Abstract
Urbanization may affect genetic differentiation among animal populations because it converts native vegetation to novel land cover types that can affect population connectivity. The effect of land cover change on genetic differentiation may vary among taxa; mobile birds may be least affected. Regardless, genetic differentiation between populations should be best predicted by measures of distance that incorporate the effect of land cover on movement. We studied the relationship between land cover and genetic differentiation in Song Sparrows (Melospiza melodia) at eighteen sites in the Seattle metropolitan region. We generated a series of hypothetical “resistance surfaces” based on land cover and development age, calculated “resistance distances” between pairs of sampling sites, and related them to pairwise genetic differentiation. Genetic differentiation was best described by a multiple regression model where resistance to gene flow (1) linearly increased with age of development and (2) was greater in high- and medium-density urbanization than in native forest land cover types (R 2 = 0.15; p = 0.003). The single variable with the highest correlation with genetic differentiation was derived from a linear relationship between development age and resistance (R 2 = 0.08; p = 0.007). Our results thus suggested that urban development reduced population connectivity for Song Sparrows. However, the relation of development age to genetic differentiation suggested that equilibrium was not yet reached. Hence, the effects of lost connectivity will increase. Our understanding of the landscape genetics of this recently anthropogenically modified landscape benefited from considering population history.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Urbanization poses a conservation challenge to animals by increasing the resistance of the landscape to dispersal, movement, and gene flow. Native vegetation is converted to new types of anthropogenically induced land cover, which alter movement rates or create barriers for animals (Whittaker and Marzluff 2012). Organisms may not travel safely through novel urban land covers where survival can be reduced by increased predation or reduced food resources (Marzluff 2001; Lepczyk et al. 2003). Moreover, the degree of connectivity among fragments is not simply a function of direct dispersers, but also includes stepping-stone movements over the course of multiple generations (Crochet 1996). Population connectivity is necessary for viability of metapopulations (With 2004), as it reduces local extinctions, accelerates recolonization (Hanski 1999), and limits the deleterious effects of inbreeding (Saccheri et al. 1998). Conservation of native species thus requires the consideration of connectivity among populations and not simply preservation of existing habitats (Marzluff and Rodewald 2008).
The degree to which population connectivity is affected by changes in land cover may be related to a species’ dispersal ability (Bohonak 1999). Movement by nonvolant terrestrial species is often severely impeded by barriers created by anthropogenic land cover change (Trombulak and Frissell 2000; Andrews and Gibbons 2005), but it is less clear how volant species such as birds might be affected. Birds can fly over barriers, but they may also exhibit behavioral responses affecting movement (Harris and Reed 2002). Furthermore, the amount of breeding habitat within the range of common dispersal distances facilitates stepping-stone gene flow over multiple generations, so reduced habitat availability could result in less population connectivity despite birds’ high mobility.
Movement of birds among populations is difficult to monitor directly, but can indirectly be studied using genetic methods (Raybould et al. 2002). Genetic differentiation among populations can be used to infer their isolation or connectivity (Hedrick 2000), which in turn can be related to landscape characteristics in order to make inferences about processes affecting connectivity (Manel et al. 2003). Resistance surfaces are maps that represent the relative resistance to movement for organisms moving through a landscape. Multiple resistance surfaces can be developed for different hypotheses of the relative resistances of different land cover types. By examining the relationship between genetic differentiation and distances derived from resistance surfaces, factors that influence population connectivity can be identified (e.g., Cushman et al. 2006; Lee-Yaw et al. 2009; Schwartz et al. 2009; Garroway et al. 2011).
We were interested in understanding how land cover change due to urbanization might affect population connectivity of forest songbirds in the greater Seattle region. We chose Song Sparrows as our focal species for this research for several reasons. They are locally dense in appropriate habitat (Arcese et al. 2002), have short generation times (average breeding age is 1.9–2.2 years, derived from Arcese et al. 2002), are non-migratory, having the shortest dispersal distance of common resident birds in western Washington (median = 200 m; Sutherland et al. 2000; Whittaker and Marzluff 2012), and can have fine-scale genetic structure in populations separated by less than 10 km, and even less than 2 km in island populations (Wilson et al. 2011), traits which increase the likelihood of observing differentiation at the relatively small spatial and temporal scales of our urban study system. Song Sparrows are commonly found in forest fragments throughout the region (Donnelly and Marzluff 2006), but use residential areas for foraging, particularly in the non-breeding season (C Templeton, personal communication). While it is possible for Song Sparrows to utilize and travel through urban land cover (Whittaker and Marzluff 2009, 2012), it does not have the dense shrub understory structure and native vegetation of the forests typically used for breeding. The reduction of breeding habitat availability in urban landscapes may inhibit the stepping-stone dispersal movements necessary for widespread gene flow. Furthermore, radio-tracking of juvenile Song Sparrows showed that their movements were slowed by high-density urbanization, and somewhat by low-density urbanization (Whittaker and Marzluff 2012).
For these reasons, we hypothesized that urban land cover would reduce Song Sparrow population connectivity. To investigate this hypothesis, we examined the relationship between genetic differentiation and distances derived from resistance surfaces reflecting a range of hypotheses for relative resistances of land cover types. We predicted that the relationship would be strongest for distances derived from resistance surfaces in which urban land cover types had high resistances. We predicted that high density urban land cover would create the highest resistance to movement, while lower density urban land cover would have intermediate resistance to movement, and land cover types such as forests, grassland, and agriculture would have the least resistance to movement. If land cover types did not influence connectivity, resistances of all land cover types would be essentially the same, genetic differentiation would be most related to geographic distance, and isolation by distance (Wright 1943; Rousset 1997) would be observed.
Additionally, population differentiation increases with duration of population isolation. Differentiation begins increasing when gene flow is reduced, but it may take generations to reach a new migration-drift equilibrium in an altered landscape (Varvio et al. 1986; Whitlock and McCauley 1999; Landguth et al. 2010). Therefore, if urbanization inhibits connectivity, the presence of urbanization between populations for longer time would cause more differentiation between those populations. The age of development should be positively related to resistance, purely via the reduction of gene flow over more time causing more differentiation, and independent from a potential relationship between development age and urban habitat quality, which we found no evidence for affecting connectivity. Thus we expected that resistance distances calculated from development ages should also be better predictors of genetic differentiation than geographic distance. Finally, we hypothesized that land cover type and development age would each affect population differentiation through their separate but related mechanisms—via differences in rates of gene flow and via duration of reduced gene flow, respectively. We predicted that a multiple regression model that had resistance distances based on both land cover and development age would outperform single variable models. A positive relationship between genetic differentiation and distances derived from resistance surfaces in which urbanization had high resistance or in which development age was related to resistance would suggest that urbanization reduced gene flow among the Song Sparrow populations.
Materials and methods
Study area
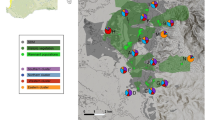
We located study sites in a 1,700 km2 study area including the lowlands (<500 m elevation) around Seattle, Washington, USA (47°36′N, 122°19′W; Fig. 1; Online Resources 1 & 2). The native vegetation is mostly moist temperate forest, much of which has been converted to urban land cover as the metropolitan region has expanded over the past 50 years (Robinson et al. 2005; Puget Sound Regional Council 2007). We selected 18 forested study sites with abundant Song Sparrows (Melospiza melodia), spanning a broad gradient of urbanization and a broad range of geographic distances in between them (median: 22.4 km; range 1.4–55.4 km). Sixteen study sites were part of a long-term study of the effects of ongoing urbanization on forest songbirds, selected to provide a random sample of landscapes stratified by urban patch size, urban land cover, and forest aggregation (Donnelly 2002; Marzluff et al. 2007). Two additional sites in Seattle were included to extend the urban end of the gradient. Song Sparrows were relatively common in our study area: 50-m radius point count surveys at our sites averaged 0.50 Song Sparrows per point (SD = 0.31; Unfried 2009), a pattern corroborated by three Breeding Bird Survey (Sauer et al. 2011) routes closest to our study sites that averaged 0.65 Song Sparrows per survey point (SD = 0.28).
Maps of the study area and of sampling locations. a Five-class land cover derived from 14-class land cover classification of 2002 Landsat imagery (Online Resource 2). Vegetation land cover includes deciduous-mixed forest, coniferous forest, regenerating forest, clear-cut, grass, and non-forested wetland; low-density urban land cover has <50 % impervious surface; high- and medium-density urban land cover has >50 % impervious surface; non-habitat includes agriculture, snow/rock/ice, and shoreline. b Average age of developments within 500 m radius derived from county assessor parcel databases
Sample collection
We captured 469 Song Sparrows during the breeding seasons (March–August) of 2004–2007 in mist nets placed throughout the forested areas of the eighteen study sites. Nets were placed both arbitrarily throughout the study site and also targeting specific birds by placing nets within their territories and attracting them with the aid of song playback and decoys. Once in hand, we collected either a feather or a blood sample or both for use in genetic analysis. We collected blood by pricking the ulnar vein with sterile needle (Arctander 1988), and gathered the resulting blood droplet (~50–100 μl) with a capillary tube, which we then transferred to a storage medium. We stored blood on an FTA® card (Whatman® BioScience), or in 1 ml 95 % ethanol or in 1 ml sterile isotonic buffer (1× SSC: 0.15 M NaCl; 15 mM trisodium citrate, pH 7.0; 10 mM EDTA, pH 7.4) kept cold in a portable ice box until transfer to long-term storage at −20 °C (Bruford et al. 1992). For feathers, we pulled one or two rectrices or five to ten breast feathers from the bird and stored them in a paper envelope at room temperature with desiccants.
DNA was extracted from blood on FTA® cards following Whatman’s® protocol for blood. Purified FTA® discs were then placed in 50 μl water and heated for 10 min at 90 °C to suspend the DNA in water for ease of sample processing thereafter (Smith and Burgoyne 2004). We extracted DNA from blood stored in ethanol, SSC, and feathers using the DNeasy™ Tissue Kit (QIAGEN), following the manufacturer’s protocol for cultured cells, with some modifications in the lysis steps (Unfried 2009).
Sample analysis
We genotyped each bird at 12 polymorphic microsatellite loci that have been used in genetic research on Song Sparrows: Mme1, Mme2, Mme7, Mme8 (Jeffery et al. 2001), GF2.35, Escu1 (Chan and Arcese 2002), Sosp01, Sosp02, Sosp04, Sosp07, Sosp08, and Sosp14 (Sardell et al. 2010). Polymerase chain reaction (PCR) conditions for Mme1, Mme2, Mme7, Mme8, Escu1, and GF2.35 were optimized in our laboratory from published protocols (Chan and Arcese 2002; details in Unfried 2009; Online Resource 3). We added “tails” to the 5′ end of the reverse primers for some problematic loci to reduce scoring errors resulting from single base pair addition caused by adenylation (GTTTCTT for Mme2, Mme7 and GF2.35; GCTTCT for Mme8; Brownstein et al. 1996). We used a M13 fluorescent labeled primer protocol for Sosp01, Sosp02, Sosp04, Sosp07, Sosp08, and Sosp14. M13 protocols allow flexibility and affordability in screening loci (Schuelke 2000; Boutin-Ganache et al. 2001). Genotyping was conducted on ABI 3100, 3130xl, and 3730xl automated sequencers (Applied Biosystems Inc., Foster City, California), and allele reads were made in GeneMapper 3.7 software and verified manually. We used samples from eight birds with a wide range of allele fragment sizes to calibrate adjustment curves for potential differences in fragment size reads among sequencers. After calibration, we compared 356 additional alleles scored on more than one sequencer (132 comparisons between ABI 3100 and ABI 3130xl, 172 comparisons between ABI 3100 and ABI 3730xl, and 52 comparisons between ABI 3130 and ABI 3730xl), of which 339 (95.2 %) were identical. Differences between sequencers were not due to misreading the fragment lengths; of the 17 alleles that were scored differently, 14 discrepancies were due to a misidentified fragment peak rather than fragment length. The 4.8 % genotyping error rate between sequencers was comparable to the 4.9 % genotyping error rate within sequencers (36 discrepancies out of 736 duplicate allele calls). In general, genotypes from blood extractions were scored more easily, though one locus (Mme1) better amplified from feather extractions. We genotyped 373 of the birds at all 12 loci, and 445 were genotyped at >10 loci. The number of birds genotyped per sampling site ranged from 17 to 52 (mean = 26.1, median = 23.5).
Landscape analysis
Our analytical approach was to generate resistance surfaces that reflected our hypotheses for the effects of land cover on population connectivity, then compute either the circuit theory resistance distance or least cost distance (hereafter collectively referred to as resistance distances, see below) between each pair of sampling sites, and finally compare the resistance distance to the genetic differentiation between each sampling site pair. This process was repeated for a series of different resistance surfaces, each reflecting a different set of hypothetical resistances for land covers. We were most interested in the effects of urban land cover types relative to more natural land cover types. Hence, most hypotheses involved variation in urban land cover resistances (Table 1). In addition to the null hypothesis that all land covers had equal resistance (isolation by distance), we had five sets of hypotheses in which high- (>80 % impervious surface), medium- (50–80 % impervious surface), and low-density (20–50 % impervious surface) land cover resistances varied in relation to one another (Table 1). We also included resistances of non-inhabitable land cover: agriculture, water, and snow/rock/ice. For each category of hypotheses, we varied the land cover in question over several orders of magnitude, namely twice to 1,000 times the resistance of more natural land cover. We suspected that non-inhabitable water, snow/rock/ice, agriculture and shoreline would deter dispersal, so high resistance values for these were included in some of the urban hypotheses sets.
In order to calculate resistance distances between pairs of sites, we created resistance landscape maps based on classified land cover geographic information system (GIS) grids and county parcel shapefiles that included year-built data fields. Using a 14-class land cover map with 30-m resolution derived from 2002 Landsat TM images (Fig. 1a; Online Resource 2) as a starting point, we reclassified pixel values of various land cover types to hypothetical resistance values in ArcView 3.3 (ESRI 2002). We also developed a development-age GIS grid, derived from King County and Snohomish County Assessors’ parcel databases by subtracting the year-built from 2008. Non-developed parcels were assigned the age of 0.1. We hypothesized that resistance would be linearly related to development age, so resistance at each pixel equaled the development age. To avoid the problem of having areas within developed areas with no development year, such as roads, the development age for each pixel was calculated as the mean of the pixels within a 500 m circle (Fig. 1b). Two development age resistance surfaces were created to account for hypotheses about the effect of water in relation to development age-related resistance: one hypothesis in which water was given a resistance of 1,000, the resistance of ancient development, and a second hypothesis in which water’s resistance was 0.1, as if water influenced dispersal like an undeveloped area.
We used two methods for determining land cover resistance distances between sampling points based on the resistance surfaces. Least cost cumulative distance was calculated with the ArcView extension pathmatrix (Ray 2005) which determines the single path that minimizes the sum of the accumulated cost distance between the sampling points. Circuit theory landscape resistance calculated in Circuitscape v.3.0.1 (McRae et al. 2008) represents resistance between two points over the entire landscape, treating the landscape as grid of resistors creating a circuit. The two methods produce correlated results, but their differences are important. Circuitscape’s landscape resistance values incorporate all possible routes between two points in a landscape, while pathmatrix just finds the shortest landscape distance between two sites, even if there is little connectivity elsewhere. Sampling site coordinates were calculated as the centroids of the capture locations, territory centers, or resighting locations of all the genotyped birds at a sampling site. Due to computational limitations, GIS grid pixels were aggregated to 60 m resolution on a subset of nine resistance surfaces for analysis with Circuitscape.
Statistical analysis
Genetic summary statistics H e, F IS, and allelic richness were calculated in F stat v.2.9.3.2 (Goudet 2001). Hardy–Weinberg equilibrium and linkage disequilibrium of loci per sampling location was tested with a probability test in Genepop v.4.0 (Rousset 2008), with 10,000 dememorization steps followed by 20 batches of 5,000 iterations per batch. We also checked for sex linkage of the 12 loci by coding sex as a locus and testing for linkage disequilibrium in Genepop.
To examine differentiation between pairs of sampling locations, we calculated pairwise F ST-values by the estimator θ (Weir and Cockerham 1984) for all sampling location pairs and tested their significance with an exact G-test in Genepop. Multiple comparisons were controlled for with a sequential Bonferroni adjustment (Holm 1979). The magnitude of F ST estimators of population differentiation are limited by within-population heterozygosity (Hedrick 2005), so population differentiation estimates may be influenced by the loss of heterozygosity (H E) in small populations. To check for this effect, we recoded the data so that there was no overlap in allele sizes among populations in RecodeData v.0.1 (Meirmans 2006) and then calculated the maximum possible F ST-value accounting for within-population heterozygosity. The maximum possible pairwise F ST-values determined from recoding data ranged from 0.222 to 0.256, and the standardized θ = 0.042. The correlation between pairwise θ and pairwise θ standardized for within-population heterozygosity was 0.996 (p < 0.001, df = 151), so raw pairwise θ was used for analysis. Furthermore, heterozygosity was similar among populations (range of mean H E: 0.734—0.783), so it would have little effect on our models. To further investigate the potential for population size to influence F ST, we estimated the effective population size (N e) at each sampling location using a sibship method in Colony 2.0 (Wang 2004) that can accommodate genotyping error and allelic dropout during sibship assignments, which we set at 0.035 and 0.014 per locus, respectively.
We modeled a linear regression of genetic differentiation, θ/(1 − θ) (Rousset 1997), on resistance distances between sites for each hypothesis in R (R Development Core Team 2008) to calculate R 2 and slope coefficients. We then performed model selection on Akaike’s An Information Criterion (AIC; Burnham and Anderson 2002) to determine which hypotheses for land cover resistances best fit the genetic data. We regressed genetic differentiation on resistance distances derived from least cost paths for 43 land cover hypotheses and two development age hypotheses, and resistance distances derived from circuit theory in Circuitscape for nine land cover hypotheses and one development age hypothesis. Finally, to test the hypothesis that both land cover type and development age affect gene flow, we performed multiple regression of genetic differentiation on the linear combination of development-age circuit theory resistance distance with each of 52 land cover hypothesis distances with their interaction. We tested the significance of regressions by permuting the rows and columns of the response variable distance matrix 2000 times to generate a distribution of the R 2 statistic for comparison with observed values (Anderson 2001). For the top ten models, with p values close to 1/2,000, the permutation test was run for 10,000 iterations to obtain a precise p-value.
Results
Genetic diversity was generally high at all loci (mean H O = 0.56–0.90, mean H E 0.66–0.91; Online Resource 4), with the exception of locus Sosp08 (H O = 0.20 and H E = 0.22; Online Resource 4). Mean allelic richness was similar among sites, ranging from 6.4 to 7.5 alleles per locus (Online Resource 4; SE: 0.77–1.01, two-factor ANOVA F 17,187 = 1.38, p = 0.15). Of 216 locus-site combinations, 31 had significantly positive F IS, suggesting that some loci were not in Hardy–Weinberg equilibrium in particular populations. However, after sequential Bonferroni adjustment for multiple tests just three of those locus-site combinations’ F IS remained significant. There were no consistent patterns among loci or sites to suggest that any particular locus or site was out of Hardy–Weinberg equilibrium. Four of 1,151 tests for linkage disequilibrium remained significant after sequential Bonferroni adjustment. Tests for linkage with sex showed eleven of the 12 loci were not sex-linked; the exception was Mme7, which is Z-linked, so female and unknown sex birds were coded as missing data for the missing allele (Chan and Arcese 2002). Effective population sizes were similar among sites (mean N e = 38.1, range: 25–50; Online Resource 1). The similarities among effective population sizes as well as among within-population heterozygosities, the fact that observed F ST-values were much smaller than the maximum possible F ST-values, and the high correlation between raw F ST-values and F ST-values corrected for within-population heterozygosity led us to conclude that population size and heterozygosity would have minimal effect on F ST-values, so we used raw F ST-values in our analysis.
For the entire study area, genetic differentiation was low, but significant (θ = 0.010; 99 % CI: 0.005–0.016). Pairwise θ between sampling locations ranged from −0.004 to 0.028 (Table 2). Differentiation was significant between 125 of 153 pairs at p < 0.05, and 81 of those tests for differentiation remained significant after sequential Bonferroni adjustment. The sites that were most differentiated from the others tended to be those in older or denser urban developments (Table 2; Fig. 1): SS (significantly differentiated from 16 other sites), DP (15 significant differences), and BE (15 significant differences). In contrast, the sites that were differentiated from the fewest other sites were those in areas most recently developed, least developed, or in or near large forest reserves: CW (one significant difference), BL (two significant differences), and SM (three significant differences).
The hypothesis that resistance was linearly related to age of development and that water’s resistance was like that of ancient development was the best single-variable model predicting F ST (DevelopmentAge=Resistance,Water×1000_csr; R 2 = 0.08; p = 0.007; Fig. 2a; Table 3). The single-variable land cover type hypothesis that best predicted differentiation was Urban×1000, meaning that all urban land cover types were 1,000 times more resistant to dispersal than all other land cover types. There was a positive relationship between resistance distance and genetic differentiation, though marginally non-significant (Urban×1000; ΔAIC = 11.68; R 2 = 0.06; p = 0.08; Fig. 2b; Table 3). There was no clear relationship between population differentiation and the null model of all-equal resistance (isolation by distance), and this non-significant trend was negative (Null; ΔAIC = 19.60; Fig. 2d; Table 3). Hypotheses involving only water or agriculture did not predict genetic differentiation (Online Resource 5).
Linearized genetic differentiation versus resistance distance. a Resistance distance as a linear function of development age, hypothesis DevelopmentAge=Resistance,Water×1000 (R 2 = 0.08; p = 0.007). b Resistance distance as a function of land cover, hypothesis Urban×1000, meaning that all urban land cover types are 1000 times more resistant than all other land cover types (R 2 = 0.06; p = 0.08). c Resistance distance from hypothesis Low-Urban×1,HighMedium-Urban&Non-habitat×100 (R 2 = 0.01; p = 0.429). d Resistance distance based on the null hypothesis that all land covers have equal resistance to dispersal (isolation by distance; R 2 = 0.01; p = 0.389). Pairwise resistance distances calculated in Circuitscape are given in a and c and those from least cost paths are given in b and d
Genetic differentiation was most strongly related to the multiple regression hypothesis that resistance increased with development age (DevelopmentAge=Resistance,Water×1000_csr) and with high and medium urbanization (Low-Urban×1,HighMedium-Urban&Non-habitat×100; R 2 = 0.15; p = 0.003; Table 3). The hypothesis Low-Urban×1,HighMedium-Urban&Non-habitat×100 was positively correlated to differentiation in a single regression (Fig. 2c), but was negatively correlated to differentiation in a multiple regression including development age, as were many of the land cover hypotheses that had positive relationships with differentiation (Table 3; Online Resource 5).
Discussion
The positive relationship between genetic differentiation and resistance distances assuming high effects of urbanization indicated that Song Sparrow connectivity has been reduced by urbanization in the Seattle metropolitan region. Low overall genetic differentiation indicated gene flow throughout the study area, yet many of the study sites were significantly differentiated from one another. Furthermore, the relationship between geographic distance and genetic differentiation was non-significant, suggesting that a distance measure other than Euclidian distance contributed to genetic differentiation, or some other factors were involved. Given equal migration rates and time since divergence, population differentiation is higher in small populations because they are more affected by genetic drift. However, estimates of effective population sizes and genetic diversity were similar among study sites, so population sizes should not have had a major influence on differentiation. Development age and urban land cover together best explained genetic differentiation, accounting for 15 % of the variation in F ST-values. It appears that urban land cover affects bird connectivity, even at the small scale of the Seattle metropolitan region. Although birds have the ability to fly over and beyond unsuitable habitat, factors such as their behavioral avoidance of crossing habitat gaps (Harris and Reed 2002), increased mortality, and reduced breeding habitat (Marzluff 2001) might contribute to decreased movement and gene flow through urban landscapes. The fact that urban landscapes reduce population connectivity for a relatively mobile, disturbance-tolerant species such as the Song Sparrow may not bode well for less mobile, disturbance-intolerant species.
The age of developments in the landscape was related to genetic differentiation in Song Sparrows, as predicted. Development-age derived landscape resistance was correlated with genetic differentiation, providing evidence that urbanization restricted gene flow. Genetic differentiation accumulates over time in the absence of migration, so more time should result in more differentiation until equilibrium is reached (Whitlock and McCauley 1999; Landguth et al. 2010). Theoretically, reaching equilibrium in F ST-values can take very long with low migration rates (Whitlock and McCauley 1999), but if equilibrium is achieved, by definition F ST-values should not change much over time, and the relationship between genetic differentiation and duration of reduced gene flow should disappear. We found that older development age between populations was related to more differentiation, consistent with the expectation that longer periods of reduced gene flow should cause increased genetic differentiation. Thus, very low F ST-values may have been due to recent separation rather than ongoing gene flow, and we observed a time lag between the land cover change and its effect on population differentiation (Landguth et al. 2010). However, even the sites that have been separated by older development had relatively small F ST-values, suggesting that some gene flow still occurred, though reduced by urbanization. As Song Sparrow populations continue to exist in urbanized landscapes, population connectivity and gene flow will likely be further reduced, resulting in small isolated populations being more subject to deleterious effects of inbreeding. A comparison of naturally and anthropogenically fragmented Song Sparrow populations found that genetic diversity was relatively lower in urban Song Sparrows, despite similar levels of differentiation, suggesting that ecological factors in the urban landscape such as different sources of mortality may further affect population genetics (MacDougall-Shackleton et al. 2012).
Our results suggest that some gene flow persists in urban environments despite small median dispersal distances. Song Sparrows contain many subspecies across nearly all of North America and the level of differentiation across their range can be much larger (F ST = 0.00–0.40; Pruett et al. 2008a) than what was found in our study. Many of those larger differences are related to long term biogeographic effects, such as small populations on islands, and subspecific reproductive isolation, rather than recent anthropogenic change. Yet small scale genetic structuring has been observed previously in Song Sparrows (Wilson et al. 2011). The range of pairwise F ST-values we observed were comparable to those seen in other regions, such as the San Francisco Bay area and coastal British Columbia, particularly when accounting for subspecific differentiation (Chan and Arcese 2003; Wilson et al. 2011). Also similar to our results, Song Sparrows did not exhibit isolation by distance over the San Francisco Bay area and coastal British Columbia (Chan and Arcese 2003; Wilson et al. 2011). Yet morphological and behavioral differences occurred among five subspecies in the Bay area, despite relatively high levels of gene flow suggested by the low range of pairwise F ST-values among subspecies (Chan and Arcese 2003; Pruett et al. 2008b). Local population adaptation may have been facilitated by reduced gene flow, but selection by environmental conditions was the driving factor for morphological differentiation (Chan and Arcese 2003). Similarly, reduced gene flow, combined with selective pressure from the urban environment, has the potential to enable local adaptation in urban Song Sparrows in our study system. House Finch (Carpodacus mexicanus) populations in urban areas have experienced reproductive isolation and differentiation as a result of natural selection on bill morphology due to altered food availability (Badyaev et al. 2008). Song Sparrows are somewhat tolerant of urban development, venturing into vegetated suburban neighborhoods to use bird feeders in the non-breeding season, so reduced gene flow might allow selection for eating the novel urban food resources to result in beneficial adaptations. However, the benefits of such adaptations might not outweigh the declines and extirpations of small populations that no longer receive the immigration necessary to sustain them. Urbanization has the potential to impact population connectivity even for vagile birds, which may result in positive or negative consequences.
Year-round resident populations such as these Song Sparrows may suffer greater effects from habitat fragmentation, compared with annual long-distance migrants. Migratory birds have more opportunity for dispersal than sedentary species, through the process of breeding site selection during annual migration. For example, more gene flow was found amongst migratory populations of House Wrens (Troglodytes aedon) than sedentary populations (Arguedas and Parker 2000). Other migratory species such as Cerulean Warblers (Dendroica cerulea) and Tree Swallows (Tachycineta bicolor) have no genetic differentiation or population structure over continental scales (Veit et al. 2005; Stenzler et al. 2009). Although the sampling schemes in those studies were on a different scale, the lack of differentiation suggests widespread gene flow across their ranges. In contrast, we found differentiation between populations as little as 2.4 km apart, which is similar to the 2–10 km genetic patch sizes found in Song Sparrows in San Francisco Bay and small islands in British Columbia (Wilson et al. 2011). Delaney et al. (2010) found reduced gene flow among habitat fragments over a similarly small scale for Wrentits (Chamaea fasciata) in urban southern California, a phenomenon corroborated by similar patterns of gene flow reduction in three lizard species. Migratory populations of Song Sparrows have higher dispersal rates and are more likely to have occurrences of long distance dispersal than sedentary populations (Pruett et al. 2008b), which should enable better population connectivity despite urbanization. These results along with ours suggest that gene flow reduction due to urbanization may be a conservation concern for year-round resident birds.
While results from a single species cannot be generalized for other taxonomic groups, the results from Song Sparrows are illustrative for other species of resident songbirds that breed in forests surrounded by urbanized areas, because they share many life history qualities. Our results suggest that urbanization does restrict population connectivity and causes population differentiation. Other resident birds native to Pacific Northwest forest vary in their degree of use of and movement through urban landscapes. Species that are restricted to larger, older forests such as Pacific Wren (Troglodytes pacificus) may be expected to have further loss of connectivity and local extinctions may increase due to reduced immigration or the effects of inbreeding. The effect of urbanization on other species connectivity will depend on their specific requirements for breeding and migration, and so further research on other species will reveal more generally applicable patterns.
References
Anderson MJ (2001) Permutation tests for univariate and multivariate analysis of variance and regression. Can J Fish Aquat Sci 58:626–639
Andrews KM, Gibbons JW (2005) How do highways influence snake movement? Behavioral responses to roads and vehicles. Copeia 4:772–782
Arcese P, Sogge MK, Marr AB, Patten MA (2002) Song Sparrow (Melospiza melodia). In: Poole A, Gill F (eds) The birds of North America, vol 704. The Birds of North America, Inc., Philadelphia
Arctander P (1988) Comparative studies of avian DNA by restriction fragment length polymorphism analysis: convenient procedures based on blood samples from live birds. J Ornithol 129:S205–S216
Arguedas N, Parker PG (2000) Seasonal migration and genetic population structure in house wrens. Condor 102:517–528
Badyaev AV, Young RL, Oh KP, Addison C (2008) Evolution on a local scale: developmental, functional, and genetic bases of divergence in bill form and associated changes in song structure between adjacent habitats. Evolution 62:1951–1964
Bohonak AJ (1999) Dispersal, gene flow and population structure. Quart Rev Biol 74:21–45
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31:25–28
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by tag DNA polymerase: primer modifications that facilitate genotyping. Biotechniques 20:1004–1010
Bruford MW, Hanotte O, Brookfield JFY, Burke T (1992) Single-locus and multilocus DNA fingerprinting. In: Hoelzel AR (ed) Molecular genetic analysis of populations: a practical approach. Oxford University Press, Oxford, pp 225–269
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer, New York
Chan Y, Arcese P (2002) Subspecific differentiation and conservation of Song Sparrows (Melospiza melodia) in the San Francisco Bay region inferred by microsatellite loci analysis. Auk 119:641–657
Chan Y, Arcese P (2003) Morphological and microsatellite differentiation in Melospiza melodia (Aves) at a microgeographic scale. J Evol Biol 16:939–947
Crochet P-A (1996) Can measures of gene flow help to evaluate bird dispersal? Acta Oecol 17:459–474
Cushman SA, McKelvey KS, Hayden J, Schwartz MK (2006) Gene flow in complex landscapes: testing hypotheses with causal modeling. Am Nat 168:486–499
Delaney KS, Riley SPD, Fisher RN (2010) A rapid, strong, and convergent genetic response to urban habitat fragmentation in four divergent and widespread vertebrates. PLoS ONE 5(9):e12767. doi:10.1371/journal.pone.0012767
Donnelly RE (2002) Design of habitat reserves and settlements for bird conservation in the Seattle metropolitan area. University of Washington, Dissertation
Donnelly R, Marzluff JM (2006) Relative importance of habitat quantity, structure, and spatial pattern to birds in urbanizing environments. Urban Ecosyst 9:99–117
Environmental Systems Research Institute (2002) ArcView 3.3. Environmental Systems Research Institute, Redlands, CA
Garroway CJ, Bowman J, Wilson PJ (2011) Using a genetic network to parameterize a landscape resistance surface for fishers, Martes pennanti. Mol Ecol 20:3978–3988
Goudet J (2001) FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www.unil.ch/izea/softwares/fstat.html
Hanski I (1999) Metapopulation ecology. Oxford University Press, Oxford
Harris RJ, Reed JM (2002) Behavioral barriers to non-migratory movements of birds. Ann Zool Fenn 35:275–290
Hedrick PW (2000) Genetics of populations, 2nd edn. Jones & Bartlett, Sudbury, MA
Hedrick PW (2005) A standardized genetic differentiation measure. Evolution 59:1633–1638
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65–70
Jeffery KJ, Keller LF, Arcese P, Bruford MW (2001) The development of microsatellite loci in the Song Sparrow, Melospiza melodia (Aves) and genotyping errors associated with good quality DNA. Mol Ecol Notes 1:1–13
Landguth EL, Cushman SA, Schwartz MK, McKelvey KS, Murphy M, Luikart G (2010) Quantifying the lag time to detect barriers in landscape genetics. Mol Ecol 19:4179–4191
Lee-Yaw JA, Davidson A, McRae BH, Green DM (2009) Do landscape processes predict phylogeographic patterns in the wood frog? Mol Ecol 18:1863–1874
Lepczyk CA, Mertig AG, Liu JI (2003) Landowners and cat predation across rural-to-urban landscapes. Biol Conserv 115:191–201
MacDougall-Shackleton EA, Clinchy M, Zanette L, Neff BD (2012) Songbird genetic diversity is lower in anthropogenically versus naturally fragmented landscapes. Conserv Genet 12:1195–1203
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18:189–197
Marzluff JM (2001) Worldwide urbanization and its effects on birds. In: Marzluff JM, Bowman R, Donnelly R (eds) Avian ecology and conservation in an urbanizing world. Kluwer, Norwell, MA, pp 19–47
Marzluff JM, Rodewald AD (2008) Conserving biodiversity in urbanizing areas: nontraditional views from a bird’s perspective. Cities Environ 1:6
Marzluff JM, Withey JC, Whittaker KA, Oleyar MD, Unfried TM, Rullman S, Delap J (2007) Consequences of habitat utilization by nest predators and breeding songbirds across multiple scales in an urbanizing landscape. Condor 109:516–534
McRae BH, Dickson BG, Keitt TH, Shah VB (2008) Using circuit theory to model connectivity in ecology and conservation. Ecology 10:2712–2724
Meirmans PG (2006) Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60:2399–2402
Pruett CL, Arcese P, Chan YL, Wilson AG, Patten MA, Keller LF, Winker K (2008a) Concordant and discordant signals between genetic data and described subspecies of Pacific Coast Song Sparrows. Condor 110:359–364
Pruett CL, Arcese A, Chan YL, Wilson AG, Patten MA, Keller LF, Winker K (2008b) The effects of contemporary processes in maintaining the genetic structure of western Song Sparrows (Melospiza melodia). Heredity 101:67–74
Puget Sound Regional Council (2007) Population change and net migration. Puget Sound Trends no. D7. Puget Sound Regional Council, Seattle, WA. http://www.psrc.org/assets/785/d7feb07.pdf. Accessed June 2010
Ray N (2005) PATHMATRIX: a GIS tool to compute effective distances among samples. Mol Ecol Notes 5:177–180
Raybould AF, Clarke RT, Bond JM, Welters RE, Gliddon CJ (2002) Inferring patterns of dispersal from allele frequency data. In: Bullock JM, Kenward RE, Hails RS (eds) Dispersal ecology. Blackwell, Oxford, pp 89–112
Robinson L, Newell JP, Marzluff JM (2005) Twenty-five years of sprawl in the Seattle region: growth management responses and implications for conservation. Landsc Urban Plan 71:51–72
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Rousset F (2008) GENEPOP ‘007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Notes 8:103–106
Saccheri I, Kuussaari M, Kankare M, Vikman P, Fortelius W, Hanski I (1998) Inbreeding and extinction in a butterfly metapopulation. Nature 392:491–494
Sardell RJ, Keller LF, Arcese P, Bucher T, Reid JM (2010) Comprehensive paternity assignment: genotype, spatial location and social status in song sparrows, Melospiza Melodia. Mol Ecol 19:4352–4364
Sauer JR, Hines JE, Fallon JE, Pardieck KL, Ziolkowski DJ Jr, Link WA (2011) The North American Breeding Bird Survey, results and analysis 1966–2010. Version 12.07.2011. Accessed 5 Sept 2012
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Schwartz MK, Copeland JP, Anderson NJ, Squires JR, Inman RM, McKelvey KS, Pilgrim KL, Waits LP, Cushman SA (2009) Wolverine gene flow across a narrow climatic niche. Ecology 90:3222–3232
Smith LM, Burgoyne LA (2004) Collecting, archiving and processing DNA from wildlife samples using FTA® databasing paper. BMC Ecol 4:4. http://www.biomedcentral.com/1472-6785/4/4. Accessed June 2010
Stenzler LM, Makarewich CA, Coulon A, Ardia DR, Lovette IJ, Winkler DW (2009) Subtle edge-of-range genetic structuring in transcontinentally distributed North American Tree Swallows. Condor 111:470–478
Sutherland GD, Harestad AS, Price K, Lertzman KP (2000) Scaling of natal dispersal distances in terrestrial birds and mammals. Conserv Ecol 4:16
R Development Core Team (2008) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org. Accessed June 2010
Trombulak SC, Frissell CA (2000) Review of ecological effects of roads on terrestrial and aquatic communities. Conserv Biol 14:18–30
Unfried TM (2009) Urban landscape relationships with Song Sparrow (Melospiza melodia) population structure and connectivity and human health and behavior. Dissertation, University of Washington
Varvio S-L, Chakraborty R, Nei M (1986) Genetic variation in subdivided populations and conservation genetics. Heredity 57:189–198
Veit SL, Robertson RJ, Hamel PB, Friesen VL (2005) Population genetic structure and dispersal across a fragmented landscape in cerulean warblers (Dendroica cerulea). Conserv Genet 6:159–174
Wang J (2004) Sibship reconstruction from genetic data with typing errors. Genetics 166:1963–1979
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: F ST ≠ 1/(4Nm + 1). Heredity 82:117–125
Whittaker KA, Marzluff JM (2009) Species-specific survival and relative habitat use in an urban landscape during the postfledging period. Auk 126:288–299
Whittaker KA, Marzluff JM (2012) Post-fledging mobility in an urban landscape. In: Lepczyk CA, Warren PS (eds) Urban bird ecology and conservation. Stud Avian Biol (no. 45). University of California Press, Berkeley, CA, pp 183–198
Wilson AG, Arcese P, Chan YL, Patten MA (2011) Micro-spatial genetic structure in song sparrows (Melospiza melodia). Conserv Genet 12:213–222
With KA (2004) Metapopulation dynamics: perspectives from landscape ecology. In: Hanski I, Gaggiotti OE (eds) Ecology, genetics, and evolution of metapopulations. Elsevier, London, pp 23–44
Wright S (1943) Isolation by distance. Genetics 28:114–138
Acknowledgments
M. Alberti and J. Hepinstall of the University of Washington’s Urban Ecology Research Laboratory provided the classified Landsat TM imagery. Sample collection was aided by M. D. Oleyar, S. Rullman, K. Whittaker, J. Delap, M. Dickerson, and C. Templeton. J. Lawler provided computing resources. Molecular laboratory and analytical assistance was provided by T. R. Seamons, M. Baird, L. Newton, B. Godfrey, and H. D. Bradshaw. The research was conducted under an animal use protocol overseen by the University of Washington IACUC and permits issued by the Washington Department of Fish & Wildlife and USGS Bird Banding Laboratory. This material is based upon work supported under a National Science Foundation Graduate Research Fellowship to TMU. Project funding was also provided by NSF Integrative Graduate Education and Research Traineeship for Urban Ecology (National Science Foundation awards DEB-9875041, BCS0120024, BCS 0508002 and IGERT0114351), Achievement Rewards for College Scientists Seattle Chapter, and University of Washington College of Forest Resources. We thank R. Holderegger and four anonymous reviewers for helpful comments on this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Unfried, T.M., Hauser, L. & Marzluff, J.M. Effects of urbanization on Song Sparrow (Melospiza melodia) population connectivity. Conserv Genet 14, 41–53 (2013). https://doi.org/10.1007/s10592-012-0422-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-012-0422-2