Abstract
Studies of small-scale spatial genetic structure are crucial to the implementation of successful conservation and management plans as they enhance our capacity to predict a population’s ability to respond to changing environments. This is especially true in the tropics, where high rates of fragmentation and land-use change create potential barriers to genetic exchanges. We examined the genetic diversity of wild populations of the economically important palm, Mauritia flexuosa (Arecaceae) in and around the Aripo Savannas Scientific Reserve on the island of Trinidad. In a small collection area of roughly 10 km2 we found both spatial and inter-generational, or temporal, genetic differentiation and high levels of nearest-neighbor mating. We suggest that these spatial and temporal genetic patterns reflect recent and drastic anthropogenic changes to the landscape of the study sites, which we hypothesize have created barriers to gene flow through animal-mediated seed dispersal and pollination.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
For Angiosperms, both pollen and seed dispersal contribute to gene flow. Gene flow is integral to the perpetuation of resilient natural communities because it prevents genetic structuring and loss of diversity that could form as a result of inbreeding, which in turn may lead to local extinction (Young et al. 1996). Understanding mechanisms for the maintenance of gene flow in tropical floras is especially important because of their natural tendency towards inbreeding, as demonstrated by Hamrick et al. (1993). Today, with ever increasing rates of tropical deforestation and urbanization, it is essential that wild populations maintain sufficient genetic diversity to be able to respond to rapid landscape changes, thereby reducing the likelihood of local extinctions. Seed dispersal in particular is an effective strategy of ensuring that offspring can take advantage of growing sites removed from their parents, reducing competition among related individuals and decreasing the likelihood of inbreeding (Janzen 1970; Connell 1971).
For plants relying on animal-mediated seed dispersal, anthropogenic habitat fragmentation could influence gene flow by imposing barriers to dispersal such as the construction of roads and an increase in hunting. With reduced dispersal, greater numbers of seeds should germinate near their parent trees, eventually resulting in high rates of inbreeding among geographically proximate individuals (Janzen 1970; Connell 1971; Pacheco and Simonetti 2000). These patterns were observed in the tree Inga ingoides, when its mammalian disperser was heavily hunted (Pacheco and Simonetti 2000). Here we explore the impacts of hunting and habitat fragmentation on gene flow in the Amazonian palm Mauritia flexuosa (Arecaceae) in Trinidad’s Aripo Savanna Scientific Reserve (ASSR).
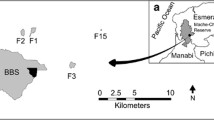
The seasonally flooded flora of the ASSR savannas is dominated by a border between 3–20 m wide of the long-lived diploid, dioecious palm M. flexuosa. M. flexuosa in turn serves as an important source of food and shelter to its dispersers, the red-bellied macaw (Orthopsittaca manilata) and red brocket deer (Mazama americana) (Zona and Henderson 1989). M. flexuosa pollination appears to be primarily wind-mediated (Rosa and Koptur 2013). Originally, these savannas were surrounded by marsh forest, making them analogous to small M. flexuosa dominated islands (Fig. 1) (EMA 2007). While some marsh forest remains, it is patchy in its distribution due to significant anthropogenic disturbances beginning in the 1930s until the ASSR was declared a prohibited area in 1987 and later, in 2007, an Ecologically Sensitive area (Fig. 1) (EMA 2007).
Map indicating land-use and M. flexuosa collection sites in and around the Aripo Savannas Scientific Reserve (ASSR). The ASSR’s placement on Trinidad is indicated by the black box in the grey silhouette of the island in the bottom right-hand corner. In this roughly 10 km2 collection site, there have been high levels and many forms of disturbance including clear cuts, burning, and quarrying, as indicated on the map. Hunting, another form of disturbance not indicated on the map has occurred throughout the reserve area
Historically, the savannas were used as a US military base during WWI, during which time roads were constructed that bisected the savannas. After WWI, the roads remained and parts of the savannas were used as quarries (EMA 2007). Additionally, settlements arose around the perimeter of the savannas, and hunting of the large animals was a common practice (EMA 2007). Today, some of the villages remain and busy paved roads surround reserve. Furthermore, much of the protected area is still subject to illegal hunting of large animals, including M. flexuosa’s dispersers the red-bellied macaws and red brocket deer. Hunting continues to such an extent that some of the savannas were unsafe to collect in during the course of this study. This historic fragmentation combined with continued illegal hunting could pose significant barriers to seed dispersal, even on the relatively small spatial scale of the ASSR and nearby forests (Fig. 1).
Limited seed dispersal is likely to lead to genetic structuring on small spatial scales. Dick et al. (2008) review instances where large-scale spatial genetic structure is due to pollinators, while seed dispersal shapes small-scale genetic structuring. Choo et al. (2012) examined small-scale spatial genetic structuring in Attalea phalerata palms, which are also dispersed by large mammals. Because pollination occurred on a larger scale than A. phalerata’s dispersal, Choo et al. (2012) attributed the observed spatial genetic structure to seed dispersal. Being primarily wind pollinated, M. flexuosa has a broader pollination syndrome than A. phalerata (Rosa and Koptur 2013). Additionally, its seed-dispersers act on scales analogous to those examined in Choo et al. (2012), relatively short distances well within the range of the study area (Bodmer 1991). Thus, while pollination plays undoubtedly plays an important role in the genetic structure of M. flexuosa on a large spatial scale, previous work indicates that on the small spatial scale of the Aripo Savannas seed dispersal should be the primary determinant of M. flexuosa’s genetic structure (Dick et al. 2008; Choo et al. 2012).
To investigate the impact of sustained habitat fragmentation and the hunting of dispersal vectors on M. flexuosa communities, we examined gene flow and dispersal in four M. flexuosa stands in the Aripo Savannas area using 13 microsatellite loci specific to M. flexuosa (Federman et al. 2012). In areas with a history of habitat fragmentation and hunting we expected to see high levels of nearest-neighbor mating due to barriers to gene flow (Pacheco and Simonetti 2000; Born et al. 2008). We also expected to observe temporal differences in genetic structuring, because, following the declaration of ASSR as a prohibited in area in 1987, park ranger protection could have fostered higher levels of gene flow, thus reducing genetic structuring among juveniles as compared to adults. This investigation is relevant not just to the Aripo Savannas area, but for understanding the impacts of fragmentation on communities of M. flexuosa, which is one of the most widely used palms in Amazonia (Padoch 1988; Peters et al. 1989). This is the first study to examine the population genetic structure of M. flexuosa.
Methods
Sampling and data collection
Our sampling area consisted of four localities: three seasonally flooded savannas in the ASSR (Savanna 1, 2 and 3) and 1 year-round flooded swamp ~1 km outside of the reserve (Fig. 1). The four sites comprised a collection area of ~10 km2, with the greatest distance between sites being ~6.5 km (Fig. 1). Our collection permit was obtained through the University of the West Indies and the Department of Natural Resources and the Environment, while our import permit was provided by the USDA and the New York Botanical Garden. All necessary permits were obtained for the described study, which complied with all relevant regulations.
At each site, we collected leaf samples of roughly 25 adult and juvenile individuals respectively (see Table 1 for exact collection numbers at each site), and geo-referenced collections using a Garmin 60csx GPS. For each individual sampled, 10–15 g of leaf tissue were dried in silica in the field and stored at −80 °C upon returning from the field. For each adult collection, we calculated approximate age by dividing the number of leaves and leaf scars along the stem by 2.5, following Sampaio et al. (2008). M. flexuosa has a generation time of roughly 30 years, so we defined juveniles as individuals ~28 years of age and below (Holm et al. 2008).
In seasonally flooded savannas, M. flexuosa borders the savanna in stands between 3–20 m wide. Therefore, to obtain a representative sampling of M. flexuosa in savannas, we circled each savanna and sampled M. flexuosa individuals within the border at 20–30 m intervals (Table 1). In the swamp, as M. flexuosa blanketed the area and we could therefore not employ the same sampling methodology that was used in the savannas, so sampling was performed as a random point process (Table 1).
For DNA extractions, 15–20 mg of dried leaf tissue was homogenized using a FastPrep®-24 instrument (MP Biomedicals), and DNA was extracted, following the manufacturer’s protocol, using the Qiagen DNeasy Plant Kit (Qiagen Inc.). The 13 microsatellite loci used in this study are described in Federman et al. (2012). For each M. flexuosa individual, microsatellites were amplified and scored as described in Federman et al. (2012); all fragment analysis was performed at the Yale DNA Analysis Facility, and genotypes were assigned using GeneMarker v1.91 (SoftGenetics, State College, PA, USA).
Genetic analyses
All 13 microsatellite loci were tested for within-collection-site deviation from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) using the online version of GENEPOP (Raymond and Rousset 1995). Markov chain parameters were set to 10,000 dememorizations, 1000 batches, and 10,000 iterations per batch for both HWE and LD. All HWE and LD results were assessed using sequential Bonferroni corrections. Using GENEPOP we assessed overall genetic structure by calculating F ST for all collection site pairs. We used the same program to calculate overall levels of inbreeding with F IS . Observed and expected heterozygosities were calculated for each collection site using ARLEQUIN 3.5 (Excoffier and Lischer 2010).
We assessed patterns of population structure and assigned individuals to inferred genetic clusters using the Bayesian clustering method in the program STRUCTURE v. 2.3 (Pritchard et al. 2000). STRUCTURE identifies genetic clusters and assigns individuals to those clusters without a priori information regarding their geographic origin. To determine the most likely number of clusters (K), we followed Pritchard et al. (2000) and conducted ten independent runs for each K = 1–5. For all runs, we incorporated admixture and correlated allele frequencies in the model. We used a burn-in of 100,000 iterations followed by 500,000 Markov chain Monte Carlo replications. To determine the number of clusters we also calculated ∆K, the second-order rate of change of the log-likelihood between values of K (Evanno et al. 2005). We complemented the Bayesian analysis of population structure with a multivariate statistical approach using K-means clustering followed by discriminant analysis of principal components (DAPC) in R (R development Core Team 2012) with the package adegenet (Jombart 2008). We determined optimal cluster numbers using the Bayesian information criterion (BIC). In contrast to the Bayesian method, DAPC does not assume Hardy–Weinberg or linkage equilibrium (Jombart et al. 2010).
We assessed temporal stability across generations of the DAPC-defined genetic clusters by carrying out tests of genic (allelic) and genotypic differentiation between adults and juveniles using GENEPOP. These analyses assess differences in allele and genotype frequencies between subsets of populations, here adults and juveniles. To assess relatedness among individuals, as a proxy for levels of nearest-neighbor mating, we used the program ML-Relate to calculate the log-likelihood of relatedness for all collected individuals and within the DAPC-defined genetic clusters (Kalinowski et al. 2006). We examined the correlation between relatedness and distance, with kinship-distance curves and a concurrent Monte Carlo simulation to develop confidence intervals in R.
In order to investigate the impact of disturbance on genetic clusters, we extracted satellite forest cover data for collection sites as a proxy for disturbance from the global land cover facility (DeFries et al. 2000), using the R package raster (Hijmans and van Etten 2012). Imagery was processed at a 1 km scale from 1992 to 1993, the rough time of germination for the majority of collected juveniles. To see if disturbance correlated with any patterns in our genetic analyses, we created a matrix containing all of the collected individuals, collection sites, DAPC-defined genetic clusters, and forest cover data to perform a multiple correspondence analysis (MCA), using the R package ca (Nenadic and Greenacre 2007). Using the same matrix, we performed a logistic regression in R of disturbance and genetic clusters to determine if the genetic clusters were significantly different in terms of forest cover.
Results
Of 52 collection site-by-locus specific F IS values, only 9.6 % deviated significantly from Hardy–Weinberg expectations after sequential Bonferroni correction (Online Resource T1). No locus-by-locus test for LD was significant. Pairwise F ST values were consistent with the DAPC and Bayesian analyses (Online Resource T2). There were no significant differences between observed and expected heterozygosities in any collection site, and thus there were no significant F IS values for any collection site (Table 1).
The Bayesian analysis of population structure clearly indicated the most likely number of clusters was three, when evaluated using ∆K (Fig. 2a). DAPC concurred with the Bayesian analysis (Fig. 2b). Cluster 1 comprised individuals collected at Savanna 2, Cluster 2 included primarily collections from Savanna 1 and some individuals from Savanna 3, and Cluster 3 included individuals from the swamp, the remaining collections from Savanna 3 and a small number of individuals from Savanna 1 (Fig. 2).
Discriminant analysis of principle components (DAPC) with 3 K-means clusters. The 206 individuals collected from four sites clearly describe three distinct genetic groups with the majority falling within the 95 % confidence interval of assignment represented by the elipses. Each ine represents an individual and the shades of grey and shapes represents its assigned genetic cluster, where the grey triangles are Cluster 1, the black squares are Cluster 2, and the grey circles represent Cluster 3 (a). Results of the Bayesian analysis of group membership for all individuals. Each line represents an individual and the colors represent genetic clusters, such that the proportion of color for each individual indicates its probability of assignment to that genetic cluster (b)
In order to make inferences on temporal differences in genetic structure, we compared genic and genotypic differentiation between adults and juveniles at each site (Table 1). Given that generation times are ~30 years, collections were separated by up to two generations. While there was no genic or genotypic differentiation between adults and juveniles in Clusters 1 and 2, we found a significant difference between the two age classes in Cluster 3, indicating inter-generational differences in genetic structuring (Table 1). Assessments of the log-likelihood of relatedness within each genetic cluster indicated that nearly every individual in a site was related as either parent-offspring or sibling. The kinship distance curve for all individuals showed a strong correlation of kinship and distance up to ~3 km (Online Resource 1). Clusters 1 and 2 displayed a similar pattern, while Cluster 3 samples showed a strong kinship-distance correlation across its entire area of ~3 km2 (Online Resource 1).
The MCA analysis shows that genetic Clusters 3 and 2 are defined by the same axis of inertia, meaning that they are best explained by similar environmental variables. However, Cluster 3 corresponds to higher levels of forest cover, or lower levels of disturbance, than Cluster 2 (Fig. 3). Genetic Cluster 1 is in its own quadrant with Savanna 2, with mid-range levels of forest cover. Complementing the MCA results, the logistic regression indicates that genetic clusters 2–1 and 3–1 are significantly different from one another, while clusters 3–2 are similar (Fig. 3).
The multiple correspondence analysis of collection sites, genetic clusters, and disturbance. The grey circles correspond to percent forest cover, which ranges from 19 to 31 %; the grey stars correspond to the DAPC-defined genetic Clusters 1, 2 and 3; the grey triangles correspond to the collection sites with 1 being the Swamp, 2 being Aripo Savanna 1, 3 being Aripo Savanna 3, and 4 being Aripo Savanna 2
Discussion
With increased fragmentation and disturbance, we expect that dispersers, and most likely pollinators as well, are less likely to move freely among savannas, resulting in increased kinship levels correlated to distance (Pacheco and Simonetti 2000). Though our study was conducted at a small scale both spatially and temporally, relative to others of its ilk (Pacheco and Simonetti 2000; Born et al. 2008), we observed fine scale genetic structuring along with evidence of high levels of nearest-neighbor mating in M. flexuosa. Genetic cluster membership and kinship-distance curves indicate that, though gene flow occurs among collection sites, nearest-neighbor mating predominates over out-crossing through seed dispersal or pollination and that kinship is strongly correlated with distance (Fig. 2; Online Resource 1). These observations combined with our knowledge of the ASSR’s history suggest that genetic structuring in M. felxuosa is due to barriers to animal mediated seed dispersal in the form of roads, settlements, and high levels of hunting. Our small-scale approach could be broadly applicable, as many tropical reserves are on analogous scales to the ASSR.
The second genetic cluster, composed primarily of individuals collected at Savanna 2, displayed high levels of kinship-distance correlation (Online Resource 1). Site-specific group membership and high levels of kinship imply high levels of nearest-neighbor mating rather than gene flow among sites; this pattern appears temporally stable as indicated by non-significant tests for genic and genetic (Table 1). One of the most isolated sites, Savanna 2 is also the furthest removed from the park ranger station and relatively close to a road (Fig. 1). We infer that these high levels of nearest-neighbor mating are due, in part, to anthropogenic barriers to gene flow such as hunting of M. flexuosa dispersers and the construction of roads and settlements. However, it is also possible that the observed low rates of gene flow into and out of this site could be due to natural isolation.
The impacts of recent land-use changes on genetic structure are highlighted in Clusters 1 and 3. The first is comprised primarily of individuals from the highly disturbed Savanna 1, and half of the individuals from the relatively undisturbed Savanna 3, while the second half of Savanna 3 and the Swamp comprise Cluster 3 (Fig. 2). In the past 80 years, Savanna 1 has been a military training area and hunting ground, in addition to having been adjacent to a quarry (EMA 2007). Currently, Savanna 1 is near a village, adjacent to a park ranger station, and is the primary site for tourism and environmental education in the ASSR (EMA 2007) (Fig. 1). The swamp is located in a forest reserve outside of the ASSR near a main road and receives less official protection (Fig. 1). Though hunting probably occurs in the forest surrounding the swamp at similar rates to the ASSR, we saw no obvious signs of anthropogenic disturbance within the swamp itself.
In the case of the Swamp and Savannas 1 and 3, we can infer that what had once been a single genetic group is now fragmented into two. This pattern of genetic structuring in Clusters 1 and 3 combined with significant levels of inter-generational structuring (Table 1) implies that Clusters 1 and 3 became differentiated due to lack of gene flow among the three sites, most likely due to habitat fragmentation. Temporal instability, as indicated by measures of genic and genotypic differentiation (Table 1), indicates that levels of gene flow among the three sites differ across generations; this implies that though M. flexuosa has a generation time of 30 years, disturbances impacting the genetic structure of populations can be noticeable on a relatively short ecological time-scale. This observation of intergenerational differentiation is not without precedent as recent studies have also noted rapid change in Arecaceae as well as mimosoid trees due to habitat fragmentation and hunting (Galetti et al. 2013; Pacheco and Simonetti 2000). We hypothesize this difference is due to a historical restriction of their genetic neighborhood, namely reduced gene flow from the highly disturbed Savanna 1 to the Swamp. We believe that sustained disturbances beginning in the 1930s, such as the construction of a military base and hunting of dispersal vectors, are responsible for the observed disjointed pattern of gene flow from Savanna 3 to both the Swamp and Savanna 1, with no gene flow occurring between Savanna 1 and the Swamp, which are physically much closer to each other.
To more directly investigate the impact of anthropogenic fragmentation on genetic structure, we used forest cover as a proxy for disturbance, since the loss of forest is a common and quantifiable event during the course of urbanization and land use change. Cluster 1, primarily composed of the highly isolated Savanna 2, remains separate from the other two clusters, which is consistent with our other analyses. When regressed against forest cover, Clusters 2 and 1 and 3 and 1 are significantly different from one another, while Clusters 3 and 2 are not. The MCA analysis lends support to our hypothesis that anthropogenic disturbance is the causal factor behind the disruption in gene flow among the Swamp and Savannas 1 and 3. The genetic Clusters 2 and 3, which represent those sites, correspond strongly to low levels of forest cover and therefore high levels of disturbance (Figs. 1, 3). That we were able to identify spatial temporal shifts in population structure on such a small scale could be of high utility to the planning and implementation of small-scale conservation and management initiatives, where heuristic site knowledge could complement genetic data.
Conclusions
Although our study was conducted on a very small scale both spatially and temporally, we were able to detect patterns of genetic differentiation using microsatellite markers. Our collections accounted for three generations of M. flexuosa, and provided adequate resolution to infer the impacts of historical and current land use practices on the genetic structure of an important species within the ASSR. In the future, control sites lacking any disturbance would provide a potentially clearer picture of the issue, but none were available in Trinidad. Despite the limited nature of our sampling, these data could provide insight for managers to design dispersal corridors and monitoring programs among the savannas in this region. It is clear that for plants reliant on animal-mediated seed dispersal, barriers to dispersal constrain their genetic world and reduce them to nearest-neighbor matings, which lead to reductions in genetic diversity. Furthering the import of this study, M. flexuosa is one of the most widely used palms in Amazonia, and many local communities rely on its harvest for their livelihoods (Padoch 1988; Peters et al. 1989). As Amazonia is currently undergoing rapid land-use change and deforestation, a better understanding of M. flexuosa’s natural history is integral to formulating meaningful and effective sustainable management plans. This study is the first to examine population genetic structure in M. flexuosa, and it was performed on the scale of most local harvesting operations. We hope that the scale of our study will be useful to conservation workers and M. flexuosa harvesters in addressing similar scales of management.
References
Bodmer RE (1991) Seed dispersal and seed predation in amazonian ungulates. Biotropica 23(3):255–261
Born C, Hardy OJ, Chevallier MH, Ossari S, Atteke C, Wickings J, Hossart-Mckey M (2008) Small-scale spatial genetic structure in the Central African rainforest tree species Aucuomea klaineana: a stepwise approach to infer the impact of limited gene dispersal, population history and habitat fragmentation. Mol Ecol 17:2041–2050
Choo J, Juenger E, Simpson B (2012) Consequences of frugivore-mediated seed dispersal for the spatial and genetic structures of a neotropical palm. Mol Ecol 21:1019–1031
Connell JH (1971) On the role of natural enemies in preventing competitive exclusion in some marine animals and in rain forest trees. In: Den Boer PJ, Gradwell GR (eds) Dynamics of populations. Pudoc, Wageningen, pp 298–312
DeFries R, Hansen M, Townshend JRG, Janetos AC, Loveland TR (2000) 1 kilometer tree cover continuous fields, 1.0. Department of Geography, University of Maryland, College Park, Maryland, 1992–1993
Dick CW, Hardy OJ, Jones AF, Petit RJ (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop Plant Biol 1:20–33
Environmental Management Authority (EMA) (2007) Aripo savannas environmentally sensitive area literature review to facilitate the preparation of management plans. Prepared by the Caribbean Natural Resources Institute (CANARI), Port Au Prince, Trinidad
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Federman S, Hyseni C, Clement W, Caccone A (2012) Isolation of 13 novel and highly polymorphic microsatellite loci for the Amazonian palm Mauritia flexuosa L.f. (Arecaceae). Conserv Genet Res 4(2):355–357
Galetti M, Guevara R, Cortes MC, Fadini R, Von Matter S, Leite AB, Labecca F, Ribeiro T, Carvalho CS, Collevatti RG, Pires MM, Guimaraes PR Jr, Brancalion PH, Ribeiro MC, Jordano P (2013) Functional extinction of birds drives rapid evolutionary changes in seed size. Science 340(6136):1086–1090
Hamrick JL, Murawski DA, Nason JD (1993) The influence of seed dispersal mechanisms on the genetic structure of tropical tree populations. Vegetation 107(108):281–297
Hijmans RJ, van Etten J (2012) raster: geographic analysis and modeling with raster data. R package version 2.0-12
Holm JA, Miller C, Cropper WP (2008) Population dynamics of the dioecious amazonian palm Mauritia flexuosa: simulation analysis of sustainable harvesting. Biotropica 40(5):550–558
Janzen DH (1970) Herbivores and the number of tree species in tropical forests. Am Nat 104:501–528
Jombart T (2008) adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Balloux F (2010) Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet 11:94
Kalinowski ST, Wagner AP, Taper ML (2006) ML-Relate: a computer program for maximum likelihood estimation of relatedness and relationship. Mol Ecol Notes 6:576–579
Nenadic O, Greenacre M (2007) Correspondence Analysis in R, with two- and three-dimensional graphics: The ca package. J Stat Softw 20(3):1–13
Pacheco LF, Simonetti JA (2000) Genetic structure of a mimosoid tree deprived of its seed disperser, the spider monkey. Conserv Biol 14(6):1766–1775
Padoch C (1988) Aguaje (Mauritia flexuosa L. f) in the economy of Iquitos. Peru Adv Eco Bot 6:214–224
Peters CM, Balick MJ, Kahn F, Anderson AB (1989) Oligarchic forests of economic plants in Amazonia: utilization and conservation of an important tropical resource. Conserv Biol 3:341–349
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
R Development Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0, URL http://www.R-project.org
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Heredity 86:248–249
Rosa RK, Koptur S (2013) New findings on the pollination biology of Mauritia flexuosa (Arecaceae) in Roraima, Brazil: linking dioecy, wind, and habitat. Am J Bot 100(3):613–621
Sampaio MB, Schmidt IB, Figueiredo IB (2008) Harvesting effects and population ecology of the buriti palm (Mauritia flexuosa L.f. Arecaceae) in the Jalapao region, Central Brazil. Econ Bot 62(2):171–181
Young AG, Boyle T, Brown T (1996) The population consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Zona S, Henderson A (1989) A review of animal-mediated seed dispersal of palms. Selbyana 11:6–21
Acknowledgments
We thank F. Cooper and Sydney for help collecting, J. Brown, A. Reuben, D. Edwards, and R. Eyton for their comments and instruction. S.F. was supported by the Carpenter-Sperry Grant at the Yale School of Forestry and Environmental Studies, The Yale Institute for Biospheric Studies, and The New York Botanical Garden’s Cullman Fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Federman, S., Hyseni, C., Clement, W. et al. Habitat fragmentation and the genetic structure of the Amazonian palm Mauritia flexuosa L.f. (Arecaceae) on the island of Trinidad. Conserv Genet 15, 355–362 (2014). https://doi.org/10.1007/s10592-013-0543-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0543-2