Abstract
Our current research aimed to decipher the role and underlying mechanism with regard to miR-29b-3p involving in myocardial ischemia/reperfusion (I/R) injury. In the present study, cardiomyocyte H9c2 cell was used, and hypoxia/reoxygenation (H/R) model was established to mimic the myocardial I/R injury. The expressions of miR-29b-3p and pentraxin 3 (PTX3) were quantified deploying qRT-PCR and Western blot, respectively. The levels of LDH, TNF-α, IL-1β and IL-6 were detected to evaluate cardiomyocyte apoptosis and inflammatory response. Cardiomyocyte viability and apoptosis were examined employing CCK-8 assay and flow cytometry, respectively. Verification of the targeting relationship between miR-29b-3p and PTX3 was conducted using a dual-luciferase reporter gene assay. It was found that miR-29b-3p expression in H9c2 cells was up-regulated by H/R, and a remarkable down-regulation of PTX3 expression was demonstrated. MiR-29b-3p significantly promoted of release of inflammatory cytokines of H9c2 cells, and it also constrained the proliferation and promoted the apoptosis of H9c2 cells. Additionally, PTX3 was inhibited by miR-29b-3p at both mRNA and protein levels, and it was identified as a direct target of miR-29b-3p. PTX3 overexpression could reduce the inflammatory response, increase the viability of H9c2 cells, and inhibit apoptosis. Additionally, PTX3 counteracted the function of miR-29b-3p during the injury of H9c2 cells induced by H/R. In summary, miR-29b-3p was capable of aggravating the H/R injury of H9c2 cells by repressing the expression of PTX3.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acute myocardial infarction (AMI), is one of the most life-threatening diseases (Yin et al. 2019). At present, reperfusion, the classical treatment strategy for AMI, can be clinically achieved by antiplatelet or antithrombotic therapy, bypass surgery and percutaneous coronary intervention (PCI) balloon angioplasty (Maneechote et al. 2017). Nevertheless, ischemia/reperfusion (I/R) may lead to secondary myocardial injury (Tan et al. 2018). Unfortunately, there is no effective therapeutic scheme exists in clinical practice for protecting heart I/R injury (Intachai et al. 2018).
MicroRNA (miRNA) is a category of endogenous non-coding small RNA molecules capable of regulating cell differentiation, proliferation, apoptosis and other biological processes, with a length of about 21–25 nucleotides. The occurrence and development of myocardial I/R injury is etiologically associated with aberrant expression of miRNAs (Hu et al. 2018). For example, after induction of myocardial I/R injury in rats, a significant down-regulation of miR-346 expression is observed, and miR-346 has the potential of reducing myocardial infarction area, impedes myocardial apoptosis by repressing Bax (Lv et al. 2020). MiR-29b-3p is identified as an important regulator in myocardial injury. It has been demonstrated that, miR-29b-3p, capable of targetedly down-regulating the expression of cIAP-1 in senescent cardiomyocytes, aggravates myocardial injury induced by hypoxia/reoxygenation (H/R) (Zhang et al. 2019). Nonetheless, the detailed role and mechanism of miR-29b-3p in myocardial I/R injury remain unclear.
Pentraxin 3 (PTX3), identified as a homogeneous molecule of CRP, is a multifunctional protein, regulating inflammation and extracellular matrix remodeling, and it has been reported to be associated with the pathogenesis of myocardial infarction, heart failure and cardiac arrest (Ristagno et al. 2019; Shimizu et al. 2015; Salio et al. 2008). PTX3 produced by bone marrow-derived cells is capable of protecting myocardium from I/R injury by reducing neutrophil infiltration, ROS and inflammatory cytokine production (Shimizu et al. 2015). In a mice model with myocardial I/R injury, the significant up-regulation of PTX3 expression in myocardium and peripheral blood is demonstrated, and the area of myocardial infarction was significantly expanded after PTX3 was knocked out in the mice, indicative of the crucial role of PTX3 in heart protection (Salio et al. 2008).
Herein, cardiomyocytes H9c2 cells were administrated with H/R to mimic the myocardial I/R injury (Cai and Li 2019), and then we investigated the expression change and function of miR-29b-3p/PTX3 axis in this model.
Materials and methods
Bioinformatics analysis
Gene Expression omnibus (GEO) datasets GSE74951 (Feng et al. 2017) and GSE108940 (Willis 2019) were utilized for exploring potential molecules which may participate in myocardial I/R injury. C57/BL6 mice received I/R injury with coronary artery ligation or only underwent the left thoracotomy, and plasma samples were collected and miRNA expression characteristics were detected in GSE74951. GSE108940 contains gene expression information in cardiac tissues of wild type C57BL/6 mice with or without I/R. In the present study, GEO2R analysis was performed, and differentially expressed miRNAs or genes with P < 0.05 and |log2FC|> 1.5 were screened. Animal data aforementioned were produced by past studies. The present study re-analyzed datasets which have been published. No animals were enrolled in the research.
Cell culture and transfection
The rat cardiomyocyte H9c2 cell line was purchased from the Cell Center of Chinese Academy of Sciences (Shanghai, China). The cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Thermo Fisher Scientific, Waltham, MA, USA) with supplementation of 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, New York, CA, USA), and cultivated at 37 °C with 5% CO2. In order to mimic myocardial I/R injury in vitro, H9c2 cells were treated with H/R according to previous reports (Cai and Li 2019; Liu et al. 2020). To put it succinctly, H9c2 cells were inoculated into serum-free DMEM and cultivated in an incubator containing 94% N2, 5% CO2 and 1% O2 for 6 h (hypoxic environment). Subsequently, these cells were maintained in complete medium and placed in an incubator containing 95% O2 and 5% CO2 for 12 h for reoxygenation.
The PTX3 overexpression plasmid, miR-29b-3p mimics, miR-29b-3p inhibitors and their corresponding negative controls were collectively constructed by GenePharma (Shanghai, China). They were transfected into H9c2 cells using Lipofectamine 3000 (Invitrogen, New York, CA, USA) in conformity with the supplier’s protocol.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The RNA of H9c2 cells was extracted using TRIzol reagent (Invitrogen, New York, CA, USA) (for total RNA extraction) or mirPremier® microRNA Isolation Kit (Sigma, St. Louis, MO, USA) (for miRNA extraction) according to manufacturers’ protocols. Reverse transcription was performed with First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Rockford, IL, USA). In Bio-Rad PCR system (Bio-Rad, Hercules, CA, USA), with Light Cycler Fast Start DNA MasterPlus SYBR Green I kit (Roche Diagnostics, Burgess Hill, UK), qRT-PCR was employed for quantifying the expressions of miR-29b-3p, TNF-α, IL-6, IL-1β and PTX3. U6 and β-actin were employed as the endogenous controls. The relative expression was calculated employing 2−ΔΔCT method, and the sequence of each primer was shown in Table 1.
Western blot
The cells in each group were added with RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) and incubated, and then centrifuged (4 °C, 12,000 r/min for 15 min), and the supernatant was collected. Protein quantification was conducted implementing BCA method. Followed by SDS-PAGE, the protein was transferred onto the PVDF membrane (Millipore, Bedford, MA, USA). The membranes were blocked with 5% skim milk at room temperature for 1 h, added with primary antibody (anti-PTX3 antibody, rabbit anti-human monoclonal antibody, ab125007, Abcam, Shanghai, China, diluted at 1:1000), and incubated overnight at 4 °C. After that, the membranes were rinsed with TBST, and HRP-labeled secondary antibody (goat anti-rabbit antibody IgG, ab205718, Abcam, Shanghai, China, diluted at 1:2500) was added to incubate in room temperature for 1 h. Next, ECL (Beyotime Biotechnology, Shanghai, China) was added on to the membrane, and the protein bands were developed. ImageJ software was used to analyze the gray values of the bands. β-actin was used as the endogenous control.
Cell viability assay
The transfected cells were inoculated into a 96-well plate with 1 × 103 cells/well, and incubated for ensuing 24 h. Next, 10 μL of Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan) solution was dripped to each well, and the cells were incubated at 37 °C for consecutive 1 h, and then the value of optical density (OD) at 450 nm in each well was measured on a spectrophotometer (Bio-Rad, Hercules, CA, USA).
Flow cytometry analysis
H9c2 cells were rinsed with PBS and trypsinized, and about 1 × 106 cells from each group were collected by centrifugation. 200 μL of binding buffer (Haoranbio. Co., Ltd, Shanghai, China) was used to re-suspend the cells. Next, 5 μL of propidium iodide (PI) staining solution (Yeasen Biotech Co., Ltd., Shanghai, China) and 10 μL of FITC-Annexin-V staining solution (Haoranbio. Co., Ltd, Shanghai, China) were supplemented, mixed, and incubated for 15 min at room temperature in the dark. Afterwards, 300 μL of binding buffer was supplemented, and apoptosis rate of the cells was measured with a flow cytometer (Beckman Coulter, Fullerton, CA, USA) within 1 h.
Detection of lactate dehydrogenase content
The cells of each group were centrifuged (1000g, 10 min), followed by the collection of supernatant. The lactate dehydrogenase (LDH) release of H9c2 cells was determined employing LDH detection kit (Beyotime. Co., Ltd, Shanghai, China) according to manufacturers’ protocol.
Dual-luciferase reporter assay
The binding site of miR-29b-3p on PTX3 3′ untranslated region (3′UTR) was predicted utilizing TargetScan database. The fragments were subsequently inserted into pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA) for construction of PTX3 wild type (PTX3 WT) and PTX3 mutant (PTX3 MUT) reporter plasmids. Then the reporter plasmids and miR-29b-3p mimics or mimics-NC were co-transfected into H9c2 cells, and the luciferase activity of each group was detected deploying luciferase detection kit (Invitrogen, New York, CA, USA) after 36 h.
Statistical analysis method
The data in this study were analyzed with SPSS 22.0 statistical software (SPSS Inc., Chicago, IL, USA), and measurement data were expressed by the mean ± standard deviation (x ± s). Comparison between two groups was achieved utilizing independent sample T-test, and comparison among multiple groups was performed utilizing one-way ANOVA. The difference was deemed to be statistically significant with P < 0.05.
Results
MiR-29b-3p aggravated the injury of H9c2 cells treated with H/R
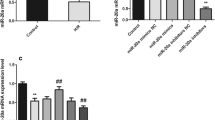
According to GEO dataset GSE74951, miR-29b-3p was noted to be significantly up-regulated in arterial blood samples of patients with cardiac I/R injury by comparison with those of healthy people (Fig. 1a, b). Therefore, we hypothesized that miR-29b-3p could probably exhibit a certain role in the process of myocardial I/R injury. Next, miR-29b-3p mimics and miR-29b-3p inhibitors were transfected into H9c2 cells for establishing cell models with miR-29b-3p overexpression and low expression, respectively (Fig. 2a). Then the mRNA expressions of inflammatory factors TNF-α, IL-6, and IL-1β in H9c2 cells were examined, and it was observed that miR-29b-3p significantly promoted inflammatory response of cardiomyocytes induced by H/R (Fig. 2b). Through CCK-8 experiment, H/R was noted to significantly suppress the viability of H9c2 cells, and the transfection of miR-29b-3p mimics enhanced the effect of H/R (Fig. 2c, d). Moreover, H9c2 cells treated with H/R released higher level of LDH (compared with the control group), and the release of LDH in H/R + miR-29b-3p mimics group was increased than that in H/R + mimics NC group (Fig. 2e). Furthermore, experimental data procured from flow cytometry analysis manifested that miR-29b-3p significantly elevated apoptosis rate of H9c2 cells (Fig. 2f). Conversely, miR-29b-3p inhibitors markedly ameliorated the inflammatory response, increased the viability, reduced the release of LDH, and inhibited the apoptosis of H9c2 cells (Fig. 2b–f). Altogether, our above experiments authenticated that miR-29b-3p aggravated the damage of H/R to H9c2 cells.
Expression of miR-29b-3p in H9c2 cells and its effect on H9c2 cells. a Expression of miR-29b-3p in normal H9c2 cells and H9c2 cells treated with H/R. b Expressions of TNF-α mRNA, IL-6 mRNA, IL-1β mRNA in H9c2 cells were detected by qRT-PCR. c, d Effect of miR-29b-3p overexpression or inhibition on the proliferation of H9c2 cells was detected employing CCK-8 assay. e LDH detection kit was employed for detecting effect of miR-29b-3p overexpression or inhibition on LDH release by H9c2 cells. f The effect of over-expression or inhibition of miR-29b-3p on the apoptosis of H9c2 cells was detected by flow cytometry analysis. *** denotes p < 0.001. In c and d, ** and *** represent p < 0.01 and p < 0.001 respectively in comparison with H/R group. In c, & represents p < 0.05 between H/R + mimics NC group and H/R + miR-29b-3p mimics group. In d, & and && represent p < 0.05 and p < 0.01 respectively by comparing H/R + inhibitors NC group and H/R + miR-29b-3p inhibitors group
Biological function of PTX3 in H9c2 cells
According to GEO dataset GSE108940, we noted that as opposed to sham operation group, the expression of PTX3, which was reported to have myocardial protective effect (Shimizu et al. 2015), was significantly augmented in the myocardial tissues of mice with myocardial I/R injury (Fig. 3a, b). Interestingly, PTX3 was predicted by TargetScan database as one of the downstream target genes of miR-29b-3p (Fig. 3c). A hypothesis that miR-29b-3p might exert its role on aggravation of injury of cardiomyocytes through PTX3 was proposed. For substantiating this hypothesis, pcDNA3.1-PTX3 and sh-PTX3 were transfected into H9c2 cells, respectively (Fig. 4a). The results of qRT-PCR indicated that, PTX3 overexpression significantly attenuated the expression of TNF-α, IL-6 and IL-1β mRNA (Fig. 4b). In CCK-8 experiment, overexpressed PTX3 was observed to significantly potentiate the viability of H9c2 cells (Fig. 4c, d). Additionally, the apoptosis rate and LDH release in pcDNA3.1-PTX3 group were significantly decreased (Fig. 4e, f). Conversely, PTX3 knockdown aggravated the injury of H9c2 cells (Fig. 4b–f). These results depicted that PTX3 was capable of ameliorating H/R induced injury of H9c2 cells.
Expression of PTX3 in GEO data set GSE108940. a Expression of miRNAs in GSE108940. MiRNAs with Log2FC < − 1.5 and p < 0.05 were marked in purple, miRNAs with Log2FC > 1.5 and p < 0.05 were marked in red and miR-29b-3p was marked in green. b Expression of miRNAs with p < 0.05 and Log2FC > 1.5 or < − 1.5 in GSE108940. c Binding sequence between PTX3 and miR-29b-3p predicted by TargetScan database. (Color figure online)
Function of PTX3 in H9c2 cells. a pcDNA3.1-PTX3 and sh-PTX3 were transfected into H/R H9c2 cells to establish a cell model of overexpression and low expression of PTX3. b qRT-PCR was employed for quantifying expression of TNF-α mRNA, IL-6 mRNA, IL-1β mRNA in H9c2 cells. c, d CCK-8 was employed for detecting effect of overexpression or low expression of PTX3 on the proliferation of H9c2 cells. e LDH detection kit was utilized for detecting the effect of overexpression or low expression of PTX3 on LDH release by H9c2 cells. f The effect of overexpression or low expression of PTX3 on apoptosis of H9c2 cells was detected by flow cytometry. *** denotes p < 0.001. In c and d, ** and *** represent p < 0.01 and p < 0.001 respectively in comparison with H/R group. In c, & and && represent p < 0.01 and p < 0.001 respectively in comparison with pcDNA3.1-PTX3 group. In d, & and && represent p < 0.01 and p < 0.001 respectively by comparing sh-NC group and sh-PTX3 group
Targeted regulation of PTX3 by miR-29b-3p
Then the targeting relationship between miR-29b-3p and PTX3 predicted by TargetScan databases (Fig. 3c) was verified by dual-luciferase reporter assay. In comparison with mimics NC group, luciferase activity of wild-type PTX3 reporter vector in miR-29b-3p mimics group was attenuated significantly, whereas no alteration was observed in the luciferase activity of mutant PTX3 reporter vector (Fig. 5a). Additionally, qRT-PCR and Western blot suggested that miR-29b-3p was capable of suppressing PTX3 expression at both mRNA and protein levels (Fig. 5b, c). These data illuminated that miR-29b-3p was capable of targetedly repressing the expression of PTX3.
MiR-29b-3p targetedly regulated PTX3. a Targeted binding relationship between miR-29b-3p and PTX3 was verified with dual-luciferase reporter gene assay. b Expression of PTX3 mRNA in miR-29b-3p overexpression and inhibition models was quantified employing qRT-PCR. c Expression of PTX3 in miR-29b-3p overexpression and inhibition models was detected by Western blot. *** denotes p < 0.001
Augmentation of PTX3 counteracted the effect of miR-29b-3p
Furthermore, miR-29b-3p mimics and pcDNA3.1-PTX3 were co-transfected into H9c2 cells for verification of functional interaction between miR-29b-3p and PTX3. Expression of miR-29b-3p and PTX3 was quantified utilizing qRT-PCR and Western blot, the results of which indicated that expression of PTX3 was significantly promoted after transfection of pcDNA3.1-PTX3, whereas overexpression of PTX3 had no significant effect on the expression of miR-29b-3p, suggesting that miR-29b-3p regulated PTX3 in a unidirectional way (Fig. 6a, b). Through qRT-PCR, CCK-8 assay, flow cytometry analysis and LDH measurements, we demonstrated that miR-29b-3p promoted the release of inflammatory factors, inhibited the viability of H9c2 cells, and aggravated the injury and apoptosis of H9c2 cells, which was reversed by the co-transfection of pcDNA3.1-PTX3 (Fig. 6c–f). These results further affirmed that miR-29b-3p was capable of aggravating the damage caused by H/R to H9c2 cells by down-regulating PTX3.
Overexpression of PTX3 abrogated the effect of miR-29b-3p on H9c2 cells. a Expression of miR-29b-3p in H/R + mimics NC, H/R + miR-29b-3p mimics and H/R + miR-29b-3p mimics + pcDNA3.1-PTX3 groups was detected by qRT-PCR. b Expression of PTX3 was detected by Western blot in H/R + mimics NC, H/R + miR-29b-3p mimics, H/R + miR-29b-3pmimics + pcDNA3.1-PTX3 groups. c Expression of TNF-α mRNA, IL-6 mRNA, IL-1β mRNA in H9c2 cells was detected by qRT-PCR. d CCK-8 was employed for detecting the impact of co-transfection of miR-29b-3p mimics and pcDNA3.1-PTX3 on inhibitory effect of miR-29b-3p on H9c2 cells proliferation. e Impact of co-transfection of miR-29b-3p mimics and pcDNA3.1-PTX3 on promoting effect of miR-29b-3p on LDH release from H9c2 cells was detected by LDH detection kit. f Impact of co-transfection of miR-29b-3p mimics and pcDNA3.1-PTX3 on promoting effect of miR-29b-3p on apoptosis of H9c2 cells was detected by flow cytometry. *, **, *** represent p < 0.05, p < 0.01 and p < 0.001 respectively. In e, ** and *** represent p < 0.01 and p < 0.001 respectively by comparing H/R + mimics NC group and H/R + miR-29b-3p mimics group. &, && represent p < 0.05 and p < 0.01 respectively by comparing H/R + miR-29b-3p mimics group and H/R + miR-29b-3p mimics + pcDNA3.1-PTX3 group
Discussion
Accumulating evidence has shown that a large number of miRNAs are aberrantly expressed in the process of myocardial I/R injury, and many of them are also crucial factors in the pathogenesis, whereas the specific role and underlying mechanism of many miRNAs in myocardial I/R injury still remain to be delineated. In recent years, mounting studies have implicated that miR-29b-3p exhibits a pivotal role in multiple cardiovascular diseases. For example, miR-29b-3p has been found to reduce the hypertrophy of H9c2 cells induced by angiotensin II (Zou et al. 2019). In cardiac fibroblasts stimulated by angiotensin II, miR-29b-3p, impeding cell proliferation and migration, possesses the potential for blocking myocardial fibrosis (Ni et al. 2019). In our current study, expression of miR-29b-3p was observed to be augmented significantly in H/R-treated H9c2 cells, and it was capable of aggravating the inflammatory responses and apoptosis of H9c2 cells, concomitantly constrained cell proliferation. Similarly, in a recent study, it has been reported that the expression of miR-29b-3p is significantly enhanced in rat heart tissue with myocardial I/R as well as in H9c2 cells treated with H/R; by repressing PI3K (p85a), miR-29b-3p participates in the apoptosis of cardiomyocytes via modulating PI3K/Akt/GSK3b pathway (Pei et al. 2020). Intriguingly, miR-29b-3p exhibits distinct effects on cardiomyocytes prior or posterior to reperfusion. The expression level of miR-29b-3p is significantly down-regulated by hypoxia treatment in human AC16 cardiomyocytes, and in this condition, miR-29b-3p significantly reduces the the apoptosis of cardiomyocytes through targeted inhibition of TRAF5 (Cai and Li 2019). Additionally, the expression of miR-29b-3p in intestinal epithelial cells of rats treated with H/R and in intestinal tissues of mice with I/R is significantly down-regulated, and by targeted inhibition of TRAF3, miR-29b-3p represses inflammatory reaction and inhibits apoptosis (Dai et al. 2019). These studies suggest that the function of miR-29b-3p has space–time speciality.
In the present study, for further dissecting the mechanism pertaining to miR-29b-3p, the binding site between miR-29b-3p and PTX3 was predicted and verified with bioinformatics analysis and dual-luciferase reporter gene assay, respectively, and it was affirmed that miR-29b-3p was capable of repressing PTX3 expression at both mRNA and protein levels. Co-transfection of pcDNA3.1-PTX3 counteracted the effects of miR-29b-3p in aggravating H/R injury and inflammatory reaction of H9c2 cells, further validating that the deleterious effect of miR-29b-3p in H/R injury was at least partially, dependent on its regulatory function on PTX3. PTX3, an octamer protein which predominantly exhibits its biological role by C-terminal, is an indispensable part of humoral immunity, which participates in recognition of microorganisms and complement cascade reaction (Bonavita et al 2015; Bottazzi et al 2016). When the body produces inflammatory factors such as TNF-α, PTX3 is rapidly synthesized and released by mononuclear phagocytes and fibroblasts, participating in the regulation of inflammatory reaction (Salio et al. 2008; Zhu et al 2014). Importantly, in human body, PTX3 is dominantly expressed in the heart; accordingly, PTX3 is associated with myocardial I/R injury, and regulates the inflammatory response (de Oliveira et al. 2019). It has been reported that in the model of I/R injury, PTX3- deficient mice suffer more severe myocardial injury, with enlargement of no-reflow area of myocardial tissue, enhanced infiltration of neutrophils and macrophages, decreased number of capillaries, and increased number of apoptotic cardiomyocytes (Salio et al. 2008). A study focusing on heart transplantation has found that PTX3 is capable of significantly inhibiting the proliferation of γδT cells and reducing the release of IL -23 and IL -17, so as to alleviate inflammatory reaction (Zhu et al. 2014). Enhanced PTX3 expression is observed in the platelets of AMI patients, inhibiting aggregation of platelet-neutrophils and further minimizing toxic effect of inflammatory reaction on the heart (de Oliveira et al. 2019). These mechanisms illustrated above are consistent with the anti-inflammatory effects of PTX3 observed in this study and the effect of PTX3 on reducing H9c2 cell injury. Additionally, PTX3 has been proven to be a valuable marker for the irreversible damage of cardiomyocytes (de Oliveira et al. 2019). Combined with our research, a conclusion could hence be drawn that regulation of miR-29b-3p/PTX3 axis might be appointed as one of effective ways to protect myocardium, whereas supplementation of animal experiments is warranted for further verification of its potential efficacy.
To recapitulate briefly, our current research verifies that miR-29b-3p aggravates the injury of cardiomyocytes and inflammatory reaction caused by H/R through targeted down-regulation of PTX3. The results provide theoretical basis for exploring diagnosis and treatment targets of myocardial I/R injury.
References
Bonavita E, Gentile S, Rubino M, Maina V, Papait R, Kunderfranco P, Greco C, Feruglio F, Molgora M, Laface I, Tartari S, Doni A, Pasqualini F, Barbati E, Basso G, Galdiero MR, Nebuloni M, Roncalli M, Colombo P, Laghi L, Lambris JD, Jaillon S, Garlanda C, Mantovani A (2015) PTX3 is an extrinsic oncosuppressor regulating complement-dependent inflammation in cancer. Cell 160:700–714. https://doi.org/10.1016/j.cell.2015.01.004
Bottazzi B, Inforzato A, Messa M, Barbagallo M, Magrini E, Garlanda C, Mantovani A (2016) The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J Hepatol 64:1416–1427. https://doi.org/10.1016/j.jhep.2016.02.029
Cai Y, Li Y (2019) Upregulation of miR-29b-3p protects cardiomyocytes from hypoxia-induced apoptosis by targeting TRAF5. Cell Mol Biol Lett 24:27. https://doi.org/10.1186/s11658-019-0151-3
Dai Y, Mao Z, Han X, Xu Y, Xu L, Yin L, Qi Y, Peng J (2019) MicroRNA-29b-3p reduces intestinal ischaemia/reperfusion injury via targeting of TNF receptor-associated factor 3. Br J Pharmacol 176:3264–3278. https://doi.org/10.1111/bph.14759
de Oliveira THC, Souza DG, Teixeira MM, Amaral FA (2019) Tissue dependent role of PTX3 during ischemia-reperfusion injury. Front Immunol 10:1461. https://doi.org/10.3389/fimmu.2019.01461
Feng Y, Zou L, Yan D, Chen H, Xu G, Jian W, Cui P, Chao W (2017) Extracellular microRNAs induce potent innate immune responses via TLR7/MyD88-dependent mechanisms. J Immunol 199:2106–2117. https://doi.org/10.4049/jimmunol.1700730
Hu S, Cao S, Tong Z, Liu J (2018) FGF21 protects myocardial ischemia-reperfusion injury through reduction of miR-145-mediated autophagy. Am J Transl Res 10:3677–3688
Intachai K, Chattipakorn SC, Chattipakorn N, Shinlapawittayatorn K (2018) Revisiting the cardioprotective effects of acetylcholine receptor activation against myocardial ischemia/reperfusion injury. Int J Mol Sci 19:2466. https://doi.org/10.3390/ijms19092466
Liu B, Wei H, Lan M, Jia N, Liu J, Zhang M (2020) MicroRNA-21 mediates the protective effects of salidroside against hypoxia/reoxygenation-induced myocardial oxidative stress and inflammatory response. Exp Ther Med 19:1655–1664. https://doi.org/10.3892/etm.2020.8421
Lv X, Lu P, Hu Y, Xu T (2020) miR-346 inhibited apoptosis against myocardial ischemia-reperfusion injury via targeting Bax in rats. Drug Des Dev Ther 14:895–905. https://doi.org/10.2147/dddt.s245193
Maneechote C, Palee S, Chattipakorn SC, Chattipakorn N (2017) Roles of mitochondrial dynamics modulators in cardiac ischaemia/reperfusion injury. J Cell Mol Med 21:2643–2653. https://doi.org/10.1111/jcmm.13330
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y, Shen G, Wang F (2019) Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol 292:188–196. https://doi.org/10.1016/j.ijcard.2019.04.006
Pei YH, Chen J, Wu X, He Y, Qin W, He SY, Chang N, Jiang H, Zhou J, Yu P, Shi HB, Chen XH (2020) LncRNA PEAMIR inhibits apoptosis and inflammatory response in PM2.5 exposure aggravated myocardial ischemia/reperfusion injury as a competing endogenous RNA of miR-29b-3p. Nanotoxicology 26:1–16. https://doi.org/10.1080/17435390.2020.1731857
Ristagno G, Fumagalli F, Bottazzi B, Mantovani A, Olivari D, Novelli D, Latini R (2019) Pentraxin 3 in cardiovascular disease. Front Immunol 10:823. https://doi.org/10.3389/fimmu.2019.00823
Salio M, Chimenti S, De Angelis N, Molla F, Maina V, Nebuloni M, Pasqualini F, Latini R, Garlanda C, Mantovani A (2008) Cardioprotective function of the long pentraxin PTX3 in acute myocardial infarction. Circulation 117:1055–1064. https://doi.org/10.1161/circulationaha.107.749234
Shimizu T, Suzuki S, Sato A, Nakamura Y, Ikeda K, Saitoh S, Misaka S, Shishido T, Kubota I, Takeishi Y (2015) Cardio-protective effects of pentraxin 3 produced from bone marrow-derived cells against ischemia/reperfusion injury. J Mol Cell Cardiol 89:306–313. https://doi.org/10.1016/j.yjmcc.2015.10.013
Tan H, Qi J, Fan BY, Zhang J, Su FF, Wang HT (2018) MicroRNA-24-3p attenuates myocardial ischemia/reperfusion injury by suppressing RIPK1 expression in mice. Cell Physiol Biochem 51:46–62. https://doi.org/10.1159/000495161
Willis MS (2019) Cardiac Muscle Ring Finger-1 (MuRF1) and transcriptional regulation of ischemia-reperfusion injury in vivo. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE108940. Public on Jan 05, 2019
Yin Y, Lv L, Wang W (2019) Expression of miRNA-214 in the sera of elderly patients with acute myocardial infarction and its effect on cardiomyocyte apoptosis. Exp Ther Med 7:4657–4662. https://doi.org/10.3892/etm.2019.7464
Zhang X, Cheng L, Xu L, Zhang Y, Yang Y, Fu Q, Mi W, Li H (2019) The lncRNA, H19 mediates the protective effect of hypoxia postconditioning against hypoxia-reoxygenation injury to senescent cardiomyocytes by targeting microRNA-29b-3p. Shock 52:249–256. https://doi.org/10.1097/shk.0000000000001213
Zhu H, Cui D, Liu K, Wang L, Huang L, Li J (2014) Long pentraxin PTX3 attenuates ischemia reperfusion injury in a cardiac transplantation model. Transpl Int 27:87–95. https://doi.org/10.1111/tri.12197
Zou X, Wang J, Tang L, Wen Q (2019) LncRNA TUG1 contributes to cardiac hypertrophy via regulating miR-29b-3p. In Vitro Cell Dev Biol Anim 55:482–490. https://doi.org/10.1007/s11626-019-00368-x
Acknowledgements
We thank Hubei Yican Health Industry Co., Ltd for its linguistic assistance during the preparation of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The procedures of animal experiments were reviewed and approved by Wuhan Asia Heart Hospital and in compliance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and Animal Welfare Act.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
He, D., Yan, L. MiR-29b-3p aggravates cardiac hypoxia/reoxygenation injury via targeting PTX3. Cytotechnology 73, 91–100 (2021). https://doi.org/10.1007/s10616-020-00446-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-020-00446-z