Abstract
Background
Embryonic ectoderm development (EED) protein is involved in multiple cellular protein complexes. EED mediates the repression of gene activity through histone deacetylation, and it may act as a specific regulator of integrin’s function. This gene was identified as a candidate gene for the susceptibility to IBD by our previous cDNA microarray analysis.
Aim
The present study aimed to validate the expression level of the EED gene in patients with IBD by performing RT-PCR, and we investigated whether the polymorphisms in the EED gene are associated with the susceptibility to UC, and whether a functional EED promoter polymorphism is related to UC.
Methods
Genotype analysis of the EED SNPs was performed by single-base extension analysis. The haplotype frequencies of the EED gene for multiple loci were estimated using the expectation maximization algorithm. The promoter region of the human EED gene, including the g.-1850G>C allele, was isolated by PCR. The amplified PCR products were inserted into the pGL3-basic vector and the luciferase activity was analyzed.
Results
The expression level of the EED gene was significantly decreased in both the UC and CD patients and it was significantly higher in the liver and ileum than in the other tissues of the human digestive system. The genotype and allele frequencies of the g.-1850G>C polymorphism of the EED gene in the UC patients were significantly different from those of the healthy controls (p = 0.018 and 0.017, respectively). The luciferase activity assay showed that the promoter activity was decreased about twofold in the construct containing the g.-1850G allele compared to that of the construct containing the g.-1850C allele, which means that the allele G could produce less EED mRNA.
Conclusions
These results suggest that the g.-1850G>C polymorphism in the EED gene might be associated with the susceptibility to UC by the change of the EED expression level.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Inflammatory bowel disease (IBD) is a complex multifactorial disorder that involves genetic, environmental, and microbial factors [1, 2]. Crohn’s disease (CD) and ulcerative colitis (UC) are the two main clinical forms of IBD in humans [3]. Within these subgroups, there are many differences regarding the disease extent, localization, and behavior and the occurrence of extra-intestinal manifestations [4, 5]. While the inflammation in UC starts from the anal margin and it is limited to the mucosa of the colon, the inflammation in CD is trans-mucosal, and it can occur throughout the gastrointestinal tract, and the inflammation is characterized by a T helper 1 cell (Th1) response [6]. Multiple susceptibility loci have been implicated in IBD by a genome-wide linkage analysis in siblings with IBD or in relative-paired families [7]. Many studies have suggested the existence of at least nine IBD loci (referred to as IBD 1–9) in the human genome. Whereas some loci appear to be specific to CD (IBD1) [8, 9] or UC (IBD2) [10], others seem to confer susceptibility to both CD and UC (IBD3) [11, 12].
The embryonic ectoderm development (EED; also called WAIT-1) protein is involved in multiple cellular protein complexes, and it is a member of the superfamily of Polycomb group (PcG) proteins [13, 14] that mediate the silencing of hundreds of genes that are important for embryonic development, cell proliferation, and differentiation [15–17]. The human EED works in both the nucleus and the plasma membrane, and it can interact with several cellular proteins such as the cytoplasmic tail of the integrin β7 subunit [18] and three HIV-1 proteins, the structural protein matrix (MA) [19], the enzyme integrase (IN) [20], and the regulatory protein Nef [21]. The EED is a member of the Polycomb Repressive Complexes (PRCs) that are found in Drosophila and in mammals [22]. Several types of the PRCs, PRC1, PRC2, and PRC3 have been identified [23]. The PRC2/3 complex consists of at least five components, EED, EZH2, SUZ12, RbAp38, and AEBP2 [24, 25]. The EED play a role in targeting the regions of proviral DNA that are integrated into the host chromatin during the early phase of the HIV-1 life cycle [26]. At the late steps of the HIV-1 replication cycle, the overexpression of EED had a negative effect on virus production [27].
We previously conducted a study to find the genes that are related to IBD by performing a cDNA microarray analysis using the mRNAs isolated from healthy controls and IBD (UC and CD) patients, and several tens of genes were identified as having higher or lower mRNA levels in the IBD patients [28]. One of them, the EED gene, was identified as a candidate gene because the expression level of the EED gene was decreased in both the UC and CD patients as compared to that of the healthy controls. However, there was no information about the genetic variation in the EED gene or the role of EED in the pathogenesis of IBD. In this study, we examined the gene expression profiles in the tissues obtained from human digestive tracts, and we tested whether the single-nucleotide polymorphisms (SNPs) of the EED gene might be associated with the susceptibility to UC. To determine whether these EED SNPs are associated with the susceptibility to UC, we have analyzed and compared their frequencies in 142 UC patients and 535 healthy controls. We further investigated the haplotype frequencies by these SNPs between the UC patient group and the healthy control group. We also evaluated the expression levels in the putative functional promoter polymorphisms by performing a luciferase reporter assay to determine the change of their activity.
Materials and Methods
Patients and DNA Samples
Blood samples were obtained from 142 UC patients (77 males and 65 females) and 535 healthy controls (334 males and 201 females). The mean ages of the UC patients and the healthy controls were 40.4 and 40.7 years, respectively. Genomic DNA was extracted from the leukocytes of the peripheral blood by a standard phenol–chloroform method or by using a Genomic DNA Extraction kit (iNtRON Biotechnology, Korea) according to the manufacture’s instructions. The UC patients were recruited from the outpatient clinic at Wonkwang University Hospital. The patients were classified according to clinical features, endoscopic findings, and histopathologic examinations. The healthy controls were recruited from the general population and they had undergone comprehensive medical screening at Wonkwang University Hospital. All the subjects enrolled in this study were Koreans.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
To validate the gene expression data obtained by the cDNA microarray analysis, approximately 1 μg of total RNA was used in the first-strand cDNA synthesis with a sequence-specific primer and using M-MLV reverse transcriptase (Bioneer, Korea) for reverse transcription-polymerase chain reaction (RT-PCR). The PCR samples were prepared in a 20-μl reaction volume containing 50 ng cDNA, 10× buffer 2 μl, 10 mM dNTP 0.5 μl, EF-taq 0.2 U (Solgent, Korea) and 0.5 μM of each primer (Table 1) under the following conditions: 10 min at 95°C for initial denaturation, and than 30 cycles of PCR that consisted of denaturation for 10 s at 98°C, annealing for 15 s at 55°C, and extension for 30 s at 72°C, and a final extension step was done for 10 min at 72°C in a PCR Thermal Cycler DICE Gradient (TaKaRa, Japan). The expression assay for EED mRNA was performed using the cDNA obtained from various human digestive tract tissues (Clontech, USA).
SNP Selection
On the basis of the Genbank dbSNP database (http://www.ncbi.nlm.nih.gov), we selected three SNPs (rs7952481, rs1391221, and rs974144) in the EED gene. The information of rs7952481 (g.-1850G>C), rs1391221 (g.-993G>C), and rs974144 (g.12351C>T) SNPs was derived from the NCBI SNP database. The SNPs with unknown heterozygosity and minor allele frequencies below 5% in an Asian population were excluded. The reference sequence for the EED gene was based on the sequence of human chromosome 11, NC_000011.9.
Genotype Analysis
Genotype analysis for the SNPs of the EED gene was performed by high-resolution melting (HRM) analysis. The 10-μl reaction mixtures were made up using the 1× QuantiTect Probe PCR Kit (Qiagen, USA) and they consisted of 50 ng of genomic DNA, 100 nM of each primer (Table 1), and 1× Evagreen solution (Biotium, USA). The PCR cycling and HRM analysis was carried out using a Rotor-Gene thermal cycler RG6000 (Corbett Research, Australia). The PCR cycling conditions were as follows: one cycle of 95°C for 15 min; 45 cycles of 95°C for 10 s, and the annealing conditions were 58°C for 10 s and 72°C for 30 s; HRM analysis was performed at 77–95°C with the temperature being increasing at 0.1°C/s and the fluorescence was recorded.
Statistic Analysis
χ2 tests were applied to estimate the Hardy–Weinberg equilibrium (HWE). Pair-wise comparison of the biallelic loci was employed for the analyses of the Linkage Disequilibrium (LD). The haplotype frequencies of the EED gene for multiple loci were estimated using the expectation maximization (EM) algorithm with SNPAlyze software (DYNACOM, Japan). Logistic regression analyses (SPSS 11.5) were used to calculate the odds ratios (with the 95% confidence intervals). A p value <0.05 was considered to indicate statistical significance.
Luciferase Assay
The promoter region of the human EED gene carrying either the g.-1850G or g.-1850C allele was inserted upstream of the firefly luciferase gene in the pGL3 basic plasmid vector (Promega, Madison, WI, USA) and this was verified by DNA sequencing. To make these constructs, we amplified the target sequence by PCR using a forward primer with SacI and a reverse primer with HindIII (Table 1). These promoter plasmids were co-transfected into the 293T cells (ATCC, USA) with a pRLTK control vector (Promega), using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), according to the manufacturer’s instructions. The assays for firefly luciferase activity and Renilla luciferase activity were performed 48 h after the transfection of the cells using the Dual-Luciferase Reporter Assay System (Promega) with a microplate luminometer LB 96V (BERTHOLD, Australia). To normalize the transfection efficiency, the luminescence value of the pGL3-basic vector was standardized with the value of the pRL-TK vector. Each experiment was repeated three times, and each sample was studied in triplicate.
Results
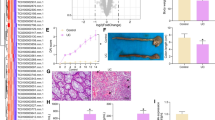
The EED gene was selected as a candidate susceptibility gene for IBD by our previous cDNA microarray analysis [28] because the expression level of the EED gene was more decreased in both the UC and CD patients than that of the healthy controls. To validate the accuracy of the microarray expression profiling data, the expressed levels of the EED gene were compared in three CD and UC patients and three healthy controls by RT-PCR. As shown in Fig. 1a, the expressed level of EED mRNA was significantly down-expressed in the peripheral blood of both the UC and CD patients as compared to that of the healthy controls. The expression pattern of EED mRNA in 12 normal tissues obtained from the digestive tract was also examined. The results showed that the expressed level of EED mRNA in the tissues from the human digestive system was highest in the liver and ileum tissues, while the expressed levels of EED mRNA in the transverse colon and rectum tissues were barely detectable (Fig. 1b).
To determine whether the EED SNPs are associated with UC susceptibility, the genotypes of the EED polymorphisms were analyzed by the HRM method, and the genotype and allelic frequencies between the UC patient and healthy control groups were compared. The genotype frequencies of all loci were in HWE, except for g.-993G>C in the healthy controls, as determined by χ2 tests (data not shown). The genotype and allelic frequencies of the g.-1850G>C SNP in the UC patient group were significantly different from those of the healthy control group (Table 2; p = 0.018 and 0.017, respectively). These results suggest that the g.-1850G>C SNP in the EED gene appear to be associated with UC susceptibility.
We further analyzed the haplotype frequencies of the SNPs in the UC patients and the healthy controls to determine the possible correlation between the haplotypes associated with g.-1850G>C, g.-993G>C and g.12351C>T of the EED gene and the susceptibility to UC (Table 3). Out of eight possible haplotypes, four major haplotypes (98.8 and 97.8%, respectively) were identified in the healthy controls and UC patients, respectively (Table 3). The distribution of the two haplotypes (GGT and CGC) were notably different in the UC patients compared to that of the healthy controls (p = 0.024 and 0.033, respectively). These results suggest that EED polymorphisms might be an important genetic factor associated with UC susceptibility.
To evaluate the putative functional polymorphisms, we assessed actual promoter activity with the luciferase reporter assay (Fig. 2). We prepared two reporter constructs that contained the G or C allele of the human EED gene g.-1850G>C polymorphisms. In the 293T cells, the luciferase activity in the cells with the C allele with g.-1850G>C polymorphism was at least twofold higher than that in the cells with the G allele. This result suggests that the g.-1850C allele might affect the increased transcriptional activity of the EED gene in vivo.
EED promoter activity between polymorphisms by luciferase reporter assay. EED g.-1850G and EED g.-1850C represent luciferase activities in two constructs with a major allele (g.-1850G) and a minor allele (g.-1850C), respectively. Control represents luciferase activity without polymorphism as a standard (pGL3 basic only). Data were obtained from three independent experiments and are represented as the mean ± SE. Data are standardized as a mean value in the control
Discussion
IBD is a chronic disease that is frequently encountered in the gastrointestinal tract and it can profoundly affect the quality of life. Although great advances have been made in the management of IBD with the introduction of immune-modulators and monoclonal antibodies, the precise etiology of IBD is unclear. However, IBD is thought to be the result of an aberrant intestinal immune response to bacterial microflora and this occurs in genetically susceptible individuals. DNA microarray analysis can be used to identify susceptibility candidate genes in complex genetic disorders and to identify novel disease-related genes. This technology has been used to study gene expression profiling in the mucosal biopsies from UC or CD patients and healthy controls, and it has identified the genes with altered expression, including the CD74, NGAL, GRO, and calcium-binding S100 genes [29, 30]. We previously reported that an exon 4 variation of the Tim-1 gene and the SNPs of the IL27 and TNFRSF17 genes were associated with UC in a Korean population [28, 31, 32].
The EED gene was previously identified as a candidate gene because the expressed level of this gene was more decreased in both the UC and CD patients than that of the healthy controls, as was determined by cDNA microarray analysis using the mRNAs isolated from healthy controls and IBD (UC and CD) patients [28]. However, there was no information on the relation of genetic variations of the EED gene and gastrointestinal disease such as IBD. In this study, we validated the expressed levels of the EED gene by RT-PCR with using the total RNAs isolated from three CD or UC patients and three healthy controls. The expression levels of the EED gene in both the UC and CD patients were shown to be similar to the results of the cDNA microarray analysis (Fig. 1a). We analyzed the genotypes of the EED gene SNPs in the UC patients and healthy controls. The genotype and allele frequencies of the g.-1850G>C polymorphism of the EED in the UC patients were significantly different from those in the controls group (Table 2). This result suggests that the g.-1850G>C polymorphism of the EED might be associated with the susceptibility to UC. The distributions of the two haplotypes (GGT and CGC) of the EED SNPs g.-1850G>C, g.-993G>C and g.12351C>T in the UC patients were also notably different from that of the healthy controls (Table 3). These results suggest that EED polymorphisms might be an important genetic factor associated with UC susceptibility.
These results led us to think it is interesting to know that EED gene polymorphism (g.-1850G>C) may have some influence on the susceptibility to UC. This may happen because the polymorphisms within the binding site of the promoter region may influence the expression level by suppression or activation of binding between the specific transcriptional binding site and transcription factor. Due to its implication, we decided to investigate the role of the g.-1850G>C polymorphism in the EED by luciferase reporter assay. Our result shows that the luciferase activity for the C allele with g.-1850G>C polymorphism was higher than that for the G allele in the 293T cells (Fig. 2). This result indicates that the g.-1850C allele might affect the increased transcriptional activity level of the EED gene in vivo, and it might have an influence on the susceptibility to UC.
In conclusion, the results of this study suggest that the EED gene might be a candidate gene associated with the pathogenesis of UC. The haplotypes of the EED polymorphisms might be one of the markers for UC susceptibility. Although it is not clear how the EED polymorphisms are related to the susceptibility of UC, our results provide useful information for further functional studies of the EED gene and gastrointestinal disease such as colorectal cancer and inflammatory responses. It will be interesting to show the expression levels of the EED in the colon tissues of UC patients and health controls by further studies. This may substantiate that the EED is involved in regulation of inflammation.
References
Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. 1998;115:182–205.
Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429.
Blumberg RS, Saubermann LJ, Strober W. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr Opin Immunol. 1999;11:648–656.
Travis S, Jewell DP. Ulcerative colitis: clinical presentation and diagnosis. In: Satsangi J, Sutherland LR, eds. Ciclosporin Use in Acute Ulcerative Colitis: A Long-Term Experience. New York: Churchill Livingstone; 2003:169–182.
Forbes A. Clinical presentation and diagnosis of Crohn’s disease. In: Satsangi J, Sutherland LR, eds. Fistulating anal Crohn’s Disease: Results of Combined Surgical and Infliximab Treatment. New York: Churchill Livingstone; 2003:183–189.
Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, et al. Disparate CD4+ lamina propria (LP) lymphocyte secretion profiles in inflammatory bowel disease. Crohn’s disease LP cells manifest increased secretion of IFN gamma, whereas, ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. 1996;157:1261–1270.
Ahmad T, Tamboli CP, Jewell D, Colombel JF. Clinical relevance of advances in genetics and pharmacogenetics of IBD. Gastroenterology. 2004;126:1533–1549.
Cavanaugh J. International collaboration provides convincing linkage replication in complex disease through analysis of a large pooled data set: Crohn’s disease and chromosome 16. Am J Hum Genet. 2001;68:1165–1171.
Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, et al. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature. 1996;379:821–823.
Parkes M, Barmada MM, Satsangi J, Weeks DE, Jewell DP, et al. The IBD2 locus shows linkage heterogeneity between ulcerative colitis and Crohn’s disease. Am J Hum Genet. 2000;7:1605–1610.
Hampe J, Shaw SH, Saiz R, Leysens N, Lantermann A, et al. Linkage of inflammatory bowel disease to human chromosome 6p. Am J Hum Genet. 1999;65:1647–1655.
van Heel DA, Fisher SA, Kirby A, Daly MJ, Rioux JD, Lewis CM. Inflammatory bowel disease susceptibility loci defined by genome scan meta-analysis of 1952 affected relative pairs. Hum Mol Genet. 2004;13:763–770.
Schumacher A, Lichtarge O, Schwartz S, Magnusson T. The murine Polycomb-group EED and its human orthologue: functional implications of evolutionary conservation. Genomics. 1998;54:79–88.
Sewalt RGAB, van der Vlag J, Gunster MJ, Hamer KM, den Blaauwen JL, et al. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595.
Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353.
Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genomewide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136.
Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313.
Crowe DT, Chiu H, Fong S, Weissmann IL. Regulation of the avidity of integrin alpha4 beta7 by the beta7 cytoplasmic domain. J Biol Chem. 1994;269:14411–14418.
Peytavi R, Hong SS, Gay B, d’Angeac AD, Selig L, et al. H-EED, the product of the human homolog of the murine EED gene, binds to the matrix protein of HIV-1. J Biol Chem. 1999;274:1635–1645.
Violot S, Hong SS, Rakotobe D, Petit C, Gay B, et al. The human Polycomb group EED protein interacts with the integrase of human immunodeficiency virus type 1. J Virol. 2003;77:12507–12522.
Witte V, Laffert B, Rosorius O, Lischka P, Blume K, et al. HIV-1 Nef mimics an integrin receptor signal that recruits the Polycomb group protein EED to the plasma membrane. Mol Cell. 2004;13:179–190.
Otte AP, Kwaks THJ. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–454.
Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, et al. Silencing of human Polycomb target genes is associated with methylation of histone H3 Lys27. Genes Dev. 2004;18:1592–1605.
Kuzmichev A, Jenuwein T, Tempst P, Reinberg D. Different Ezh2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol Cell. 2004;14:183–193.
Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EEDEZH2 complex. Mol Cell. 2004;15:57–67.
Rakotobe D, Violot S, Hong SS, Gouet P, Boulanger P. Mapping of immunogenic and protein-interacting regions at the surface of the seven-bladed beta-propeller domain of the HIV-1 cellular interactor EED. Virol J. 2008;27:5–32.
Rakotobe D, Tardy JC, André P, Hong SS, Darlix JL, et al. Human Polycomb group EED protein negatively affects HIV-1 assembly and release. Retrovirology. 2007;4:4–37.
Chae SC, Yu JI, Oh GJ, Choi CS, Choi SC, et al. Identification of single nucleotide polymorphisms in the TNFRSF17 gene and their association with gastrointestinal disorders. Mol Cell. 2010;29:21–28.
Lawrance IC, Fiocchi C, Chakravarti S. Ulcerative colitis and Crohn’s disease: distinctive gene expression profiles and novel susceptibility candidate genes. Hum Mol Genet. 2001;10:445–456.
Warner EE, Dieckgraefe BK. Application of genome-wide gene expression profiling by high-density DNA arrays to the treatment and study of inflammatory bowel disease. Inflamm Bowel Dis. 2002;8:140–157.
Park YR, Choi SC, Lee ST, Kim KS, Chae SC, et al. The association of eotaxin-2 and eotaxin-3 gene polymorphisms in a Korean population with ulcerative colitis. Exp Mol Med. 2005;37:553–558.
Li CS, Zhang Q, Lee KJ, Cho SW, Lee KM, et al. Interleukin 27 polymorphisms are associated with inflammatory bowel diseases in a Korean population. J Gastroenterol Hapatol. 2009;24:1692–1696.
Acknowledgments
The biospecimens for this study were provided by the Biobank of Wonkwang University Hospital, a member of the National Biobank of Korea, which is supported by the Ministry of Health, Welfare and Family Affairs. This research was supported by Wonkwang University in 2010.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ji-In Yu and In-Hong Kang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Yu, JI., Kang, IH., Seo, GS. et al. Promoter Polymorphism of the EED Gene Is Associated with the Susceptibility to Ulcerative Colitis. Dig Dis Sci 57, 1537–1543 (2012). https://doi.org/10.1007/s10620-012-2045-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-012-2045-3