Abstract
Relative quantification of in vitro gene expression using real-time PCR requires stably expressed reference gene for normalisation. In this study, total RNA from MCF7, HCT116 and HepG2 cells were extracted and converted to cDNA using commercially available kit, and real-time PCR was then performed to analyse the expression levels of twelve reference genes to select the most ideal reference gene for accurate normalisation in gene expression study. geNorm and NormFinder software were used to analyse the stabilities of the reference genes, which showed a wide range of Ct values. The geNorm analysis showed the following ranking for stability of genes: UBC, YWHAZ > RPLP > TBP > ACTB > HPRT1 > PPIA > GAPDH > GUSB > B2M > TUBB > RRN18S. A similar ranking of reference genes was obtained by NormFinder, and the four most stable reference genes were identical using both approaches. UBC and YWHAZ were proposed to be the two most suitable reference genes based on the above analyses. To further assess the stabilities of the UBC and YWHAZ in a formal experiment, MCF7, HCT116 and HepG2 cell lines were subjected to treatments with 5-aza-dC and TSA. Both UBC and YWHAZ exhibited stable expression levels across control and treatment groups. Therefore, we propose that UBC and YWHAZ are the two most suitable reference genes for our gene expression studies using MCF7, HCT116 and HepG2 cell lines.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Real-time PCR has become the preferred tool for accurate quantification of gene expression to provide a better understanding of gene function in biological sciences (Bustin et al. 2005; Jian et al. 2008). This technology has many advantages over conventional transcript quantification methods such as Northern blot analysis, ribonuclease protection assays and in situ hybridisation (Vandesompele et al. 2002; Garson et al. 2005). The advantages of this technology include high sensitivity, high speed, wide dynamic quantification range and high throughput. However, several considerations such as starting target material, PCR efficiency and inter-sample variation still need to be taken into account when performing gene expression analysis using real-time PCR (Ginzinger 2002; Vandesompele et al. 2002; Bustin et al. 2005). Moreover, normalisation of target gene expression with a reference gene is absolutely necessary for eliminating sampling difference and identifying accurate gene-specific variation (Vandesompele et al. 2002).
Several genes, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB), β-2-microglobulin (B2M), cyclophilin, 18S rRNA and ubiquitin, have been used as references for gene expression studies (de Kok et al. 2005; Jian et al. 2008). These genes play crucial roles in regulating and maintaining basal cellular functions in cells and tissues (de Kok et al. 2005). They are expected to act as endogenous controls as they are consistently expressed in samples tested (Bustin 2000; Jung et al. 2007; Jian et al. 2008). However, studies have reported that the expression levels of the reference genes may vary among cells and tissues (de Kok et al. 2005). Variability may also occur when cellular signalling pathways and basic cell metabolism are disturbed (Singh and Green 1993). In addition, the expression of these reference genes may be influenced by experimental conditions (Thellin et al. 1999; Selvey et al. 2001; Tricarico et al. 2002; Arukwe 2006; Valenti et al. 2006; Tanic et al. 2007), drug and hormone treatments (Bustin 2000; Gorzelniak et al. 2001; Huggett et al. 2005). Therefore, appropriate reference genes need to be identified and validated for every experimental design. Indeed, several studies have provided evidence that the common practice of using single reference gene for normalisation has led to inaccurate or biased target gene expression profiling (Tricarico et al. 2002; Vandesompele et al. 2002). Therefore, the geometric mean of multiple reference genes should be carefully selected as the internal controls for accurate and reliable normalisation in gene expression study using real-time PCR (Vandesompele et al. 2002). The use of the geometric mean controls for possible outlying values and differences in abundance between the different genes. Therefore, the geometric mean should be used, instead of the arithmetic mean, for accurate and reliable normalisation in gene expression study.
In the present study, the expression profiles of twelve reference genes (GAPDH, TUBB, PPIA, ACTB, YWHAZ, RRN18S, B2M, UBC, TBP, RPLP, GUSB and HPRT1) in MCF7, HCT116 and HepG2 cells were determined and evaluated using real-time PCR. Thus far, the suitable reference genes for reliable normalisation using these cell lines have never been reported. MCF7, HCT116 and HepG2 cells were used in this study because they are the most widely used human cancer cell lines for in vitro gene expression study (Krishnan et al. 1994; Zajchowski et al. 2001; Fukuda et al. 2003; Gao et al. 2006; Su et al. 2008; Dong et al. 2010). The expression stability of genes and the optimal number of the reference genes required for accurate normalisation in this study were assessed using geNorm (Vandesompele et al. 2002) and NormFinder (Andersen et al. 2004). To further assess the stability of the selected reference genes in a formal experiment, MCF7, HCT116 and HepG2 cell lines were subjected to treatments with a DNA demethylating agent, 5-aza-deoxycytidine (5-aza-dC), and a histone deacetylase inhibitor, Trichostatin A (TSA). The 5-aza-dC has been widely used to treat leukaemia (Rivard et al. 1987), and TSA has been suggested as a new target for the therapy of lung cancer (Huang et al. 2003). We present a practical approach to identify multiple reference genes out of twelve candidates with the most stable expression for a reliable gene expression profiling study using MCF7, HCT116 and HepG2 cells.
Materials and methods
Culture of MCF7, HCT116 and HepG2 cells
MCF7 (ATCC no. HTB-22), HCT116 (ATCC no. CCL-247) and HepG2 (ATCC no. HB-8065) are human breast adenocarcinoma, colon carcinoma and hepatocellular carcinoma cell lines, respectively, available in our laboratory. All three cell lines were grown in Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco) containing l-glutamine, 4.5 g/L d-glucose and 110 mg/L sodium pyruvate, supplemented with 10% foetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Gibco). The cell lines were maintained at 37 °C with 95% humidity and 5% carbon dioxide. After reaching 90% confluence, the cells were detached from the culture flasks, and RNA was extracted as described below.
Extraction of total cellular RNA
Cells were detached from culture flasks using trypsin–EDTA (Gibco) and collected into a centrifuge tube. The number of cells was counted prior to centrifugation. Total RNA was extracted from 1 × 106 cells with the RNeasy total RNA kit (Qiagen), according to the manufacturer’s instruction. RNase-Free DNase I (Qiagen) was applied to eliminate any contaminating genomic DNA. The yield and purity of the total RNA was assessed by measuring the absorbance of the sample at OD260 and OD280 using the BioPhotometer Plus (Eppendorf). Additionally, the integrity and size distribution of the total RNA were verified by electrophoresis on an ethidium bromide-stained 2% agarose gel. The extracted total RNA was then used for cDNA synthesis as described below.
Synthesis of cDNA
The first strand of cDNA was synthesised from the extracted total RNA using the RevertAid™ H Minus First Strand cDNA Synthesis kit (Fermentas) and a MyCycler Thermal Cycler (Bio-Rad). First, 1 μg of total RNA was added to a primer mix consisting of 50 μM of oligo(dT)18 and 50 μM of random hexamers. The mixture was brought up to 12 μL using DEPC-treated water and was then incubated at 65 °C for 5 min. The reaction was then initiated by adding 4 μL of 5× reaction buffer, 1 μL of RiboLock™ RNase Inhibitor (20 U/μL), 2 μL of dNTP mix (10 mM) and 1 μL of RevertAid™ H Minus M-MuLV Reverse Transcriptase (200 U/μL), incubated at 25 °C for 5 min and followed by incubation at 42 °C for 1 h. The reaction was terminated by incubation at 70 °C for 5 min. The concentration of the reaction was then diluted to 25 ng/μL, and an aliquot was used for real-time PCR. The success of cDNA synthesis and genomic DNA contamination were assessed with GAPDH gene primers for conventional PCR. The cDNA was then used for subsequent relative quantification.
Design of PCR primers for reference genes
Primers for the twelve reference genes (GAPDH, TUBB, PPIA, ACTB, YWHAZ, RRN18S, B2M, UBC, TBP, RPLP, GUSB and HPRT1) used in this study were obtained from the Human Endogenous Control Gene Panel, developed by TATAA Biocenter (Table 1). According to the manufacturer, these primers were designed to span exon boundaries and were optimised to reduce the amount of primer-dimer formation during the PCR reaction.
Quantitative real-time PCR
Quantitative real-time PCR was performed using the ABI PRISM 7000 Sequence Detection System (SDS, Applied Biosystems). Each sample was run in triplicate in a 96-well reaction plate (Axygen Scientific). Five serial dilutions of each transcript were used to construct a standard curve for each reference gene to determine the amplification efficiency. For each primer pair, control containing no template and control DNA was processed in parallel to detect positive reaction and possible cross contamination, respectively. Each reaction was performed in a 25 μL volume containing 12.5 μL of Power SYBR Green I Master Mix (Applied Biosystems), 1 μL of specific primer (TATAA Biocenter) and 1 μL of the 1:2 diluted cDNA template. Cycling was performed using the default settings of the ABI PRISM 7000 SDS software 1.0 as follows: 2 min at 50 °C, 10 min at 95 °C, followed by 40 cycles of 10 s at 95 °C and 1 min at 60 °C. Melting curve analysis was carried out immediately after the amplification, ranging from 60 to 95 °C in 0.1 °C increments, to verify the PCR specificity. A control reaction without reverse transcription was always performed prior to reference gene transcriptional profiling.
Analysis of reference gene expression stability
Expression stability of the twelve reference genes was analysed using geNorm ver. 3.4 (Vandesompele et al. 2002) and further validated by NormFinder (Andersen et al. 2004) software. For the geNorm approach, the expression stability value (M) of the least to the most stable reference gene was analysed using the cycle threshold (Ct) values generated from the real-time PCR. The Ct values were converted into relative quantities before analysis was done using the geNorm software. A chart displaying the M values of the reference genes during stepwise exclusion of the analysis was then shown. The optimal number of the reference gene for normalisation was also determined using the geNorm software, where the pairwise variation (V) between 2 continuous normalisation factors containing an increasing number of reference genes was calculated. A large variation indicated that the reference gene had significant effects and should preferably be included for the calculation of a reliable normalisation factor (Vandesompele et al. 2002). A cut-off point of 0.15 was recommended by Vandesompele et al. (2002) for this analysis. The NormFinder approach was used to further evaluate the expression stability of the reference genes analysed using the geNorm approach. The NormFinder approach compared the variation of every reference gene in different groups by calculating both intra- and inter-group expression variation and then ranked the reference genes, according to the reference gene stability (Andersen et al. 2004).
Treatment of the cells with drugs
MCF7, HCT116 and HepG2 cells were separately plated at a density of 1 × 104 cells per well in a 24-well plate and allowed to recover for 24 h in DMEM supplemented with 10% FBS. For the first experiment, the cells were treated with 10 μM 5-aza-dC (Sigma) for 96 h. After that, cultures treated with 5-aza-dC were either co-treated with dimethylsulfoxide [DMSO (Sigma), as control] or 100 ng/mL TSA (Sigma) for another 24 h for the second experiment. On the fifth day, all cells either treated with 5-aza-dC alone or with 5-aza-dC and TSA were harvested. Both 5-aza-dC and TSA were prepared by dissolving the drugs in DMSO and diluting with fresh DMEM supplemented with 10% FBS. Controls for each cell line were seeded at the same cell density and similarly treated with only DMSO. After the treatments, total RNA was extracted, converted into cDNA and used for the quantitative real-time experiment as described above.
Statistical analysis of data
The Mann–Whitney U Test was used to analyse the significance of differences between the control and drug-treated groups using SPSS 18.0 software. A p value of <0.05 was considered to be statistically significant. The geometric mean was calculated by transforming the Ct value into quantity using the GenEx Light software (TATAA Biocenter), where the highest relative quantity for the reference genes was set as 1.00.
Results
The quality and purity of extracted total RNA
The ratio of OD260/280 for all RNA samples was between 1.8 and 2.0, which indicated that the quality and purity of the extracted total RNA was adequate for subsequent cDNA synthesis. Moreover, the ethidium bromide-stained gels showed clear 28S and 18S ribosomal RNA bands, indicating that the intact RNA was of good quality (Fig. 1). Additionally, when PCR without reverse transcriptase was performed, no amplification was observed. This phenomenon indicated that no genomic DNA contamination existed in the RNA samples (data not shown).
High quality and pure total RNA are crucial for subsequent real-time PCR experiment, as it directly determines the reproducibility and biological relevance of the outcome of the experiment. Moreover, it is essential to ensure that the extracted total RNA is free of genomic contamination. The presence of genomic DNA in a quantitative real-time PCR assay would lead to incorrect quantification of mRNA expression and therefore to erroneous results.
The expression levels of the reference genes
For gene expression studies, two quantification strategies can be applied: absolute or relative. Absolute and relative quantifications produce similar analysis outcomes, although several studies have reported that relative quantification is more accurate than measuring the absolute level of a gene expression (Livak and Schmittgen 2001; Eleaume and Jabbouri 2004). In our effort to select suitable reference genes for relative quantification study, the transcriptional profiling of the reference genes in MCF7, HCT116 and HepG2 cells showed that the reference genes had overall mean Ct values ranging from 14 to 24, except for RRN18S, which was the most abundant and had a mean Ct value of less than 10 (6.52 ± 0.72; Fig. 2a). On the contrary, TUBB was the least abundant, with a mean Ct value above 20 (23.02 ± 1.72). Other Ct values of the reference genes obtained were very similar across the cell lines, indicating low transcriptional variability of these reference genes in the cell lines (Fig. 2b). Visualisation of single peak in the melt curve analysis showed the specificity of each reference gene amplification (data not shown). Standard curves generated from 10-fold serial dilutions for each reference gene in MCF7, HCT116 and HepG2 cells gave correlation coefficients greater than 0.98 and efficiencies greater than 90%. These reaction efficiencies were then used to transform the Ct values into raw data for subsequent expression stability analyses using the geNorm and NormFinder software.
The expression stability and optimal number of reference gene
geNorm analysis was then performed, according to the protocol described by Vandesompele et al. (2002) to determine the expression stability value (M) of the twelve reference genes in the MCF7, HCT116 and HepG2 cells. The geNorm analysis relies on the principle that the expression ratio of two ideal reference genes is constant in all samples regardless of the experimental condition and cell type (Vandesompele et al. 2002). In general, the M values of the twelve reference genes were ranked as follows: UBC, YWHAZ < RPLP < TBP < ACTB < HPRT1 < PPIA < GAPDH < GUSB < B2M < TUBB < RRN18S. A reference gene with a lower M value is more stable in expression, whereas a higher M value indicates less stable expression of the reference gene (Vandesompele et al. 2002). The M value showed non-significant differences from the most stable to the least stable of the reference genes in this analysis. As shown in Fig. 3a, when the M values for all reference genes in MCF7, HCT116 and HepG2 cells were pooled together, the M values of UBC and YWHAZ were the lowest, followed by RPLP and TBP, while the M value of RRN18S was the highest, demonstrating that UBC, YWHAZ, RPLP and TBP had the most stable expression and that RRN18S was the most variable.
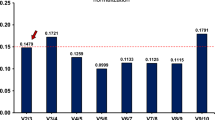
a Average expression stability (M value) of the reference genes analysed using geNorm software. The M value during stepwise exclusion analysis excluded the least stable reference gene (left) to the most stable reference gene (right). b Pairwise variation (V) analysis between two sequential normalisation factors with stepwise exclusion of the most stable reference gene determined the number of reference genes to use for normalisation. V = 0.15 was used as a cut-off threshold value. Based on the pairwise variation, V2/3 = 0.011 indicated that two most stable reference genes were sufficient for accurate normalisation in this study
The optimal number of reference genes that should be used for a reliable normalisation of gene expression was then determined using geNorm software. In this case, the normalisation factor (NFn) was calculated based on the geometric mean of the expression level of the best reference gene, as suggested by Vandesompele et al. (2002). The pairwise variations Vn/Vn+1 between the two sequential normalisation factors (NFn and NFn+1) was used to determine the necessity of adding the next reference gene for a reliable normalisation. A large variation indicates that the added gene has a significant effect and should probably be included for calculation with a reliable normalisation factor (Vandesompele et al. 2002). As shown in Fig. 3b, the two most stable reference genes, UBC and YWHAZ, were found to be sufficient for accurate normalisation. The inclusion of an additional reference gene for the normalisation would not have any significant effect because V3/4 showed the same value as V2/3, and the value was much lower than the cut-off value of 0.15 as suggested by Vandesompele et al. (2002).
NormFinder analysis was then performed to validate the outcomes of the geNorm analyses. The NormFinder approach calculates the M value based on simultaneous estimation of the overall expression variation and variation between subgroups, including intra- and inter-group variations of the reference gene (Andersen et al. 2004). As described above, the M value indicated the expression stability, and generally, a gene with a lower M value had higher expression stability. Table 2 shows the ordered arrangement of the M values for the twelve reference genes in MCF7, HCT116 and HepG2 cells as calculated using NormFinder. NormFinder analysis listed UBC and YWHAZ among the top four most stably expressed reference genes. This confirmed the results of the geNorm analyses, which suggested that UBC and YWHAZ were the two most suitable reference genes for studies using the above mentioned cell lines.
The stability of the selected reference genes post drug treatments
The stability of UBC and YWHAZ in MCF7, HCT116 and HepG2 cells was further validated by subjecting the cell lines to treatments with a DNA demethylating agent, 5-aza-dC, and a histone deacetylase inhibitor, TSA. These two drugs play important roles in the study of epigenetic aberrations, and their contribution to malignant transformation and progression of cancer are well demonstrated (Dobosy et al. 2007). An ideal reference gene, according to the previous study, should have minimal variation in gene expression across different experimental conditions or treatments (Bustin 2000). Indeed, our results showed that the geometric means of the mRNA levels of UBC and YWHAZ in MCF7, HCT116 and HepG2 cells were not significantly different in the 5-aza-dC-treated group compared to the control group (Fig. 4a). Similarly, treatment of MCF7, HCT116 and HepG2 cells with 5-aza-dC + TSA did not significantly alter the geometric means of the mRNA levels of UBC and YWHAZ when compared to the control group (Fig. 4b). This result indicated that UBC and YWHAZ were the stable reference genes in MCF7, HCT116 and HepG2 cells and were not influenced by the drug treatments to be used in our future studies.
Relative quantity or geometric mean of mRNA levels in drug-treated and control cells. a Geometric mean of UBC and YWHAZ mRNA levels in the 5-aza-dC-treated and control groups of MCF7, HCT116 and HepG2 cells. b Geometric mean of UBC and YWHAZ mRNA levels in the 5-aza-dC + TSA-treated and control groups of MCF7, HCT116 and HepG2 cells. The values were expressed as the mean ± SD. No significant difference in the geometric mean was found between the drug-treated and control groups
Discussion
Our study shows the following: (1) the quality and purity of total RNA extracted from MCF7, HCT116 and HepG2 cells using the commercially available kit; (2) the expression levels of twelve reference genes determined in the above cell lines using real-time PCR; (3) the expression stability of the reference genes in the above cell lines analysed using geNorm and NormFinder; (4) the optimal number of reference genes required and the most suitable reference genes for reliable normalisation of gene expression using the above cell lines; and (5) the stability of the selected reference genes in a formal drug treatment experimental design.
Overall, our study demonstrates that UBC, YWHAZ, RPLP and TBP, were the most suitable reference genes to be used for gene expression studies using MCF7, HCT116 and HepG2 cells. Although GAPDH and ACTB are used as reference genes in most in vitro studies using these cell lines (Bracke et al. 1993; Tanic et al. 2007; Cicinnati et al. 2008; Mori et al. 2008), these two reference genes did not exhibit stable expression levels across MCF7, HCT116 and HepG2 cells, as demonstrated by our current results as well as previous studies (Schmittgen and Zakrajsek 2000; Gorzelniak et al. 2001). The GAPDH and ACTB genes are variably expressed when subjected to serum-stimulation (Schmittgen and Zakrajsek 2000) and hormone treatments (Gorzelniak et al. 2001). The expression of GAPDH is also regulated differently in various human cancer tissues such as breast cancer, prostate cancer, pancreatic adenocarcinoma and bladder cancer (Rondinelli et al. 1997; Schek et al. 1988; Revillion et al. 2000; Ohl et al. 2006). Variable expression of GAPDH in different experimental conditions may be due to its multiple participation in cellular processes, including as a component of glycolytic pathway (Jian et al. 2008). Additionally, the ACTB mRNA is abundantly expressed but less stable in most cells types (Bustin 2000). This gene encodes for a cytoskeleton structural protein, and it has also been shown to exhibit variations in transcriptional levels in response to different experimental conditions in certain tissues (Carlyle et al. 1996; Foss et al. 1998; Krussel et al. 1998).
Taken together, these data suggest that the use of a single reference gene for normalisation could lead to relatively large errors (Tricarico et al. 2002; Vandesompele et al. 2002). Hence, this study was designed to identify two or more reference genes for more accurate normalisation. Our results showed that two reference genes, UBC and YWHAZ, were sufficient for reliable normalisation when using these cell lines. The inclusion of a third reference gene (RPLP) for the normalisation would have no significant effect as indicated by the V2/3 value of 0.011, which was negligible compared to the cut-off point of 0.15, as recommended by Vandesompele et al. (2002). The high stability of UBC and YWHAZ and the insignificant change in the normalisation factor even if a third reference gene was included are evidence that these two reference genes only were required for accurate gene expression normalisation in the cell lines tested.
Moreover, additional data from drug treatments also demonstrated that the expression levels of UBC and YWHAZ were relatively stable under the treatment conditions, suggesting that UBC and YWHAZ are suitable as reference genes in a wider range of experiments to study gene expression in MCF7, HCT116 and HepG2 cell lines. However, it must be noted that the stability of the two reference genes proposed in this study should be properly validated in other experimental designs when treatment agents besides 5-aza-dC and TSA are used.
Conclusion
Our study identified UBC and YWHAZ as the two most suitable reference genes among the twelve investigated once in this study using geNorm software. These two reference genes are sufficient for reliable normalisation of gene expression using the MCF7, HCT116 and HepG2 cell lines. Additionally, they were among the top-ranked reference genes analysed by NormFinder software and exhibited relatively stable expression even after 5-aza-dC and TSA treatments. Thus, UBC and YWHAZ are suggested for use as reference genes in gene expression studies using MCF7, HCT116 and HepG2 cell lines. However, proper validation of suitable reference genes for other experimental conditions remains necessary.
References
Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250
Arukwe A (2006) Toxicological housekeeping genes: do they really keep the house? Environ Sci Technol 40:7944–7949
Bracke ME, Vyncke BM, Bruynee EA, Vermeulen SJ, De Bruyne GK, Van Larebeke NA, Vleminckx K, Van Roy FM, Mareel MM (1993) Insulin-like growth factor I activates the invasion suppressor function of E-cadherin in MCF-7 human mammary carcinoma cells in vitro. Br J Cancer 68:282–289
Bustin SA (2000) Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol 25:169–193
Bustin SA, Benes V, Nolan T, Pfaffl MW (2005) Quantitative real-time RT-PCR—a perspective. J Mol Endocrinol 34:597–601
Carlyle WC, Toher CA, Vandervelde JR, McDonald KM, Homans DC, Cohn JN (1996) Changes in beta-actin mRNA expression in remodeling canine myocardium. J Mol Cell Cardiol 28:53–63
Cicinnati VR, Shen Q, Sotiropoulos GC, Radtke A, Gerken G, Beckebaum S (2008) Validation of putative reference genes for gene expression studies in human hepatocellular carcinoma using real-time quantitative RT-PCR. BMC Cancer 8:350
de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN (2005) Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest 85:154–159
Dobosy J, Roberts J, Fu V, Jarrard D (2007) The expanding role of epigenetics in the development, diagnosis and treatment of prostate cancer and benign prostatic hyperplasia. J Urol 177:822–831
Dong H, Lin W, Wu J, Chen T (2010) Flavonoids activate pregnane X receptor mediated CYP3A4 gene expression by inhibiting cyclin-dependent kinases in HepG2 liver carcinoma cells. BMC Biochem 11:23
Eleaume H, Jabbouri S (2004) Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J Microbiol Meth 59:363–370
Foss DL, Baarsch MJ, Murtaugh MP (1998) Regulation of hypoxanthine phosphoribosyltransferase, glyceraldehyde-3-phosphate dehydrogenase and beta-actin mRNA expression in porcine immune cells and tissues. Anim Biotechnol 9:67–78
Fukuda R, Kelly B, Semenza GL (2003) Vascular endothelial growth factor gene expression in colon cancer cells exposed to prostaglandin E2 is mediated by hypoxia-inducible factor 1. Cancer Res 63:2330–2334
Gao Y, Hannan NR, Wanyonyi S, Konstantopolous N, Pagnon J, Feng HC, Jowett JB, Kim KH, Walder K, Collier GR (2006) Activation of the selenoprotein SEPS1 gene expression by pro-inflammatory cytokines in HepG2 cells. Cytokine 33:246–251
Garson JA, Grant PR, Ayliffe U, Ferns RB, Tedder RS (2005) Real-time PCR quantitation of hepatitis B virus DNA using automated sample preparation and murine cytomegalovirus internal control. J Virol Methods 126:207–213
Ginzinger DG (2002) Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol 30:503–512
Goossens K, Van Poucke M, Van Soom A, Vandesompele J, Van Zeveren A, Peelman LJ (2005) Selection of reference genes for quantitative real-time PCR in bovine preimplantation embryos. BMC Dev Biol 5:27
Gorzelniak K, Janke J, Engeli S, Sharma AM (2001) Validation of endogenous controls for gene expression studies in human adipocytes and preadipocytes. Horm Metab Res 33:625–627
Huang H, Zhang ZX, Xu YJ, Shao JF (2003) HDAC1 expression and effect of TSA on proliferation and apoptosis of A549 cells. Ai Zheng 22:922–926
Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6:279–284
Jian B, Liu B, Bi Y, Hou W, Wu C, Han T (2008) Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol Biol 9:59
Jung M, Ramankulov A, Roigas J, Johanssen M, Ringsdorf M, Kristiansen G, Jung K (2007) In search of suitable reference genes for gene expression studies of human renal cell carcinoma by real-time PCR. BMC Mol Biol 8:47
Krishnan V, Wang X, Safe S (1994) Estrogen receptor-Spl complexes mediate estrogen-induced cathepsin D gene expression in MCF-7 human breast cancer cells. J Biol Chem 269:15912–15917
Krussel JS, Huang HY, Simon C, Behr B, Pape AR, Wen Y, Bielfeld P, Polan ML (1998) Single blastomeres within human preimplantation embryos express different amounts of messenger ribonucleic acid for beta-actin and interleukin-1 receptor type I. J Clin Endocrinol Metab 83:953–959
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔ C(T)) method. Methods 25:402–408
Lyng MB, Laenkholm AV, Pallisgaard N, Ditzel HJ (2008) Identification of genes for normalization of real-time RT-PCR data in breast carcinomas. BMC Cancer 8:20
Mori R, Wang Q, Danenberg KD, Pinski JK, Danenberg PV (2008) Both actin and GAPDH are useful reference genes for normalization of quantitative RT-PCR in human FFPE tissue samples of prostate cancer. Prostate 68:1555–1560
Ohl F, Jung M, Radonic A, Sachs M, Loening SA, Jung K (2006) Identification and validation of suitable endogenous reference genes for gene expression studies of human bladder cancer. J Urol 175:1915–1920
Perez S, Royo LJ, Astudillo A, Escudero D, Alvarez F, Rodriguez A, Gomez E, Otero J (2007) Identifying the most suitable endogenous control for determining gene expression in hearts from organ donors. BMC Mol Biol 8:114
Revillion F, Pawlowski V, Hornez L, Peyrat JP (2000) Glyceraldehyde-3-phosphate dehydrogenase gene expression in human breast cancer. Eur J Cancer 36:1038–1042
Rivard GE, Momparler RL, Demers J, Benoit P, Raymond R, Lin K-T, Momparler LF (1987) Phase I study on 5-aza-2′-deoxycytidine in children with acute leukemia. Leuk Res 11:319–325
Rondinelli RH, Epner DE, Tricoli JV (1997) Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in late pathological stage human prostate cancer. Prostate Cancer Prostatic Dis 1:66–72
Schek N, Hall BL, Finn OJ (1988) Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human pancreatic adenocarcinoma. Cancer Res 48:6354–6359
Schmittgen TD, Zakrajsek BA (2000) Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J Biochem Biophys Methods 46:69–81
Selvey S, Thompson EW, Matthaei K, Lea RA, Irving MG, Griffiths LR (2001) Beta-actin—an unsuitable internal control for RT-PCR. Mol Cell Probes 15:307–311
Singh R, Green MR (1993) Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science 259:365–368
Su RY, Chi KH, Huang DY, Tai MH, Lin WW (2008) 15-deoxy-Delta12, 14-prostaglandin J2 up-regulates death receptor 5 gene expression in HCT116 cells: involvement of reactive oxygen species and C/EBP homologous transcription factor gene transcription. Mol Cancer Ther 7:3429–3440
Tanic N, Perovic M, Mladenovic A, Ruzdijic S, Kanazir S (2007) Effects of aging, dietary restriction and glucocorticoid treatment on housekeeping gene expression in rat cortex and hippocampus-evaluation by real time RT-PCR. J Mol Neurosci 32:38–46
Thellin O, Zorzi W, Lakaye B, De Borman B, Coumans B, Hennen G, Grisar T, Igout A, Heinen E (1999) Housekeeping genes as internal standards: use and limits. J Biotechnol 75:291–295
Tricarico C, Pinzani P, Bianchi S, Paglierani M, Distante V, Pazzagli M, Bustin SA, Orlando C (2002) Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem 309:293–300
Valenti MT, Bertoldo F, Dalle Carbonare L, Azzarello G, Zenari S, Zanatta M, Balducci E, Vinante O, Lo Cascio V (2006) The effect of bisphosphonates on gene expression: GAPDH as a housekeeping or a new target gene? BMC Cancer 6:49
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034
Zajchowski DA, Bartholdi MF, Gong Y, Webster L, Liu HL, Munishkin A, Beauheim C, Harvey S, Ethier SP, Johnson PH (2001) Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res 61:5168–5178
Zhang X, Ding L, Sandford AJ (2005) Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol Biol 6:4
Acknowledgments
This work was funded by a Fundamental Research Grant (203/PPSK/6171103) from the Ministry of Higher Education, and Chua Siang Ling was supported by a fellowship from the Institute of Postgraduate Studies, Universiti Sains Malaysia. We also thank the technical support from UPMS, Culture and Biomedicine Laboratories, School of Health Sciences, Universiti Sains Malaysia.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Chua, S.L., See Too, W.C., Khoo, B.Y. et al. UBC and YWHAZ as suitable reference genes for accurate normalisation of gene expression using MCF7, HCT116 and HepG2 cell lines. Cytotechnology 63, 645–654 (2011). https://doi.org/10.1007/s10616-011-9383-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-011-9383-4