targeting ITGA6. Am J Transl Res. 2019;11:4348–4357.
Wang D, Tang L, Wu H, Wang K, Gu D. MiR-127-3p inhibits cell growth and invasiveness by targeting ITGA6 in human osteosarcoma. IUBMB Life. 2018;70:411–419.
Lodyga M, Hinz B. TGF-beta1—a truly transforming growth factor in fibrosis and immunity. Semin Cell Dev Biol. 2019;101:123–139.
Liang S, Yao Q, Wei D, et al. KDM6B promotes ovarian cancer cell migration and invasion by induced transforming growth factor-beta1 expression. J Cell Biochem. 2019;120:493–506.
He XR, Han SY, Li XH, et al. Chinese medicine Bu-Fei decoction attenuates epithelial-mesenchymal transition of non-small cell lung cancer via inhibition of transforming growth factor beta1 signaling pathway in vitro and in vivo. J Ethnopharmacol. 2017;204:45–57.
Fang JK, Chen L, Lu XG, et al. Optimization of transforming growth factor-beta1 siRNA loaded chitosan-tripolyphosphate nanoparticles for the treatment of colorectal cancer hepatic metastasis in a mouse model. J Biomed Nanotechnol. 2016;12:1489–1500.
Acknowledgments
This work was supported by Jilin Science and Technology Development Plan Project (201902011224 JC).
Author information
Authors and Affiliations
Contributions
TL and YCW is the guarantor of integrity of the entire study and contributed to the concepts and design of this study; ZYS contributed to the definition of intellectual content and literature research of this study; JYL contributed to the experimental studies; MNH contributed to the data acquisition and the data analysis; CYZ took charge of the manuscript preparation; All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no potential conflicts of interest.
Ethics approval
The study was ratified by the Clinical Ethical Committee of China-Japan Union Hospital of Jilin University. Animal studies were conducted as per the principles and procedures approved by the Committee on the Ethics of Animal Experiments of China-Japan Union Hospital of Jilin University. Great efforts were made to minimize the animals and their suffering.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, T., Wan, Y., Su, Z. et al. Mesenchymal Stem Cell-Derived Exosomal microRNA-3940-5p Inhibits Colorectal Cancer Metastasis by Targeting Integrin α6. Dig Dis Sci 66, 1916–1927 (2021). https://doi.org/10.1007/s10620-020-06458-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06458-1