Abstract
Purpose
Epithelial cellular adhesion molecule (EpCAM) is an attractive immunotherapeutic target to overcome metastasis of a variety of epithelium-oriented cancers. Edrecolomab, one kind of EpCAM monoclonal antibody (Panorex®), has been approved for clinical application as postoperative adjuvant therapy in breast and colorectal cancer. However, the role of EpCAM in gastric cancer metastasis remains unclear.
Results
EpCAM was found to be more highly overexpressed in metastatic gastric cancer than in nonmetastatic samples by immunohistochemistry staining. The expression level of EpCAM in gastric cancer cell lines was determined by reverse-transcription polymerase chain reaction (RT-PCR) and Western blotting, respectively. Downregulation of EpCAM by small interfering RNA (siRNA) significantly suppressed in vitro adhesive, invasive, and migratory and in vivo metastatic abilities of gastric cancer cells.
Conclusion
We provide first evidence that EpCAM contributes to the migration of gastric cancer, suggesting that EpCAM-targeted therapy might be a promising strategy in metastatic gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In spite of intense interest and extensive investigations, the 5-year survival rate of gastric cancer, the leading cause of death from cancer in China, has not improved significantly in recent years. The formation and growth of metastases at distant sites are responsible for the death of most gastric cancer patients. Exploration into regulatory and migratory factors associated with this process might help to elucidate the mechanisms of gastric cancer metastasis and prevent these processes.
Many cell adhesion molecules (CAMs) have been characterized in recent decades and been found to be involved in tumorigenesis and metastasis of cancers [1, 2]. It has been demonstrated that loss of function of E-cadherin, an important CAM, plays a role in susceptibility to initial tumor development and is a prerequisite for tumor cell invasion and metastasis formation. Re-establishing the functional cadherin complex, e.g., by forced expression of E-cadherin, results in reversion from an invasive mesenchymal to a benign epithelial phenotype of cultured tumor cells [3, 4].
Epithelial cellular adhesion molecule (EpCAM), a CAM found in 1979, is a 39–42-kDa, 314-amino-acid, type I transmembrane glycoprotein encoded by the TACSTD1 that was mapped to chromosome 2p21 and is known to mediate Ca2+-independent homotypic cell–cell adhesion [5, 6]. Recent insights revealed a more versatile role for EpCAM, not merely limited to cell adhesion but, similar to other CAMs, including processes such as signaling, cell migration, proliferation, and differentiation, which led to its application as an adjuvant therapy for a variety of cancers [7]. Meanwhile, EpCAM has been shown to have effects on metastasis of several kinds of cancer. Its antibody, edrecolomab (Panorex®), was in licensed clinical use in Germany in 2002 as postoperative adjuvant therapy for breast cancer and rectal cancer, with increased 7-year survival of 21% and reduced recurrence rate of 30% [5, 8, 9], which indicated that EpCAM could serve as a promising antimetastatic target. However, its role in gastric cancer has not been elucidated to date.
In our present study, EpCAM was shown to be highly expressed in gastric cancer tissues and cell lines. More importantly, EpCAM expression was higher in metastatic gastric cancer than in nonmetastatic cancer tissues. Downregulation of EpCAM by EpCAM small interfering RNA (siRNA) resulted in decrease of cell proliferation in AGS and SGC7901 cells, which had high endogenous EpCAM expression. And EpCAM downregulation significantly suppressed in vitro adhesive, invasive, and migratory and in vivo metastatic abilities of gastric cancer cells. Taken together, the results described herein strongly suggest that EpCAM might be used as a potential therapeutic candidate for metastatic gastric cancer.
Materials and Methods
Patients, Cell Lines, and Animals
Specimens used in this study consisted of 100 cases of gastric adenocarcinoma. The patients underwent gastrectomy between August 2006 and August 2007 in Xijing Hospital (Xi’an, China). Data on gender, age, tumor size, histological type of neoplasm, and lymph–node–metastasis status were obtained from surgical and pathological records. None of the patients had received preoperative chemotherapy or radiation therapy. Informed consent was obtained from all patients.
Five gastric cancer cell lines preserved in our laboratory, including AGS, MKN28, MKN45, SGC7901, and 9811P, were incubated at 37°C in a humidified atmosphere of 5% CO2 in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% fetal bovine serum (FBS). Cells growing to 80–90% confluence were digested with 0.25% trypsin–0.02% ethylenediamine tetraacetic acid (EDTA) and subcultured.
BALB/c nude mice, 4–6 weeks old, were provided by Shanghai Cancer Institute for tail vein metastatic assay. The experiments were performed along established, institutional animal welfare guidelines concordant with National Institutes of Health (NIH) species criteria.
Immunohistochemistry Staining and Evaluation
Four-micrometer sections of formalin-fixed paraffin-embedded specimens were made. Immunohistochemistry staining was performed with SP-9000 Histostain-Plus kits (Santa Cruz, CA). Endogenous peroxidase was blocked by incubation with freshly prepared 0.3% H2O2 in methanol for 20 min at room temperature. After blocking with 10% normal goat serum for 30 min, the sections were incubated with mouse anti-human EpCAM monoclonal antibody (prepared and gifted by Department of Immunology, Fourth Military Medical University, 50 μg/l, diluted to 1:100) [10] overnight at 4°C. After that, goat anti-mouse serum was added. Reaction products were visualized with application of diaminobenzidine substrate chromagen solution (DAB). The slides were then counterstained with hematoxylin and mounted. Sections of colon cancer served as positive staining controls. Negative controls were performed by replacing the primary antibody with normal mouse serum. Positive reaction was indicated by a reddish-brown precipitate, mainly in membrane, partly in cytoplasm. All sections were examined independently by two investigators.
EpCAM expression was evaluated by the following formula [11]: staining score = intensity of immunoreactivity × proportion of positively staining cells. Intensity of immunoreactivity was stratified into four categories: 0, no immunoreactivity; 1, weak immunoreactivity; 2, moderate immunoreactivity; and 3, strong immunoreactivity. The proportion of positive cells was classified into four groups: 0, 0–5% of tumor cells exhibiting immunoreactivity; 0.33, 5–33% of tumor cells exhibiting immunoreactivity; 0.67, 33–67% of tumor cells exhibiting immunoreactivity; and 1, 67–100% of tumor cells exhibiting immunoreactivity.
RNA Extraction and Semiquantitative Reverse-Transcription Polymerase Chain Reaction (RT-PCR)
Total cellular RNA was isolated from cell lines with TRIZOL reagent (Invitrogen) according to the manufacturer’s instruction. RT reaction was performed using the First-Strand cDNA Synthesis kit (MBI Fermentas, Vilnius, Lithuania) according to the manufacturer’s protocol. Appropriate cycles were chosen to ensure termination of PCR amplification before reaching a stable stage in each reaction. Gene expression was presented as the yield of PCR products from target sequences relative to the yield of PCR products from the β-actin gene. PCR primers and reaction parameters were as follows: prime sequence for EpCAM: 5′-TCAGAAGAACAGACAAGGAC-3′ (forward) and 5′-ACTGCTATCACCACAACCAC-3′ (reverse); for β-actin: 5′-TGGAACGGTGAAGGTGACAG-3′ (forward) and 5′-TGTGGACTTGGGAGAGGACTG-3′ (reverse). PCR reaction conditions for EpCAM were: 94°C for 30 s, 51°C for 30 s, and 72°C for 1 min; 30 cycles were performed.
Plasmid Construction and Cell Transfection
Two pairs of EpCAM siRNA were designed according to the siRNA design guidelines of Ambion, Inc. as follows: for siRNA1, sense: 5′-GATCC ACTACAAGCTGGCCGTAAA TTCAAGACG TTTACGGCCAGCTTGTAGT TTTTTTGTCGACA-3′, antisense: 3′-G TGATGTTCGACCGGCATTT AAGTTCTGC AAATGCCGGTCGAACATCA-5′; for siRNA2, sense: 5′-GATCC GTTTGGTGATGAAGGCAGA TTCAAGACG TCTGCCTTCATCACCAAAC TTTTTTGTCGACA-3′, antisense: 3′-G CAAACCACTACTTCCGTCT AAGTTCTGC AGACGGAAGTAGTGGTTTG AAAAAACAGCTGTTCGA-5′. The targeting sequences were homologous to nt 270–388, and 375–393 of the EpCAM complementary DNA (cDNA), respectively.
pSilencer3.1 (Ambion Inc.) was used for the construction of human EpCAM siRNA vectors, according to the manufacturer’s protocol. Then siRNA plasmids of EpCAM were transfected into AGS and SGC7901 cells. Cells transfected with pSilencer3.1 vector alone served as negative control. Cells were plated in 24-well plate, allowed to grow for 24 h (until they were 80% confluent), and washed with FBS-free medium. siRNA:lipofectamine 2000 complex (100 μl) was added to each well and mixed gently. Then the cells were incubated for 5 h, after which the medium was replaced with complete medium with 20% FBS and cell incubation was continued for 48–72 h. Then cells were digested and incubated in screening medium containing G418. Expression levels of EpCAM in G418-resistant clones were evaluated by Western blotting analysis.
Western Blotting Analysis
Cells were lysed in radioimmunoprecipitation assay (RIPA) lysis buffer for 15 min on ice. An aliquot (60–70 μg) of lysate was electrophoresed on 10% sodium dodecyl sulfate (SDS) polyacrylamide gel and blotted onto a nitrocellulose membrane. Membranes were blocked in 5% fat-free milk and then incubated separately with mouse monoclonal anti-EpCAM (diluted to 1:1,000) overnight at 4°C. β-actin was used as a loading control (diluted to 1:4,000; Sigma Chemical Co.). After washing, membranes were incubated with horseradish peroxidase–conjugated anti-mouse IgG antibody (diluted to 1:2,000; Santa Cruz) for 1 h at room temperature. The bands were visualized using enhanced chemiluminescence kit (Amersham, Pittsburgh, PA). Each experiment was performed in triplicate.
Growth Assay
Briefly, cells were seeded in 96-well plate at density of 2 × 103 per well and incubated. Absorbance of cultures was assayed on day 1, 2, 3, 4, 5, 6, 7, and 8 with enzyme-linked immunosorbent assay (ELISA) reader (Bio-Rad Laboratories, Richmond, CA) at the wavelength of 490 nm. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) working solution (40 μl 5 mg/ml, Sigma) was added to each well 4 h before each assaying. Each experiment was performed in triplicate.
Adhesion Assay
The ability of gastric cancer cells to adhere to Matrigel was determined in 24-well plates as described by others [12]. The plate surface was covered with 50 μl 50 μg/ml Matrigel, incubated for 2 h at 37°C, and the supernatant was discarded. A suspension (0.5 ml) of cells (1 × 105cells/ml, suspended in RPMI 1604 medium containing 1% FBS) was added to the covered wells. After 0.5, 1, 2, and 4 h of incubation at 37°C, the adhesive cells were washed with phosphate-buffered saline (PBS) twice and then counted under a microscope at 200× magnification in ten random fields for each well. Each experiment was performed in triplicate.
Invasion and Migration Assay
Cell invasion assays were done as described by others [13] using Transwells (8-μm pore size, Corning Costar Corp.). Matrigel solution (50 μL, diluted to a concentration of 2.5 mg/ml with serum-free RPMI 1640) was placed on the upper chamber of Transwells and incubated at 37°C for 6 h and rinsed with serum-free RPMI 1640 gently. Then 600 μl RPMI 1640 containing 25% bovine serum were added to the lower compartment. Freshly trypsinized and washed cells were suspended at 2 × 105/ml in RPMI 1640 containing 1% bovine serum and added to upper compartments (100 μl/well). After incubation for 36 h, cells on the upper side of the membrane were removed by cotton swabs. Cells on the lower side of the membrane were fixed with methanol for 15 min and stained with hematoxylin for 15 min. Invasive ability was determined by counting the number of cells that migrated to the lower side of the membrane using a light microscope at 200× magnification in ten random fields for each well. Each experiment was done in triplicate.
The method of in vitro migration assay, which was analyzed as described previously [13] using Transwells (8-μm pore size, Corning Costar) without Matrigel, was similar to the invasion assay. The cells were suspended at 2 × 104/ml. The incubation time was 36 h.
Tail Vein Metastatic Assay
The tail vein metastatic assay was performed as described previously [14]. Each of 4- to 6-week-old female BALB/c nu/nu mice was injected with 1 × 106 cells in 0.1 ml PBS through tail vein. The mice were then monitored for overall health and total body weight. Four weeks after injection, the mice were sacrificed. The number of visible tumors in liver surface was counted. Experimental and control groups contained five mice, respectively.
Statistical Analysis
The SPSS statistical software package (SPSS, Inc., Chicago, IL) was used to analyze data, and P < 0.05 was considered statistically significant. Chi-square test was adopted to analyze the difference in the frequency of EpCAM overexpression in relation to metastatic status. Assays for characterizing phenotype of cells were analyzed by Student’s t test and one-way analysis of variance followed by Dunnett’s multiple-comparison tests.
Results
EpCAM Was Overexpressed in Metastatic Gastric Cancer
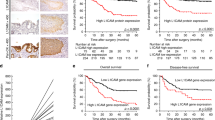
To investigate the relationship between expression of EpCAM and metastatic status of gastric cancer, we compared expression of EpCAM in primary sites from 50 cases of nonmetastatic gastric cancers with those from 50 cases of lymph-node-metastatic gastric cancers. It was found that EpCAM was primarily located on the membrane of gastric cancer cells, partly diffused into cytoplasm of cells (Fig. 1). The average staining score of EpCAM in metastatic gastric cancer was significantly higher than that in nonmetastatic gastric cancer (1.32 ± 0.18 versus 0.73 ± 0.21; P < 0.05) and rate of positive EpCAM expression in metastatic gastric cancer was 90% (45 of 50), which was higher compared with that of nonmetastatic gastric cancer (58%; 29 of 50) (Table 1). We further compared the expression of EpCAM in primary sites with corresponding metastatic lymph node from 50 patients with metastatic gastric cancers. It was shown that the average staining scores of primary sites and corresponding metastatic lymph node were 1.32 ± 0.18 and 1.43 ± 0.27, respectively. No significant difference in intensity of immunoreactivity or average staining score of EpCAM was found between the primary and metastatic sites from the same patients.
EpCAM mRNA and Protein Expression in Gastric Cancer Cell Lines
Messenger RNA (mRNA) and protein levels of EpCAM were determined in five gastric cancer cell lines. As shown in Fig. 2a, b, higher expression of EpCAM mRNA was detected in the gastric cancer cell lines AGS and SGC7901 than in other cell lines. Protein expression of EpCAM displayed a similar pattern to the mRNA level, and a 42-kDa band was detected in all cell lines. Based on the results, AGS and SGC7901 cells were chosen for the following experiments.
Expression of EpCAM in gastric cancer cell lines. a The expression level of EpCAM mRNA in five gastric cancer cell lines was determined by RT-PCR. β-Actin was used as an internal control. b Expression of EpCAM protein in five gastric cancer cell lines was determined by Western blotting. β-Actin was used as an internal control. c After stable transfection, expression of EpCAM in transfectants and parental cells was evaluated by Western blotting. β-Actin was used as an internal control
Downregulation of EpCAM Expression Suppressed Proliferation of Gastric Cancer Cells
To test the effects of EpCAM on gastric cancer cells, we firstly downregulated expression of EpCAM in gastric cancer cells by transfecting two EpCAM-specific siRNA plasmids into AGS and SGC7901 cells, which had highly expressed endogenous EpCAM as determined by RT-PCR and Western blotting. As shown in Fig. 2c, expression of EpCAM in stable transfected cells was significantly reduced and the silencing efficiency of the two siRNA was similar. Growth curve determined by MTT assay showed significant growth inhibition of EpCAM siRNA on AGS and SGC7901 cells from the fourth day onward (Fig. 3a, b); the stably transfected cell lines were used for further assay.
EpCAM siRNA Suppressed Adhesive, Invasive, and Migratory Abilities of Gastric Cancer Cells
Given the known identity of EpCAM as a member of CAMs, believed to correlate with many steps of metastasis of a variety of cancers, we assumed that EpCAM could also affect metastasis. Adhesion to extracellular matrix is the first and essential step of metastasis. The abilities of transfected cells to adhere to Matrigel were determined by adhesive assay. All the cells, parental, vector, and transfected cells, bound to Matrigel in a time-dependent manner, while compared with parental cells, the number of adhesive transfected cells was significantly lower at each time point (Fig. 4a, b). Invasion and migration are the subsequent steps in the process of tumor metastasis. We investigated the influences of EpCAM on invasive and migratory abilities of gastric cancer cells in vitro. As shown in Fig. 4c, EpCAM siRNA1 and siRNA2 significantly reduced the invasive abilities of AGS and SGC7901 cells through Matrigel, respectively. Similar results were observed in in vitro migration assay (Fig. 4d). Tail vein metastatic assay in nude mice was then performed to examine in vivo metastatic abilities of cells. Considering the properties of SGC7901, whose success rate of tumor formation was high and period of tumor formation relatively short, we chose this cell line to perform tail vein metastatic assay in nude mice. The result showed that, compared with control cells, transfected with empty vector, intravenous inoculation of SGC7901- EpCAM siRNA1 and SGC7901- EpCAM siRNA2 cells led to significantly less visible tumors in liver surface (Fig. 4e). Both in vitro invasion assay and in vivo nude mice assay demonstrated that EpCAM siRNA had the potential to inhibit metastasis of gastric cancer.
Effects of EpCAM siRNA on adhesive, invasive, and migratory abilities of gastric cancer cells. Representative of three experiments with similar results. a and b: After 0.5, 1, 2, and 4 h of incubation, AGS and SGC7901 and their transfected cells attached to Matrigel were counted under a microscope, respectively. * Statistical significance. c Invasive ability was evaluated by counting cells invading through Matrigel and membrane with 8-μm-pore Transwell. * Statistical significance. d Migratory ability was evaluated by counting cells migrating through the membrane with 8-μm-pore Transwell. * Statistical significance. e Mice were injected with cells through tail vein. Experimental and control groups contained five mice, respectively. Four weeks later, the mice were sacrificed. The number of visible tumors in liver surface was counted. * Statistical significance
Discussion
The use of immunotherapy to target tumors is well established. Successful clinical uses include the treatment of breast cancer by human epidermal growth factor receptor-2 (HER2)-specific trastuzumab and follicular non-Hodgkin B-cell lymphoma by CD20-specific rituximab [15].
EpCAM is a novel molecule that has attracted increasing attention of cancer researchers, especially those focusing on immunotherapeutic strategy, in recent years. In the present study, we explored for the first time the role of EpCAM in metastatic gastric cancer. The data presented herein provide compelling evidence that EpCAM was overexpressed in gastric cancer, as determined by immunohistochemical staining, especially in metastatic gastric cancer. To confirm our results, a series of in vitro and in vivo assays were performed. It was found that downregulation of EpCAM suppressed invasion and migration of gastric cancer cells, which suggested that EpCAM might be a prometastatic molecule in gastric cancer as well as in breast cancer.
Given the role of EpCAM as a homophilic CAM, it was initially postulated to negatively regulate metastasis of tumor as well as other CAMs, e.g., E-cadherin [4], integrin [16, 17], and selectin [18], which have been demonstrated to play an inhibitory role in tumor metastasis, although this result seemed to present a paradox. Further understanding of EpCAM-mediated adhesion shed light on this subject. Studies showed that EpCAM negatively modulated cadherin-mediated cell adhesion by disruption of the link between α-catenin and F-actin [19]. In this way, EpCAM loosened the tight cell–cell adhesions and modulated proliferation, differentiation, and tissue maintenance. Similar phenomena were already confirmed in breast and renal cancer. As such, we anticipated that, in gastric cancer, overexpression of EpCAM could disrupt cell–cell contact as well, to enable cell migration required for metastasis. Only recently, two directly interacting molecules of EpCAM, CD44v4-v7 [20] and clautin-7 [21], have been identified by immunoprecipitation (IP) and mass spectrography. CD44v4-v7 is a surface molecule that promotes tumor metastasis [22]. The latter, clautin-7, is a crucial element of tight junctions and has been found to be downregulated during invasion and metastasis in breast and esophageal carcinoma [23]. The evidence described above can also provide an explanation for the positive effect of EpCAM in metastasis of cancer.
EpCAM antibody, edrecolomab (Panorex®), has been approved for clinical application in breast cancer and rectal cancer. Some preclinical trials of EpCAM-targeted immunotherapy for intra-abdominal carcinomas have been viewed as successful. Because of the promising use of EpCAM in several kinds of cancer, it is expected that EpCAM might be used to treat gastric cancer metastasis. Indeed, with full understanding of the biological character of EpCAM and improvement of EpCAM monoclonal antibody, EpCAM-targeted immunotherapy seems exciting and may bring forth a novel approach for effective activation and harnessing of the immune system to destroy a pathological aberrance that has otherwise largely escaped attention.
In short, the experiments reported herein demonstrate for the first time that EpCAM was highly expressed in gastric cancer and even more highly in metastatic gastric cancer. In vitro and in vivo, EpCAM silencing could repress invasion and metastasis of gastric cancer cells. Our results strongly suggest that EpCAM is a promising therapeutic target in metastatic gastric cancer and provide first proof that EpCAM antibody might be used as a novel adjuvant strategy for metastasis of gastric cancer.
References
Gulubova MV. Expression of cell adhesion molecules, their ligands and tumour necrosis factor alpha in the liver of patients with metastatic gastrointestinal carcinonoms. Histochem J. 2002;34:67–77.
Ploverini P. Cellular adhesion molecules: Newly identified mediators of angiogenesis. Am J Pathol. 1996;148:1023–1029.
Huntsman DG, Carneiro F, Lewis FR, et al. Early gastric cancer in young, asymptomatic carriers of germ-line E-cadherin mutations. N Engl J Med. 2001;344:1904–1909.
Vleminckx K, Vakaet L Jr, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119.
Calabrese G, Crescenzi C, Morizio E, Palka G, Guerra E, Alberti S. Assignment of TACSTD1 (alias TROP1, M4S1) to human chromosome 2p21 and refinement of mapping of TACSTD2 (alias TROP2, M1S1) to human chromosome 1p32 by in situ hybridization. Cytogenet cell genet. 2001;92:164–165.
Balzar M, Winter MJ, de Boer CJ, Litvinov SV. The biology of the 17–1A antigen (Ep-CAM). J Mol Med. 1999;77:699–712.
Baeuere PA, Gires O. EpCAM (CD326) finding its role in cancer. Br J Cancer. 2007;96:417–423.
Riethmüller G, Schneider-Gädicke E, Schlimok G, et al. Randomised trial of monoclonal antibody for adjuvant therapy of resected Duke’s C colorectal carcinoma. Lancet. 1994;343:1177–1183.
Punt CJ, Nagy A, Douillard JY, et al. Edrecolomab alone or in combination with fluorouracil and folinic acid in the adjuvant treatment of stage III colon cancer: A randomized study. Lancet. 2002;360:671–677.
Xie X, Wang CY, Cao YX, et al. Expression pattern of epithelial cell adhesion molecule on normal and malignant colon tissues. World J Gastroenterol. 2005;11:344–347.
Liu N, Bi F, Pan Y, et al. Reversal of the malignant phenotype of gastric cancer cells by inhibition of RhoA expression and activity. Clin Cancer Res. 2004;10:6239–6247.
Thamilselvan V, Basson MD. Pressure activates colon cancer cell adhesion by inside-out focal adhesion complex and actin cytoskeletal signaling. Gastroenterology. 2004;126:8–18.
Jin H, Pan Y, He L, et al. p75 neurotrophin receptor inhibits invasion and metastasis of gastric cancer. Mol Cancer Res. 2007;5:423–433.
Pan Y, Zhao L, Liang J, et al. Cellular prion protein promotes invasion and metastasis of gastric cancer. FASEB J. 2006;20:1886–1888.
Chaudry MA, Sales K, Ruf P, Lindhofer H, Winslet MC. EpCAM an immunotherapeutic target for gastrointestinal malignancy: Current experience and future challenges. Br J Cancer. 2007;96:1013–1019.
Ito R, Oue N, Zhu X, et al. Expression of integrin-linked kinase is closely correlated with invasion and metastasis of gastric carcinoma. Virchows Arch. 2003;442:118–123.
Hippo Y, Yashiro M, Ishii M, et al. Differential gene expression profiles of scirrhous gastric cancer cells with high metastatic potential to peritoneum or lymph nodes. Cancer Res. 2001;61:889–895.
Chen JL, Chen WX, Zhu JS, et al. Effect of P-selectin monoclonal antibody on metastasis of gastric cancer and immune function. World J Gastroenterol. 2003;9:1607–1610.
Winter MJ, Nagelkerken B, Mertens AE, Rees-Bakker HA, Briaire-de Bruijn IH, Litvinov SV. Expression of Ep-CAM shifts the state of cadherin-mediated adhesions from strong to weak. Exp Cell Res. 2003;285:50–58.
Schmidt DS, Klingbeil P, Schnölzer M, Zöller M. CD44 variant isoforms associate with tetraspanins and EpCAM. Exp Cell Res. 2004;297:329–347.
Ladwein M, Pape UF, Schmidt DS, et al. The cell-cell adhesion molecule EpCAM interacts directly with the tight junction protein claudin-7. Exp Cell Res. 2005;309:345–357.
Hsieh HF, Yu JC, Ho LI, Chiu SC, Harn HJ. Molecular studies into the role of CD44 variants in metastasis in gastric cancer. Mol Pathol. 1999;52:25–28.
Kominsky SL, Argani P, Korz D, et al. Loss of the tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene. 2003;22:2021–2033.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No. 30670969).
Author information
Authors and Affiliations
Corresponding author
Additional information
This article was retracted as it plagiarized content from the following article: "EpCAM is overexpressed in gastric cancer and its downregulation suppresses proliferation of gastric cancer" by Du Wenqi, Wang Li , Cao Shanshan, Chen Bei, Zhang Yafei, Bai Feihu, Liu Jie, Fan Daiming published online on March 18, 2009 by Journal of Cancer Research and Clinical Oncology, DOI http://dx.doi.org/10.1007/s00432-009-0569-5.
The retraction note to this article can be found online at http://dx.doi.org/10.1007/s10620-013-2722-x.
Wenqi Du and Hongzan Ji contributed equally to this work.
About this article
Cite this article
Du, W., Ji, H., Cao, S. et al. RETRACTED ARTICLE: EpCAM: A Potential Antimetastatic Target for Gastric Cancer. Dig Dis Sci 55, 2165–2171 (2010). https://doi.org/10.1007/s10620-009-1033-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-009-1033-8