Abstract
Background
Clinical observations and experimental colitis models have indicated the importance of intestinal bacteria in the etiology of ulcerative colitis (UC), but a causative bacterial agent has not been identified.
Aim
To determine how intestinal bacteria are associated with UC, fecal microbiota and other components were compared for UC patients and healthy adults.
Methods
Fresh feces were collected from 48 UC patients. Fecal microbiota were analyzed by use of terminal-restriction fragment length polymorphism (T-RFLP), real-time PCR, and culture. The concentrations of organic acids, indole, and ammonia, and pH and moisture, which are indicators of the intestinal environment, were measured and compared with healthy control data.
Results
T-RFLP data divided the UC patients into four clusters; one cluster was obtained for healthy subjects. The diversity of fecal microbiota was significantly lower in UC patients. There were significantly fewer Bacteroides and Clostridium subcluster XIVab, and the amount of Enterococcus was higher in UC patients than in healthy subjects. The fecal concentration of organic acids was significantly lower in UC patients who were in remission.
Conclusion
UC patients have imbalances in the intestinal environment—less diversity of fecal microbiota, lower levels of major anaerobic bacteria (Bacteroides and Clostridium subcluster XIVab), and a lower concentration of organic acids.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ulcerative colitis (UC) is an inflammatory bowel disease that has been increasing in Japan [1]. It often occurs in the prime of life (early 30 s) and is an intractable lifelong disease. Controlling the symptoms with drugs is a heavy burden for patients physically, mentally, and socially [1]. An understanding of the pathogenic mechanism of UC is needed to lengthen remission periods and improve quality of life. Although the exact etiology is unknown, multiple causes, for example immune system disorders [2, 3], intestinal bacteria [4–6], genetics [7, 8], and environmental factors, for example lifestyle and stress, have been implicated [9].
Because inflammation is limited to the large intestine in UC, intestinal microbiota and its metabolites might be important in the etiology for UC. Interleukin (IL)-10 knockout (KO) mice and IL-2 KO mice spontaneously develop UC-like inflammation in a conventional environment, but not under germ-free conditions [2, 3, 10, 11], indicating the importance of intestinal bacteria in addition to dysfunction of the immune system. However, the involvement of intestinal bacteria in the development of UC has not been clarified. Some intestinal bacteria have been suggested as candidate agents. Bacteroides vulgatus has been found in biopsy samples of UC patients, and antibodies against this bacterium have been detected in the serum of UC patients [12]. More sulfate-reducing bacteria (SRB) reside in UC patients, producing cytotoxic hydrogen sulfide, which has been implicated in UC [13, 14]. Ohkusa et al. [15] detected Fusobacterium varium in the colonic mucosa of UC patients and reported that antibiotics against F. varium ameliorated disease symptoms. These bacteria also reside in healthy adults, however, and no bacterial species has been identified as UC-specific.
Most human microbiota belongs to four bacterial divisions: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria [16–18]. Bacteroides, Eubacterium, and Ruminococcus are the dominant genera in the adult intestine [18]. The effects of these dominant bacteria and their metabolites on the intestinal mucosa are not negligible. Intestinal microbiota metabolizes undigested carbohydrates into organic acids, for example acetic and butyric acids, in the lower intestinal tract. These acids reduce intestinal pH and suppress over-growth of pathogenic bacteria and the production of putrefactive components, for example indole and ammonia. Butyrate is an energy source for the intestinal mucosa and promotes growth and differentiation of colonocytes [19–23]. Intestinal organic acids, especially butyric acid, have immunomodulatory effects on colonic inflammation and a reinforcing effect on the intestinal defense barrier [19, 20, 24].
Microbiota colonization affects the development of the intestinal immune system, and proper immune function is regulated by the interaction between host and symbiotic bacteria [25, 26]. Normal intestinal microbiota is associated with both pro and anti-inflammation responses [25]. Fecal microbiota, innate and adaptive immune responses in the host, and the intestinal environment, including bacterial metabolites, are closely linked to gut health, and an imbalance in these factors has been implicated in intestinal disorders. Here, we investigated differences in fecal microbiota and organic acids between UC patients and healthy adults to clarify how intestinal microbiota is involved in the development of ulcerative colitis.
Materials and Methods
Reagents and Bacterial Strains
Blood liver agar (BL), Rogosa SL agar, Trypticase soy agar (TS), and desoxycholate hydrogen sulfide lactose agar (DHL) were obtained from Nissui Pharmaceuticals (Tokyo, Japan). Bacteroides fragilis JCM11019, Bifidobacterium longum JCM1217, Blautia coccoides JCM1395, Clostridium perfringens JCM1290, Desulfovibrio alkalitolerans JCM12612, Enterococcus faecalis JCM5803, Fusobacterium varium JCM3722, and Methanobrevibactor arboriphilus JCM13429 were obtained from the Japan Collection of Microorganisms, Riken Bioresource Center (Tsukuba, Japan). Clostridium difficile ATCC9689 was obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Chemicals, unless otherwise stated, were reagent-grade or higher and obtained from Sigma Chemical (St Louis, MO, USA) or Wako Pure Chemical Industries (Osaka, Japan).
Subjects and Study Design
Forty-eight UC outpatients at the Kenporen Osaka Central Hospital, Osaka, Japan, gave written consent to take part in this study. Eligible patients were aged 22–69 and had been diagnosed with UC by colonoscopy and histology (Table 1). Patients were categorized on the basis of their clinical activity index scores (CAI): remission, 0–4; mildly active, 5–7; moderately active, 8–11 [27]. Patients with fulminant UC, unable to take food orally, or treated with steroids or immunosuppressants were excluded from this study. None of the subjects was treated with antibiotics (e.g., metronidazole and ciprofloxacin) during this study. Most patients received oral or intrarectal treatment with 5-ASA compounds, for example Salazopyrin or Pentasa (Table S1). Nineteen patients were taking MiyaBM (viable Clostridium butyricum strain miyairi, 300 mg/6 tablets), and four patients were taking, daily, fermented milk supplemented with Bifidobacterium breve or a supplement containing Bifidobacterium (Table S1). All patients were omnivorous and were not on a restricted diet with one exception—one female patient (UC68) was excluded from the comparison of amount of bacteria and fecal concentrations of metabolites because she had received diet-related folk remedies (Table 1). The patients self-reported their bowel habits by using the Inflammatory Bowel Disease Questionnaire (IBDQ) with a visual analog scale (VAS). The average frequency of evacuation and time and date of the occurrence of diarrhea/loose stool was individually entered as the distance on the VAS (Table S2). The data from 36 healthy volunteers used as controls were obtained from Nemoto et al. [28]. Healthy volunteers were recruited at Tokushima University, Tokushima, Japan. All of the control subjects were omnivorous and were not taking antibiotics. All subjects were asked to provide a fresh stool sample. Patients and healthy volunteers collected samples at home and immediately placed them in an AnaeroPouch-Anaero (Mitsubishi Gas Chemical Company, Tokyo, Japan), where they were kept anaerobic and cold with refrigerants until received by the laboratory. The protocol was approved by the Ethics Committee of Kyoto Prefectural University and Osaka Chuo Hospital and the Ethics Committee of Tokushima University Hospital.
Fecal Microbiota Analysis: Culture
Bacteriological analysis was performed by use of the procedure of Mitsuoka et al. [29] with slight modification as described by Nemoto et al. [28]. Results were calculated as log10 of colony forming units (CFU) per gram (wet weight) of the initial material. Total numbers of fecal bacteria were counted after Gram staining.
Fecal Microbiota Analysis: Terminal-Restriction Fragment Length Polymorphism (T-RFLP)
Fecal and bacterial DNA was extracted for T-RFLP analysis of fecal microbiota [28, 30, 31]. Briefly, 16S ribosomal DNA was amplified by use of universal PCR primers (Table 2) and digested with HhaI or MspI (Takara). The length of the terminal-restriction fragments (T-RFs) was determined by use of Genetic analyzer and GeneScan Analysis Software. T-RF patterns were analyzed by use of BioNumerics ver.5.01 software (Applied Maths, Sint-Martens-Latem, Belgium) to construct dendrograms. Distances between samples were represented graphically by constructing a dendrogram based on Dice coefficients of the T-RFLP profiles. The Ward method was used to establish dendrogram type. Microbiota Profiler software (Infocom, Tokyo, Japan) was used to estimate the intestinal bacteria corresponding to T-RFs in UC patients and healthy subjects.
Diversity of Intestinal Microbiota
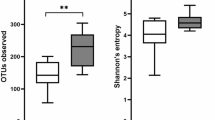
Diversity of intestinal microbiota was evaluated as the number of operational taxonomic units (OTUs) derived from T-RFLP analysis. Because 16S rDNA is highly and phylogenetically conserved, digested rDNA would yield genus and/or species-specific sizes of T-RFs because of different 16S ribosomal DNA sequences. OTUs that represent each length of T-RF might reflect single and/or phylogenetically-related bacteria. Therefore, each OTU can indicate one or more organisms, including uncultured, un-named bacteria. Peaks over 5 % of maximum peak height of the sample were used as OTUs. Diversity was represented as the sum of OTUs derived from HhaI and MspI digests.
Quantitative Analysis of Fecal Microbiota
As T-RFLP analysis is a semi-quantitative method, real time PCR was used to quantify fecal bacteria [28]. The primers and probes are shown in Table 2. SYBR Premix Ex Taq II (Takara) and Premix Ex Taq (Takara) were used with the ABI Prism 7000 and ABI Prism 7500 according to the manufacturer’s instructions. The amplification program was obtained from the references in Table 2 [32–36]. The 16S ribosomal RNA gene of each bacterial strain listed in the section “Reagents and bacterial strains” was cloned into the pCR2-TOPO vector (Invitrogen, Tokyo, Japan) or the pGEM T-Easy vector (Promega, Tokyo, Japan) according to the manufacturer’s procedure for use as a copy number standard. The results of real-time PCR for each sample were expressed as the copy number of bacterial 16S ribosomal DNA per gram feces (wet weight). The detection limits were: Clostridium I/XI/XIVab, 104 copies/10 ng; all other genera, 100 copies/10 ng fecal DNA. Gene recombination experiments for the preparation of copy number standards were approved by the University of Tokushima.
Fecal pH and Water Content
Stools (0.1–0.2 g) were homogenized with 1 ml water. Fecal pH was measured with a pH meter (Sartorius B021610 007). Water content was calculated as the weight difference before and after freeze-drying a portion of the fecal material (EYELA FDU-1000).
Measurement of Fecal Concentrations of Organic Acids, Indole, and Ammonia
Bacterial metabolites in the feces of UC patients were quantified as described elsewhere [28]. Portions (approximately 0.2 g) of homogenized stool were suspended in 1 ml water, centrifuged at 4 °C at 10 krpm for 10 min, and stored at −80 °C. Organic acids in the fecal extracts were extracted and measured by gas chromatography (C-7AG; Shimadzu, Kyoto, Japan) with external standards.
Fecal levels of indole were measured as described by Shioiri et al. [37]. Briefly, stools (approximately 0.1 g) were homogenized with nine volumes of 0.1 mM phosphate buffer (PB) and centrifuged. The supernatant was appropriately diluted with PB, and 0.2 ml diluted sample was mixed with 1 ml coloring solution (P-dimethylaminobenzaldehyde in a sulfuric acid–alcohol mixture) and kept at room temperature for 20 min. The absorbance at 570 nm was measured by use of a UV–visible spectrophotometer (Shimadzu UV-1200). As a control, 0–0.3 mM indole solution was prepared just before use. Ammonia concentration was measured by use of the Wako ammonia test.
Statistical Analysis
The Mann–Whitney U test and the Spearman correlation coefficient by rank test were performed by using Microsoft Excel 2007 and Statcel2 add-in software (OMS publishing, Tokorozawa, Japan). A probability of less than 0.05 indicated statistical significance.
Results
T-RFLP Analysis and the Diversity of Fecal Microbiota
After T-RFLP analysis of 16S rDNA (using HhaI or MspI digestion), a dendrogram was constructed by Dice correlation and use of the Ward algorithm. Four clusters of patients with UC and one of healthy subjects were roughly and weakly generated (Fig. 1). Clusters were not tied to inflammatory activity or location of UC. The number of OTUs from both the HhaI and MspI digestions, which denote the diversity of the microbiota, was compared between the UC and healthy subjects. The diversity of fecal microbiota was lower in UC patients in remission (mean OTU = 41.0, interquartile range (IQR): 35.5–49.0, P < 0.001) and with active UC (mean OTU = 44.5, IQR: 38.0–46.5, P = 0.029) than in healthy subjects (mean OTU = 50.5, IQR: 44.3–60.5).
Cluster analysis of fecal microbiota in healthy adults and patients with UC. T-RFLP patterns derived from HhaI and MspI digests were analyzed by use of the Dice coefficient and the Ward algorithm. The dendrogram comprised four clusters for UC patients and one cluster for healthy adults. The clusters of UC were generated independently by disease types and state. left left-sided colitis, RS right-sided or segmental colitis
Analysis of Fecal Microbiota by Real-Time PCR
The copy number of 16S rDNA in sulfate-reducing bacteria (SRB, represented by Desulfovibrio), bifidobacteria, enterococci, Bacteroides, and clostridia in feces was investigated by use of real-time PCR (Table 3). The fecal moisture level did not differ significantly between UC and healthy subjects (Table 4). Therefore, wet feces were used as starting materials in subsequent experiments. The copy number of Bacteroides was lower in UC patients and was less than 10 % of that in healthy subjects. Clostridium subcluster I (represented by C. perfringens) and XI (C. difficile) were not significantly different between UC and healthy subjects. However, the Clostridium subcluster XIVab (represented by B. coccoides) was significantly smaller in UC patients, as was Bacteroides. The enterococci population was significantly larger in UC patients in remission than in healthy subjects. Fewer SRB were found in patients in remission than in controls, although some samples in both groups were below the detection limit. We also investigated methane-producing archea (methanogens) and Fusobacterium, but they were rarely detected in any of the subjects (data not shown).
Culture-Based Analysis of Fecal Microbiota
Total and viable cell numbers of fecal bacteria are shown in Table 3. The total number of fecal bacteria, which was counted after Gram-staining, was significantly smaller in patients with active UC than in healthy controls. The number in those in remission was also small, but not significant. Viable cell numbers of bifidobacteria, anaerobic G(−)R, anaerobic G(+)R, anaerobic cocci, and Enterobacteriaceae in feces were no different between UC and healthy subjects. Although the number of viable aerobic G(+)C was not significantly different between UC patients in remission and healthy subjects, it was significantly larger in active UC patients than in healthy controls.
Comparison of Fecal Organic Acids, Ammonia, Indole, Moisture, and pH
The concentration of organic acids in feces is shown in Table 4. The concentrations of acetic, propionic, butyric, and lactic acids were significantly lower in the fecal material of patients in remission than in healthy subjects, but were the same for active UC and healthy subjects. The total concentration of organic acids was also significantly lower in patients in remission than in healthy controls, but was not significantly different in samples from active UC patients. Other fecal components measured in this study are shown in Table 4. The moisture level was not significantly different between UC and healthy subjects. Fecal pH was significantly higher only in those in remission. The amount of ammonia was significantly larger, and that of indole was smaller, in those with active UC than in healthy subjects, but there was no significant difference between those in remission and the controls.
Effect of Medication, Probiotics, and Bowel Habits on Intestinal Bacteria Levels
Because 5-aminosalicylic acid (5-ASA) has antimicrobial activity [38] and the sulfapyridine moiety of Salazopyrin (a conjugate of 5-ASA and sulfapyridine) is an acknowledged antibiotic because of anti-folate activity, the amount of intestinal bacteria was plotted against different doses of 5-ASA compounds (Table S1). No significant correlation was found between the amount of intestinal bacteria and the dosing of 5-ASA compounds (Fig. S1). Intake of probiotics has been known to affect intestinal microbiota. Nineteen patients in this study took MiyaBM and four ingested Bifidobacterium-containing fermented milk or supplement, as described in Table S1. However, there was no correlation between MiyaBM intake and levels of the dominant anaerobic bacteria, and viable cell number of Enterobacteriaceae was negatively correlated with intake (Fig. S2).
UC patients reported their bowel habits in a questionnaire (IBDQ) (Table S2). Statistical analysis showed that the amount of Clostridium subcluster XIVab was negatively correlated with increased evacuation frequency and the number of times a day that the patient experienced diarrhea/loose stool. In addition, the amount of Bacteroides was significantly lower than that in healthy controls, irrespective of bowel habits (Fig.S3a,b).
Discussion
The healthy gut environment is complicated and controlled by the balance of the intestinal immune system, intestinal microbiota, and microbial metabolites produced by intestinal bacteria [23, 39]. Inflammatory bowel disease could be promoted by an imbalance of these elements in genetically susceptible persons. However, the bacteria responsible for UC have not been identified, and how the intestinal microbiota affects intestinal disorders is still unclear. Therefore, fecal microbiota and microbial metabolites, as indicators of the large intestinal environment, were compared between patients with UC and healthy adults to find a clue to the roles of intestinal bacteria in UC.
Dendrogram analysis of T-RFLP profiles divided fecal microbiota into one healthy cluster and four UC clusters (Fig. 1). The UC clusters were not dependent on disease activity and location of inflammation, indicating that fecal microbiota is basically different in UC patients. Most of the same species of bacteria appeared as T-RFs in both UC patients and healthy subjects, but the amounts of the dominant genera, Bacteroides and Clostridium subcluster XIVab, tended to be less in UC patients (data not shown). Ando et al. [40] also analyzed fecal microbiota by T-RFLP and indicated that the diversity of fecal microbiota is altered in UC patients, and that unclassified bacteria, and known bacteria, contribute to the alterations. But, in our study, comparison of T-RFLP profiles revealed no increase of UC-specific bacteria, and the number of T-RFs was significantly less in UC patients. Less diversity has been reported in mucosa-associated microbiota in patients with Crohn’s disease and in fecal microbiota in patients with UC [41, 42]. Reduced diversity of fecal microbiota might be a characteristic of inflammatory bowel disease (IBD). Martinetz et al. [43] reported a time-dependent decrease of diversity (i.e., unstable microbiota) in UC patients. In this study we analyzed fresh feces at one time point. We should analyze microbiota at different times or in different places to confirm the decrease in diversity.
Consistent with the results of T-RFLP analysis, real-time PCR revealed significantly fewer Bacteroides and Clostridium subcluster XIVab in UC patients than in healthy adults. Colitis therapy with immunosuppressants and anti-inflammatory drugs, for example the 5-ASA compounds [38, 44], intake of probiotics, dietary habits, and bowel habits can affect intestinal microbiota. We investigated whether decreases of dominant anaerobes, for example Bacteroides and Clostridium subcluster XIV, in UC patients depends on these factors. Significantly lower levels of Bacteroides and Clostridium XIVab were not correlated with the 5-ASA compounds administered in this study (Fig. S1). Finegold et al. [45] showed that a mixture of typical antifolate sulfa drugs, was very poorly active against anaerobic intestinal bacteria. Therefore, a decrease of Bacteroides and Clostridium XIVab in UC patients is not because of the colitis medications, although an effect cannot be completely excluded. The intake of probiotics containing Clostridium butyricum also did not affect levels of these dominant anaerobes in UC patients (Fig. S2). However, increasing frequency of evacuation and diarrhea/loose stool were negatively correlated with the amount of Clostridium subcluster XIVab, and Bacteroides was significantly lower than in the healthy controls, irrespective of bowel habits and other factors under analysis. Among minor aerobic bacteria, both culture and real-time PCR showed an increase in enterococci in UC patients. Although enterococci are normal residents of the human intestine, monoassociation of Enterococcus faecalis reportedly enhances gene expression of proinflammatory chemokines in intestinal epithelial cells [46], and leads to the development of colorectal inflammation in IL-10 KO mice to the same extent as in conventionalized IL-10 KO mice [47]. These reports suggest that an increase of enterococci has a promoting effect on UC.
Because probiotic bifidobacteria reportedly ameliorate UC [48–50], we expected lower levels of bifidobacteria might correlate with the occurrence or relapse of colitis. Instead, the level of bifidobacteria in UC was the same as that in healthy adults. Although sulfate-reducing bacteria (SRB) has been correlated with the occurrence of UC [6], amount of SRB was smaller in UC patients than in healthy adults. Although 5-ASA can inhibit sulfate-reducing bacteria-mediated production of hydrogen sulfide in vitro, the counts and carriage rates of SRB were not significantly different from those in controls [51]. In this study, neither treatment with 5-ASA compounds nor bowel habits alone significantly affected SRB, but they may possibly exert an effect in combination.
Fecal concentration of organic acids in UC patients in remission was significantly lower than those in healthy subjects. It was slightly higher in patients with active UC than in remission patients and significantly correlated with an increase in evacuation frequency (Fig. S5). These effects on organic acid levels occur:
-
1.
because of inflamed mucosa that cannot efficiently utilize organic acids produced by intestinal bacteria; and
-
2.
because of the rapid movement of intestinal contents, for example during diarrhea.
A lower concentration of organic acids was consistent with the decrease of Bacteroides, one of the most dominant species in the human gut, which produces abundant organic acids by acidic fermentation [18, 52]. Bacteroides spp. efficiently ferment carbohydrates to acetate, propionate, and succinate. A bowel habit-dependent decrease of Clostridium subcluster XIVab, which includes butyrate-producing bacteria, might also be implicated in the lower concentration of organic acids in UC patients, but this might be hidden by the effect of increasing evacuation/diarrhea. Besides butyric acid, these organic acids also contribute to intestinal homeostasis by supplying anti-inflammatory effects and energy for colonocytes [52, 53]. As acetate and lactate are intermediates in carbohydrate fermentation by intestinal microbiota [52, 54], lower concentrations of these in UC patients indicate depressed metabolic activity of microbiota. Reduced levels of organic acids in IBD patients have also been reported by Takaishi et al. [55] and Marchei et al. [56]. Although we did not quantify Fecalibacterium prausnitzii or Clostridium subcluster IV, these bacteria have been repeatedly detected in healthy subjects and their reduction has been characterized in Crohn’s disease patients [41, 57]. Therefore, it is possible that a decrease of these commensal bacteria contributes to the lower concentration of organic acids in the feces of UC patients.
The concentration of putrefactive metabolites is significantly different between active UC and healthy controls (Table 4). Although a higher intake of protein increases the intestinal concentration of ammonia [58], dietary habits did not differ between the two groups in this study (data not shown). Considering that fecal ammonia is mainly produced by bacteria in the large intestine and has a disorganizing effect on the intestinal epithelium [59], increased ammonia might be made from proteinaceous components leaked from the inflamed mucosa of the large intestine, in turn facilitating more inflammation in active UC patients.
In summary, 16S rDNA-targeted analysis of fecal microbiota indicated less diversity, a decrease of dominant anaerobes, and reduced concentration of organic acids in UC patients. In addition, increased levels of enterococci were found as a feature of UC in this study. When and how dominant bacteria decrease during pathogenesis of UC was not clarified in this study; however, the imbalanced microbiota followed by impaired production of organic acids might make intestinal homeostasis fragile and facilitate the induction of inflammation. A deficiency of Firmicutes and Bacteroidetes has been observed in patients with antibiotic-induced Clostridium difficile-associated colitis. However, bacteriotherapy with fecal suspension prepared from a normal donor altered the microbioa of the patient to resemble that of the donor, with resolution of the symptoms [60]. Also, colonic infusion of selected UC patients with intestinal flora from a healthy donor can reverse UC [61]. These reports support our results and suggest the importance of dominant anaerobes, for example Bacteroides and commensal Clostridium, in maintaining intestinal homeostasis.
References
Asakura K, Nishiwaki Y, Inoue N, et al. Prevalence of ulcerative colitis and Crohn’s disease in Japan. J Gastroenterol. 2009;44:659–665.
Kuhn R, Lohler J, Rennick D, et al. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274.
Sadlack B, Merz H, Schorle H, et al. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75:253–261.
Nomura T, Ohkusa T, Okayasu I, et al. Mucosa-associated bacteria in ulcerative colitis before and after antibiotic combination therapy. Aliment Pharmacol Ther. 2005;21:1017–1027.
Kuehbacher T, Rehman A, Lepage P, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57:1569–1576.
Rowan FE, Docherty NG, Coffey JC, et al. Sulphate-reducing bacteria and hydrogen sulphide in the aetiology of ulcerative colitis. Br J Surg. 2009;96:151–158.
Asano K, Matsushita T, Umeno J, et al. A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet. 2009;41:1325–1329.
The UK IBD Genetics Consortium and the Wellcome Trust Case Control Consortium 2. Genome-wide association study of ulcerative colitis identifies three new susceptibility loci, including the HNF4A region. Nat Genet. 2009;41:1330–1334.
Mawdsley JE, Ramptom DS. Psychological stress in IBD: new insight into pathogenic and therapeutic implications. Gut. 2005;54:1481–1491.
Kim SC, Tonkonogy SL, Albright CA, et al. Variable phenotypes of enterocolitis in interleukin 10—deficient mice monoassociated with two different commensal bacteria. Gastroenterology. 2005;128:891–906.
Waidmann M, Bechtold O, Frick JS, et al. Bacteroides vulgatus protects against Escherichia coli—induced colitis in gnotobiotic interleukin-2—deficient mice. Gastroenterology. 2003;125:162–177.
Matsuda H, Fujiyama Y, Ando A, et al. Characterization of antibody responses against rectal mucosaassociated bacterial flora in patients with ulcerative colitis. J Gastroenterol Hepatol. 2000;15:61–68.
Gibson GR, Cumming JH, McFarlane GT. Growth and activities of sulphate-reducing bacteria in gut contents of healthy subjects and patients with ulcerative colitis. FEMS Microbiol Lett. 1991;86:103–112.
Pitcher MCL, Cummings JH. Hydrogen sulphide: a bacterial toxin in ulcerative colitis? Gut. 1996;39:1–4.
Ohkusa T, Sato N, Ogihara T, Morita K, et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002;17:849–853.
Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999;65:4799–4807.
Tap J, Mondot S, Levenez F, et al. Towards the human intestinal microbiota phylogenic core. Environ Microbiol. 2009;11:2574–2584.
Hattori M, Taylor TD. The human intestinal microbiome: a new frontier of human biology. DNA Res. 2009;16:1–12.
Wächtershäuser A, Stein J. Rationale for the luminal provision of butyrate in intestinal diseases. Eur J Nutr. 2000;39:164–171.
Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–119.
Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80.
Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118.
Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81:1031–1064.
Cavaglieri CR, Nishiyama A, Fernandes LC, et al. Differential effects of short-chain fatty acids on proliferation and production of pro- and anti-inflammatory cytokines by cultured lymphocytes. Life Sci. 2003;73:1683–1690.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323.
Hooper LV, Macpherson AJ. Immune adaptation that maintain homeostasis with the intestinal microbiota. Nat Rev Immunol. 2010;10:159–169.
Lichtiger S, Present DH, Kornbluth A, et al. Cyclosporine in severe ulcerative colitis refractory to steroid therapy. N Engl J Med. 1994;330:1841–1845.
Nemoto H, Ikata K, Arimochi H, et al. Effects of fermented brown rice on the intestinal environments in healthy adult. J Med Invest. 2011;58:235–245.
Mitsuoka T, Sega T, Yamamoto S. Improved methodology of qualitative and quantitative analysis of the intestinal flora of man and animals. Zentralbl Bakteriol Orig. 1965;195:455–469. (in German).
Morita H, Kuwahara T, Ohshima K, et al. An improved DNA isolation method for metagenomic analysis of the microbial flora of the human intestine. Microbes Environ. 2007;22:214–222.
Sakamoto M, Hayashi H, Benno Y. Terminal restriction fragment length polymorphism analysis for human fecal microbiota and its application for analysis of complex bifidobacterial communities. Microbiol Immunol. 2003;47:133–142.
Matsuki T, Watanabe K, Fujimoto J, et al. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol. 2002;68:5445–5451.
Fite A, Macfarlane GT, Cummings JH, et al. Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut. 2004;53:523–529.
Rinttilä T, Kassinen A, Malinen E, et al. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–1177.
Song Y, Liu C, Finegold SM. Real-time PCR quantitation of clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–6465.
Layton A, McKay L, Williams D, et al. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol. 2006;72:4214–4224.
Shioiri T, Yahagi K, Nakayama S, et al. The effects of a symbiotic fermented milk beverage containing Lactobacillus casei strain Shirota and transgalactosylated oligosaccharides on defecation frequency, intestinal microflora, organic acid concentrations, and putrefactive metabolites of sub-optimal health state volunteers: a randomized placebo-controlled cross-over study. Biosci Microflora. 2006;25:137–146.
Greenstein RJ, Su L, Shahidi A, et al. On the action of 5-amino-salicylic acid and sulfapyridine on M avium including subspecies paratuberculosis. PLoS ONE. 2007;2:e516.
Round JL, O’Connell RM, Mazmanian SK. Coordination of tolerogenic immune responses by the commensal microbiota. J Autoimmun. 2010;34:J220–J225.
Ando A, Sakata S, Koizumi Y, et al. Terminal restriction fragment length polymorphism analysis of the diversity of fecal microbiota in patients with ulcerative colitis. Inflamm Bowel Dis. 2007;13:955–962.
Manichanh C, Rigottier-Gois L, Bonnaud E, et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211.
Nishikawa J, Kudo T, Sakata S, et al. Diversity of mucosa-associated microbiota in active and inactive ulcerative colitis. Scand J Gastroenterol. 2009;44:180–186. (abstract).
Martinez C, Antolin M, Santos J, et al. Unstable composition of the fecal microbiota in ulcerative colitis during clinical remission. Am J Gastroenterol. 2008;103:643–648.
Greenstein RJ, Su L, Juste RA, et al. On the action of cyclosporine A, rapamycin and tacrolimus on M avium including subspecies paratuberculosis. PLoS ONE. 2008;3:e2496.
Finegold SM, John SS, Vu AW, et al. In vitro activity of ramoplanin and comparator drugs against anaerobic intestinal bacteria from the perspective of potential utility in pathology involving bowel flora. Anaerobe. 2004;10:205–211.
Hoffmann M, Kim SC, Sartor RB, et al. Enterococcus faecalis strains differentially regulate Alix/AIP1 protein expression and ERK 1/2 activation in intestinal epithelial cells in the context of chronic experimental colitis. J Proteome Res. 2009;8:1183–1192.
Balish E, Warner T. Enterococcus faecalis induces inflammatory bowel diseases in interleukin-10 knockout mice. Am J Pathol. 2002;160:2253–2257.
Ishikawa H, Akedo I, Umesaki Y, et al. Randomized controlled trial of the effect of bifidobacteria-fermented milk on ulcerative colitis. J Am Coll Nutr. 2002;22:56–63.
Furrie E, Macfarlane S, Kennedy A, et al. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with ulcerative colitis: a randomized controlled pilot trial. Gut. 2005;54:242–249.
Kato K, Misuno S, Umesaki Y, et al. Randomized placebo-controlled trial assessing the effect of bifidobacteria-fermented milk on active ulcerative colitis. Aliment Pharmacol Ther. 2004;20:1133–1141.
Pitcher MCL, Beatty ER, Cummings JH. The contribution of sulfate-reducing bacteria and 5-aminosalicylic acid to fecal sulfide in patients with ulcerative colitis. Gut. 2000;46:64–72.
Louis P, Scott KP, Duncan SH, et al. Understanding the effects of diet on bacterial metabolism in the large intestine. J Appl Microbiol. 2007;102:1197–1208.
Neish AS. Microbes in gastrointestinal health and disease. Gastroenterol. 2009;136:64–80.
Bourriaud C, Robins RJ, Martin L, et al. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. J Appl Microbiol. 2005;99:201–212.
Takaishi H, Matsuki T, Nakazawa A, et al. Imbalance in intestinal microflora constitution could be involved in the pathogenesis of inflammatory bowel disease. Int J Med Microbiol. 2008;298:463–472.
Marchesi JR, Holmes E, Khan F, et al. Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J Proteome Res. 2007;6:546–551.
Joossens M, Huys G, Cnochaert M, et al. Dysbiosis of the fecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut. 2011;60:631–637.
Geypens B, Claus D, Evenepoel P, et al. Influence of dietary protein supplements on the formation of bacterial metabolites in the colon. Gut. 1997;41:70–76.
Blachier E, Mariotti F, Huneau J, et al. Effects of amino acid-derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids. 2007;33:547–562.
Khoruts A, Dicksved J, Jansson JK, et al. Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol. 2010;44:354–360.
Borody TJ, Warren EF, Leis S, et al. Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol. 2003;37:42–47.
Acknowledgments
We express our appreciation to Dr Masako Sei for helpful advice on the statistical analysis, Dr Chiho Goto for instructive advice on the analysis of dietary habits in the subjects, Mr Shigeo Misawa for technical support with the T-RFLP analysis, Mr Hirofumi Niki for technical support with quantification of organic acids by gas chromatography, and Miss Saori Nagae and Miss Azusa Matsumoto for counting the number of Gram-stained bacteria.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below are the links to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nemoto, H., Kataoka, K., Ishikawa, H. et al. Reduced Diversity and Imbalance of Fecal Microbiota in Patients with Ulcerative Colitis. Dig Dis Sci 57, 2955–2964 (2012). https://doi.org/10.1007/s10620-012-2236-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-012-2236-y