Abstract
For species that are habitat specialists or sedentary, population fragmentation may lead to genetic divergence between populations and reduced genetic diversity within populations, with frequent inbreeding. Hundreds of kilometres separate three geographical regions in which small populations of the endangered Eastern Bristlebird, Dasyornis brachypterus, a small, ground-dwelling passerine that occurs in fire-prone bushland in eastern Australia, are currently found. Here, we use mitochondrial and microsatellite DNA markers to: (i) assess the sub-specific taxonomy designated to northern range-edge, and central and southern range-edge D. brachypterus, respectively, and (ii) assess levels of standing genetic variation and the degree of genetic subdivision of remnant populations. The phylogenetic relationship among mtDNA haplotypes and their spatial distribution did not support the recognised subspecies boundaries. Populations in different regions were highly genetically differentiated, but in addition, the two largest, neighboring populations (located within the central region and separated by ~50 km) were moderately differentiated, and thus are likely closed to migration (microsatellites, F ST = 0.06; mtDNA, F ST = 0.12, Θ ST = 0.08). Birds within these two populations were genotypically diverse and apparently randomly mating. A long-term plan for the conservation of D. brachypterus’s genetic diversity should consider individual populations as separate management units. Moreover, managers should avoid actively mixing birds from different populations or regions, to conserve the genetic integrity of local populations and avoid outbreeding depression, should further translocations be used as a recovery tool for this species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Threatened species often exist as small and fragmented populations, confronted by continuing degradation and loss of their habitat (IUCN 2010). For populations of such species, low levels or a loss of standing genetic variation is predicted to reduce the likelihood of short-term population persistence and long-term evolutionary potential. Indeed, a low or reduced number of genotypes within a population imply a reduced probability that a set of fit genotypes will be present, and thus able, to adapt in the face of future environment changes (Frankham 2005; Willi et al. 2006; Frankham et al. 2010). Moreover, inbreeding depression has been observed in small, genetically depauperate populations, where the combination of inbreeding, accumulation of deleterious recessive alleles and low levels of genetic diversity may be detrimental to fitness (Keller et al. 1994; Ralls et al. 2000; Keller and Waller 2002; Hale and Briskie 2007; Leberg and Firmin 2008).
The effects of inbreeding and loss of genetic diversity are predicted to be particularly severe for small relatively or absolutely isolated populations. Indeed, fragmentation of large continuous terrestrial populations often results in a set of disjointed, remnant populations within a highly modified matrix or environment (e.g. Laurance et al. 2002; Watson et al. 2005; Stouffer et al. 2006). Under such circumstances, connectivity among populations may be reduced, because organisms may not disperse across, or experience increased rates of predation within, the surrounding matrix (Rodriguez et al. 2001; Laurance et al. 2004; Laurance and Gomez 2005; Uezu et al. 2005; Castellon and Sieving 2006). At the extreme however, extensive habitat loss or modification with a consequent high number of local population extinctions across the range of a species, may mean that only a limited number of remnant populations persist, and that such populations occur separated by hundreds of kilometres (e.g. Reding et al. 2010). In this instance, current dispersal and subsequent gene flow is likely to be rare or indeed improbable, particularly for taxa that are sedentary or habitat specialists (Hudson et al. 2000; Brown et al. 2004; Boessenkool et al. 2007; Taylor et al. 2007).
Biodiversity strategies typically strive to conserve ecosystems and their component species as well as the genetic diversity within and among populations of individual species (e.g. Commonwealth of Australia 1996). However, for most species we lack information concerning the spatial distribution of genetic variation, thus the conservation of this level of biodiversity is rarely planned with species-specific knowledge. Nevertheless, a key goal in long-term plans for the conservation and management of threatened species in fragmented environments is the preservation of genetic diversity and the maintenance of gene flow among populations. In some instances, the only realistic way to achieve this goal is by translocation (e.g. Wolf et al. 1996). Indeed, the establishment of a new population/s within a species historical range may buffer the species against extinction by increasing the absolute number of individuals and spatial extent over which the species occurs, thus providing insurance against demographic stochasticity or environmental catastrophes that may affect local populations. Moreover, a new population may provide a stepping-stone for gene flow between nearby populations, further increasing the genetically effective population size of remnant populations (e.g. Roberts et al. 2007). For especially small and isolated populations, supplementation may enlarge effective population size, and novel genetic variation may increase adaptive potential by providing the raw material for selection to act on or ameliorating inbreeding depression (Ingvarsson 2001). Translocation programs involving the mixing of genetically distinct populations have been very successful (e.g. sea otters Bodkin et al. 1999; Larson et al. 2002). However, the scale over which the supplementation of individuals should occur depends on the scale of genetic subdivision within the species and the potential for genetic mixing to impact detrimentally on fitness e.g. through outbreeding depression (Templeton 1986; Edmands 2007; Frankham et al. 2010).
The Eastern Bristlebird, Dasyornis brachypterus, is a small, cover-dependent, ground-dwelling, semi-flightless passerine, which is endemic to southeastern Australia, occurring in a handful of isolated populations in fire-prone vegetation spread across approximately 1,000 km (Baker 1997, 2000). Radio tracking and banding of birds indicates that birds have home ranges of approximately 10 ha but may move up to 5 km through suitable habitat in 18 months (Baker and Clarke 1999, unpublished data). Schodde and Mason (1999) designated D. brachypterus located at the northern range-edge of the species, subspecies monoides, and central and southern D. brachypterus, subspecies brachypterus, based on slight differences in plumage colouration (refer to Schoode and Mason 1999 for detail). However, a preliminary phylogenetic study by Elphinstone (2008) based on mitochondrial DNA sequence variation for a small number of northern (n = 2), central (n = 27) and southern birds (n = 1) did not support this distinction because there was no obvious geographic partitioning of mtDNA haplotypes in a phylogram, indicating a lack of genetic structure. Dasyornis brachypterus is listed as endangered in all relevant jurisdictions because of the small total population, extreme fragmentation of remnant populations and threat of catastrophic fire. In an attempt to prevent the otherwise seemingly inevitable extinction of the northern range-edge population, a captive breeding programme was established, while within the central-region wild-caught birds have been translocated to two locations in an attempt to mitigate against local extinction from future catastrophic fire. Further translocations are likely to be required to contribute to the species’ recovery.
We currently know nothing about the population genetic structure of D. brachypterus, although its highly fragmented distribution and its natural demography are likely to lead to high levels of genetic divergence between populations and reduced genetic diversity within populations, with frequent inbreeding. Here we use a survey of mitochondrial DNA sequence variation and microsatellite DNA markers to assess genetic differentiation between northern and southern range-edge, and central, populations of D. brachypterus. Furthermore, we characterise levels of standing genetic variation, assess population differentiation and test for evidence of inbreeding and past genetic bottlenecks, for the two largest populations, which occur in the centre of the species range.
Methods
Study system and species history
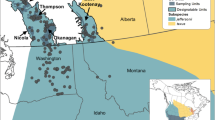
Dasyornis brachypterus was probably wide-spread in eastern Australia during the wetter Tertiary Period (Smith 1977) in the relative absence of fire. However, historic records suggest that the species was highly fragmented two centuries ago, and since then a number of local populations have gone extinct (Baker 1997). In particular, the species was extirpated at 11 of 13 locations at its southern limit and 12 of 23 locations at its northern limit (Baker 1997, 2000, 2003) and fire was implicated in many of these local extinctions. The species is currently found in a small number of locations (local populations) within three disjunct regions (Fig. 1). The southern population occurs in the Nadgee and Croajingalong wilderness areas (near the Victorian and New South Wales state border) and is in the order of 250 birds (Bramwell 2008; J. Baker unpubl. data; M. Bramwell unpubl. data). There are approximately 300 km separating the southern and central-region populations, which occur near Barren Grounds Nature Reserve and Jervis Bay (central NSW) and each supports in the order of 1,000 birds (J. Baker unpubl. data). These two central-region locations are neighboring populations, separated by only 50 km. Both of these locations have a four-decade recent history of relatively small-area, low intensity fires, but with long periods between fires, over much of the available habitat. The northern population lies approximately 700 km further north and is spread sparsely through the ranges near the NSW and Queensland state border and numbers less than 50 birds (D. Rohweder and D. Stewart pers. comm. 2010). The breeding biology and dispersal ability of D. brachypterus suggests a very limited capacity for population growth or expansion, and natural founder events to areas outside the current range of occupancy seem improbable (Baker 1997, 2000, 2001).
Suitable areas of Eastern Bristlebird (Dasyornis brachypterus) habitat and the current distribution of birds, south east Australia. Source Birds Australia (2010)
Population sampling and genetic markers
Using existing collections of DNA, pin feathers or blood samples (for DNA extraction) we compiled nuclear microsatellite genotype data (using loci: Db03; Db07; Db09; Db13; Db16 & Db17), and mitochondrial DNA sequence data (870 base pairs [bp] of the ATPase 8/6 genes using primers CoMT7828L & CoMT8720H, Elphinstone 2008), for a total of 118 and 106 birds. Overall, we genotyped, and sequenced (no. sequenced per location), 58 (48), and 47 (32) birds from Jervis Bay (JB) and Barren Grounds (BG) in the centre of D. brachypterus’s range, and 7 (13) and 6 (5) birds from the southern and northern population, respectively (Table 1 online supplementary material). Information concerning the DNA extraction protocol, microsatellite and mtDNA markers and the PCR amplification conditions can be found in Perrin and Roberts (2010) and Elphinstone (2008).
Genetic analyses
Mitochondrial DNA
The sequencing reactions were performed at Macrogen Inc. (Korea) using a Big DyeTM terminator cycle sequencing kit (Applied Biosystems). Eight hundred and seventy bp of sequence were aligned using Clustal W (Thompson et al. 1994) implemented in the software Bioedit (Hall 1999) and checked manually. We constructed a medium joining network (MJN) of the D. brachypterus mtDNA haplotypes allowing a visual representation of the pattern of molecular evolution of haplotypes, and their spatial distribution and frequencies. Haplotypes were joined using a 95% confidence criterion to form the most parsimonious network or gene tree. The analysis was performed using Network 4.2.0.1(Bandelt et al. 1999). The gene tree was compared and checked for consistency with the topology of a phylogenetic tree produced using a Bayesian analysis (in the program Mr Bayes, Huelsenbeck and Ronquist 2001). The reconstruction of the mtDNA phylogeny was based on a Generalised Time-Reversible (GTR) model of sequence evolution. The GTR model was chosen from 24 different models of sequence evolution, based on the results of hierarchical likelihood ratio tests used to evaluate the most likely mode of sequence evolution, given the observed pattern of variation in the mtDNA sequence (MrModeltest, Nylander 2004). We used default priors for the analysis in Mr Bayes, set four chains of the MCMC each for 105 generations, and sampled phylogenetic trees every 100 generations. The first 25% of trees were discarded as ‘burn in’ before calculating a consensus phylogenetic tree. To ensure that independent runs converged on a similar topology, we conducted three separate analyses. For all runs, the final average standard deviations of split frequencies were below 0.01, and scale reduction factors were close to unity for all parameters, indicating that the runs had converged. We also generated DNA sequence data for two individuals of the congener, Dasyornis broadbenti (Rufous Bristlebird). These sequences were included in the analyses to root the phylogenetic tree.
For each location, we estimated haplotype (h) and nucleotide diversity (π), and the number of pairwise sequence differences. Genetic differentiation between central-region populations (i.e., JB & BG) was estimated using F ST and Θ ST, with the level of statistical significance determined using a re-sampling approach based on 103 permutations. The analyses were based on haplotype frequencies (F ST), and on both haplotype frequencies and sequence differences (Θ ST), and were performed in Arlequin (Excoffier et al. 2005). Small sample sizes precluded the inclusion of specimens from the southern and northern population in the analyses outlined above.
Microsatellites
Here we used a subset of six of the available ten microsatellite markers for this species because genotyping a small number (n = 10) of progeny (including re-extraction and re-genotyping) produced from a single mating between a pair of birds held in captivity revealed non-Mendelian inheritance due to the presence of null alleles (Table 2 online supplementary material). PCR products were visualised with an ABI 3130 automated capillary sequencer. Assignment of allele size (bp) was achieved with reference to LIZ size standard present in each lane and scoring was performed using GeneMapper software (Applied Biosystems).
We visualised the level of genetic differentiation among D. brachypterus genotypes using a Factorial Correspondence Analysis (FCA). We performed the FCA on the matrix of allele counts with the procedure “AFC” on individuals in GENETIX 4.05 (Belkhir et al. 2002). We used an assignment test approach (implemented in the program STRUCTURE, Pritchard et al. 2000; Falush et al. 2003) to estimate the number of discrete genetic clusters, for birds sampled from across D. brachypterus’s distributional range. Under the assumptions of linkage and Hardy–Weinberg equilibrium, STRUCTURE uses observed allele frequency data (in a Bayesian framework) to estimate the likelihood of K genetic clusters or populations (where K = 1, 2, 3…n), while simultaneously estimating the ancestry of individuals i.e., the proportional membership of each individual’s genotype in one or several inferred genetic clusters. We used the admixture model to calculate individual q-values, (qi the mean posterior proportion of ancestry ±95% CI’s), as well as the log probability or likelihood of K genetic clusters, for values of between 1 and 6. An uninformative prior was used so that information concerning the sampling location of each individual was not included in the analysis. An initial burn-in period of 104 iterations preceded data collection, which was for 105 iterations, and there were 20 replicate runs for each value of K. Default values were used for all other parameters. The results were consistent across runs and the number of clusters detectable by STRUCTURE (i.e., K) for which the likelihood value was highest was taken as the number of distinct genetic clusters.
For each location, we calculated standard population genetic parameters, including the average, and standardised, number of alleles per locus, and the average observed and expected heterozygosity (using POPGENE, GenAlEx, Yeh et al. 1999; Belkhir et al. 2002; Peakall and Smouse 2006). Where sample sizes were large enough (JB & BG), we performed a linkage disequilibrium analysis (using exact tests in GENEPOP, Raymond and Rousset 1995) to determine if each locus was independent. Three pairwise tests between loci were statistically significant (P < 0.05), however, the loci in disequilibria were different at JB and BG suggesting that loci are not physically linked but rather, such non-random association of alleles reflects some aspect of the population history of each location. We proceeded to use each locus as an independent test of the extent of genetic diversity and differentiation. We assessed population subdivision using Weir and Cockerham’s (1984) estimation of Wright’s (1969) F-statistics. The analysis was performed in FSTAT (Goudet et al. 1996), with estimates based on microsatellite allele frequencies for individual loci, and as an average across loci. Bootstrapping and jackknifing over loci were used to estimate standard deviations and 95% CIs. F-statistics were judged to be statistically significant when the lower 95% confidence interval did not overlap zero. To infer the past mating system and assess inbreeding in D. brachypterus we used estimates of f, the inbreeding coefficient, F IS, and tested the statistical significance of heterozygous deficits (or excesses) using exact test implemented in GENEPOP (Raymond and Rousset 1995).
For selectively neutral loci, the number of alleles per locus and their frequency within a natural population reflect equilibrium between the creation and loss of alleles through mutation and genetic drift. The creation and loss of alleles respectively depend on the locus-specific mutation rate and the genetically effective population size. Reduction to the absolute size of a population (i.e., a demographic bottleneck) is generally predicted to have a corresponding effect on the genetically effective population size (Ne), resulting in a loss of alleles and heterozygosity. However, the loss of alleles has been shown to occur more rapidly than the loss of heterozygosity (Nei et al. 1975). Thus, if a sample from a natural population displays excess heterozygosity (across a majority of loci), relative to the heterozygosity predicted to occur within a sample that has the same size and same allele number, and where each locus is assumed to be in mutation-drift equilibrium, this may indicate that the population has experienced a recent reduction to its genetically effective population size or genetic bottleneck (Cornuet and Luikart 1996). We tested for a evidence of a recent genetic bottleneck at JB and at BG using the program BOTTLENECK (Piry et al. 1999). Although allelic variation at microsatellite loci is thought to form via step-wise mutations (SMM) of the microsatellite repeat motif, we know that microsatellite loci rarely evolve under a strict SMM (see Ellegren 2004 for review). We therefore initially conducted the analysis based on a SMM model, but then employed additional analyses using an infinite alleles model (IAM) and a two-phased mutation model (TPM) incorporating different proportions of single- and multiple-step mutations (90: 10 and 95: 5, with a variance among multiple-step mutations of 12). We used the Wilcoxon’s test (as recommended by the authors of the program in situations where <20 loci are typed), and rejected the null hypothesis of no excess heterozygosity when the P-value was <0.05.
Results
The genealogical relationship among mtDNA haplotypes, and their spatial distribution
The genealogy resolved by the MJN and Bayesian tree was complex with two clades of haplotypes; however, the distribution of haplotypes within clades was not structured by geography i.e., sampling location, and the clades were not well supported by posterior probability values (0.62 & 0.55 respectively). The mtDNAs of D. brachypterus from the northern range-edge and central populations were polyphyletic with respect to each other. The two haplotypes detected in the southern population were monophyletic with respect to each other; nevertheless, these haplotypes were more closely related to central-region haplotypes, than to haplotypes detected at the northern range-edge. Genetic distances among D. brachypterus haplotypes were shallow, relative to the distance between haplotypes of D. brachypterus and the congener, D. broadbenti. Nearly every mtDNA haplotype existed as a private haplotype, restricted to a single population; the exceptions were Db 1& Db 5, which although were geographically restricted to the central-region, were found in both populations (i.e., JB & BG). Both the MJN and Bayesian phylogenetic tree agreed (Figs. 2, 3).
A phylogram of Dasyornis brachypterus mtDNA haplotypes (Db1–Db13) (based on 870 bps of ATPase 8/6 genes). The numbers at the nodes of the consensus tree represent posterior probability values or the proportion of trees that converged on the same topology. Spatial distribution of haplotypes: open light grey circles, Jervis Bay; open black circles, Barren Grounds; southern population, open black squares; northern population, open black triangles. Corresponding sequences of the congener, Dasyornis broadbenti (Dbr 1 & Dbr 2), were included in the analysis as an outgroup taxon
Medium joining network among haplotypes (Db1–Db13) of Dasyornis brachypterus. Each circle represents a different haplotype with sizes proportional to the frequency of occurrence of the haplotype in the total sample. Shading indicates the relative frequency of each haplotype at Jervis Bay (light grey) and Barren Grounds (black) i.e., central-region populations, and within the southern population (white) and northern population (dark grey). The pattern of molecular evolution of haplotypes is represented by the connections among haplotypes formed with the solid grey lines. The black numbers in parentheses represent the number of mutational steps between haplotypes (displayed for situations where the no. mutations > 1). Inferred mutational steps (i.e., a missing haplotype, either extant, but unsampled, or extinct) are represented in the figure by an open square
Broad-scale characterisation of population subdivision based on microsatellites markers
A plot of mean ancestry (qi) representing the proportional membership of each individual’s genotype to each of four clusters (identified by STRUCTURE) revealed that D. brachypterus from JB and BG (i.e., central-region) formed relatively discrete populations. Nevertheless, there were a number of birds sampled at JB, which shared their ancestry with birds from BG (i.e., individual ancestry coefficients indicated mixed ancestry). This pattern of mixed ancestry was also true of birds sampled at BG. Interestingly, birds from JB and BG shared a large proportion of their ancestry with D. brachypterus from the southern population. Southern and northern populations similarly formed discrete genetic clusters, indicating that all populations are highly genetically distinct (Fig. 4). Factorial Correspondence Analysis (FCA) revealed that the distribution of genotypes was consistent with the genetic clusters identified by STRUCTURE, but in addition, this analysis revealed that the northern population was highly genotypically diverse, indicated by a wide scatter of points (black triangles representing individual birds) along the x-axis of the FCA (Fig. 5).
Ancestry or admixture coefficients for all 118 Dasyornis brachypterus from the central (C), southern (S) and northern (N) regions. Each bar represents an individual’s genotype; the light-grey, black, white and dark-grey shading of each bar represents the relative make-up or the inferred proportion of membership of an individual’s genotype, to each of four detectable genetic clusters. The fine vertical black lines form the bounds of the original sampling locations: central-region, C: divided into two populations, Jervis Bay (JB) (light grey) and Barren Grounds (BG) (black); S (white), southern population; N (dark grey), northern population
Factorial correspondence analysis based on the six-locus microsatellite genotype of all 118 Dasyornis brachypterus. The distribution of genotypes defined by the first three factorial axes of the analysis is presented. Spatial distribution of genotypes: Jervis Bay, open light-grey circles; Barren Grounds, open black circles; southern population, open black squares; northern population, open black triangles
Central-region Dasyornis brachypterus: genetic diversity, population differentiation, inbreeding, and tests for genetic bottlenecks based on microsatellite DNA markers
The overall sample of central-region Dasyornis brachypterus (JB & BG) comprised a genetically diverse group with evidence of moderate genetic subdivision. The number of alleles per locus ranged between 7 and 11, and the mean (±SE) was 10.5 (±0.8). We detected comparable levels of genetic variation at each location. The mean number of alleles per locus ranged between 7.8 (±0.9) and 8.6 (±0.8), while the mean observed heterozygosity was ≥0.75 for each location. Across all six loci, we detected 63 alleles, with between 74.6 and 82.5% of alleles present at each location. Private alleles (alleles unique to a single location) accounted for 42.8% of the total number of alleles detected (27/63), with private alleles generally distributed evenly between locations (JB = 11; BG = 16) (Table 1). The majority of private alleles were found at low frequency (freq. < 0.05) (see online supplementary material, Table 3).
The overall estimate of F ST (based on microsatellite allele frequency data) implied moderate, statistically significant genetic differentiation for the set of central populations (mean ± SE = 0.058 ± 0.012, lower 95% CI = 0.038). Single-locus estimates ranged from 0.024 for Db07 to 0.091 for Db03, nevertheless 5 of 6 loci showed evidence of moderate differentiation (F ST > 0.04) (Table 2).
Estimates of f, the inbreeding coefficient, F IS, used to infer the past mating system of each central population of D. brachypterus, were variable among loci and overall. For D. brachypterus from BG, a negative mean value of f indicates excesses of heterozygotes and a test for departure from Hardy–Weinberg equilibrium was significant (P < 0.05). In contrast, a slight deficit of heterozygotes (mean f ± SE = 0.010 ± 0.026) suggests inbreeding at JB, nevertheless, a test for departure from Hardy–Weinberg equilibrium was not significant (P > 0.05).
For the set of birds from each central population we examined whether a majority of the microsatellite loci displayed excess heterozygosity (relative to the expectation of mutation-drift equilibrium), an indication of a genetic bottleneck. While the results were comparable for the two locations, the analysis was clearly sensitive to the underlying mutation mechanism. Indeed, there was no indication of significant excess heterozygosity at JB or BG based on a SMM or TPM (P > 0.05), whereas the results were highly significant in both locations based on an IAM (P < 0.01), implying the possibility of a recent genetic bottleneck.
Central-region Dasyornis brachypterus: population differentiation based on mtDNA
Examination of mtDNA sequence variation revealed that central-region D. brachypterus (i.e., JB & BG) generally lack mtDNA haplotypic diversity but are genetically subdivided. Gene diversity (h) was 0.61 and 0.78 for JB and BG respectively, while π was ≤0.006 (Table 3). As for estimates of genetic subdivision based on microsatellite markers, estimates of population differentiation revealed moderate to high statistically significant genetic differentiation between these neighboring populations (F ST = 0.124, P < 0.05; Θ ST = 0.076, P < 0.05).
Comparison of mitochondrial and microsatellite genetic diversity between central, and southern and northern populations
The northern population possessed similar levels of mitochondrial haplotype (h) and nucleotide diversity (π), relative to each central population, but the southern population was clearly less diverse (Table 3). There were fewer microsatellite alleles detected per locus in southern (mean standardised allelic richness, A R ± SD = 4.0 ± 0.8) and northern (4.0 ± 0.4), than in each central population (JB 4.9 ± 0.6; BG 5.2 ± 0.3). Overall, 38 and 32% of the total number of alleles detected in the central region (n = 63) were present in the southern and northern populations, respectively. However, private alleles were detected in each of these areas, with one and four alleles respectively detected in the southern and northern populations that were not present in either population within the central-region. Expected heterozygosity was similarly lower in both northern and southern populations, with mean (±SD) values of 0.537 ± 0.010 and 0.616 ± 0.038 outside the range of values obtained for similarly sized samples from JB (0.680–0.745) and BG (0.730–0.736).
Discussion
The results of our phylogenetic survey of mtDNA sequence variation revealed two shallow clades, but the distribution of haplotypes within each clade was not geographically structured. Therefore, our data do not support the designation by Schodde and Mason (1999) of northern D. brachypterus, subspecies monoides, and central and southern D. brachypterus, subspecies brachypterus. Indeed, as in Elphinstone’s (2008) study, our data revealed a clear lack of phylogenetic distinctness or monophyly, which Moritz (1994) advocated was needed to support such a distinction. While Phillimore and Owens (2006) have shown that 36% of cases of avian sub-specific taxonomy are justified based on corresponding monophyly, our findings add to a growing number of recent studies of Australian passerines that have uncovered discordance between traditional and molecular taxonomy (Joseph et al. 2006; Lee and Edwards 2008; Donnellan et al. 2009). For example, Lee and Edwards (2008) revealed incorrect subspecies boundaries in the Red-backed Fairy-wren (Malurus melanocephalus complex), while Donellan et al. (2009) found that the boundaries of eight subspecies of the Southern Emu-wren (Stipiturus malachurus) were not concordant with the pattern of molecular evolution of haplotypes, and their spatial distribution.
Despite the potential for D. brachypterus to form genetically depauperate, inbred populations suggested by small population sizes and the species’ highly fragmented distribution, our analysis of microsatellite data show that the two largest groups (i.e., populations at JB & BG) of this endangered species form randomly mating, genotypically diverse, and genetically differentiated populations. The persistence of these populations and the maintenance of their genetic diversity indicate a capacity to recover from small population size following fire. Set against this positive, our survey of microsatellite variation suggests the possibility that both central populations have been affected by a recent genetic bottleneck, although the apparent excess heterozygosity (relative to the expectation of mutation-drift equilibrium) may be explained by some aspect of the mating system of D. brachypterus, particularly if individuals display positive assortative mating (e.g., Bitton et al. 2008) or if heterozygotes have a selective advantage (Hansson et al. 2001; MacDougall-Shackleton et al. 2005; Fossoy et al. 2009). Nevertheless, D. brachypterus is fire sensitive, and fire-history is thought to be a major determinant of local population size (Baker 2000). Although JB and BG have a recent history free from catastrophic fire, this was not always the case. For instance, all but 4 ha of Barren Grounds Nature Reserve were burned in a wildfire in 1968 (Baker 1998 and references therein). Habitat refugia seem likely to have ensured both the persistence and continuing reproduction (Bain et al. 2008; Lindenmayer et al. 2009), and hence recovery of population size thereby limiting the erosion of genetic diversity. Indeed, standing genetic variation within these populations is within the range reported for or greater than, a number of similarly threatened, sedentary or habitat specialist birds, with comparable life histories (Hudson et al. 2000; Brown et al. 2004; Boessenkool et al. 2007).
The distribution of nuclear allelic and mtDNA haplotypic variation for the set of central populations (JB & BG) indicate that D. brachypterus essentially forms two isolated, self-seeding populations, which nevertheless have been interconnected by past gene flow. Although each of these populations may now be more isolated by anthropogenic related habitat fragmentation, it seems probable that these largely neighboring populations would historically have been more strongly interconnected, even though the populations may have always been restricted to discrete areas of suitable habitat. In partial contrast, while all populations surveyed share microsatellite alleles, the distribution of mtDNA haplotypic variation provides compelling evidence for long-term restriction of dispersal and gene flow, particularly among populations in different regions. Indeed, even in the absence of current or recent gene flow among regions, shared polymorphism at nuclear microsatellite loci is, as we would predict, given nuclear DNA has a four-fold greater effective population size than mtDNA and so is less susceptible to the effects of genetic drift (Frankham et al. 2010). It therefore seems probable that shared microsatellite polymorphism among regions reflects recent common ancestry (i.e., retention of ancestral polymorphism), not recent gene flow, with all populations now effectively isolated and closed to migration.
Our estimates of genetic diversity for D. brachypterus from the extreme northern and southern range-edge of the species distribution demonstrate that these peripheral populations are less diverse (measured in terms of the average number of microsatellite alleles per locus and heterozygosity) than the larger central populations, although this may be an artifact of small sample sizes. However, although northern and southern populations of D. brachypterus retain only a small proportion of the microsatellite variation present within central populations, they nevertheless contain both nuclear allelic and genotypic, as well as haplotypic (mtDNA), variation not detected in the central populations. Thus, active conservation will be needed specific to each region, to mitigate further loss of genetic variation, including the potential loss of unique variation, particularly from the critically small southern and northern populations.
The maintenance of evolutionary potential via the protection of a species’ genetic diversity (i.e., genetic variation within and among populations) is a core objective of biodiversity conservation strategies worldwide (e.g., Commonwealth of Australia 1996). Based on our findings, we recommend that the managers of D. brachypterus should aim to conserve the genetic integrity of all four remnant populations. While genetic divergence at neutral marker loci is generally only weakly correlated with divergence at quantitative trait loci affecting fitness and thus underlying adaptation (i.e., quantitative variation) (reviewed by Reed and Frankham 2001; Willi et al. 2006; Goudet and Martin 2007), our data for D. brachypterus reveals broad-scale genetic subdivision (i.e., among regions), but in addition, moderate genetic differentiation for neighboring populations separated by approximately 50 km. This implies prolonged evolution of lineages in separate locations and the possibility that birds are locally adapted. Thus, we caution against mixing birds from different populations or regions, should captive breeding, supplementation or re-introduction be further used as a management tool, to avoid the potentially deleterious effects associated with outbreeding depression. If captive breeding continues to be used to recover the northern population, pedigrees should be tracked and the fitness offspring assessed prior to release to the wild. However, a strategy that aimed at both conserving D. brachypterus’s genetic diversity and buffering existing local populations against future environmental or demographic catastrophes would ideally involve seeding a number of additional areas (of suitable habitat) via translocation of wild-caught birds from local populations. Ideally, this would be combined with demographic surveys and monitoring of allele frequencies to assess: (i) levels of inbreeding, since the establishment of new populations will likely rely on a limited number of founders; and (ii) the parentage of offspring, to track the fate (or fitness) of subsequent generations.
References
Bain DW, Baker J, French KO, Whelan RJ (2008) Post-fire recovery of eastern Bristlebirds (Dasyornis brachypterus) is context-dependent. Wildl Res 35:44–49
Baker J (1997) The decline, response to fire, status and management of the eastern Bristlebird. Pac Conserv Biol 3:235–243
Baker J (1998) Ecotones and fire and the conservation of the endangered Eastern Bristlebird. PhD dissertation, University of Wollongong, Wollongong, Australia
Baker J (2000) The Eastern Bristlebird: cover-dependent and fire-sensitive. Emu 100:286–298
Baker J (2001) Population density and home range estimates for the eastern Bristlebird at Jervis Bay, southeastern Australia. Corella 25:62–67
Baker J (2003) Fire-sensitive birds and adaptive management. In: Bushfires: managing the risks. Proceedings of the 2002 NCC Bushfire Conference. Nature Conservation Council of NSW, Sydney, pp 59–65
Bandelt HJ, Forster P, Rohl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2002) GENETIX 4.03, Logiciel Sous WindowsTM Pour la Genetique Des Populations., Laboratoire Genome, Populations, Interactions: CNRS UMR:5000. Universite Montpellier 2, Montpellier, France
Birds Australia (2010) Birdata website. www.birdata.com.au/iba.vm. Accessed May 2010
Bitton PP, Dawson RD, Ochs CL (2008) Plumage characteristics, reproductive investment and assortative mating in tree swallows Tachycineta bicolor. Behav Ecol Sociobiol 62:1543–1550
Bodkin JL, Ballachey BE, Cronin MA, Scribner KT (1999) Population demographics and genetic diversity in remnant and translocated populations of sea otters. Conserv Biol 13:1378–1385
Boessenkool S, Taylor SS, Tepolt CK, Komdeur J, Jamieson IG (2007) Large mainland populations of South Island robins retain greater genetic diversity than offshore island refuges. Conserv Genet 8:705–714
Bramwell MD (2008) The Eastern Bristlebird Dasyornis brachypterus in East Gippsland, Victoria, 1997–2002. Austral Field Ornithol 25:2–11
Brown LM, Ramey RR, Tamburini B, Gavin TA (2004) Population structure and mitochondrial DNA variation in sedentary Neotropical birds isolated by forest fragmentation. Conserv Genet 5:743–757
Castellon TD, Sieving KE (2006) An experimental test of matrix permeability and corridor use by an endemic understory bird. Conserv Biol 20:135–145
Commonwealth of Australia (1996) National strategy for the conservation of Australia’s biological diversity. Commonwealth of Australia, Canberra, Australia
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Donnellan SC, Armstrong J, Pickett M, Milne T, Baulderstone J, Hollfelder T, Bertozzi T (2009) Systematic and conservation implications of mitochondrial DNA diversity in emu-wrens, Stipiturus (Aves:Maluridae). Emu 109:143–152
Edmands S (2007) Between a rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16:463–475
Ellegren H (2004) Microsatellites: simple sequences with complex evolution. Nat Rev Genetics 5:435–445
Elphinstone MS (2008) The development and application of molecular methods for the analysis of habitat and population fragmentation in birds. PhD dissertation, Southern Cross University
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform 1:47–50
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fossoy F, Johnsen A, Lifjeld JT (2009) Cell-mediated immunity and multi-locus heterozygosity in bluethroat nestlings. J Evol Biol 22:1954–1960
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Frankham R, Ballou JD, Briscoe DA (2010) Introduction to conservation genetics. Cambridge University Press, New York
Goudet J, Martin G (2007) Under neutrality, Q ST ≤ F ST when there is dominance in an island model. Genetics 176:1371–1374
Goudet J, Raymond M, de Meeus T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Hale KA, Briskie JV (2007) Decreased immunocompetence in a severely bottlenecked population of an endemic New Zealand bird. Anim Conserv 10:2–10
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In: Nucleic Acids Symposium Series, 41, pp 95–98
Hansson B, Bensch S, Hasselquist D, Akesson M (2001) Microsatellite diversity predicts recruitment of sibling great reed warblers. In: Proceedings of the Royal Society of London Series B-Biological Sciences, 268, pp 1287–1291
Hudson QJ, Wilkins RJ, Waas JR, Hogg ID (2000) Low genetic variability in small populations of New Zealand kokako Callaeas cinerea wilsoni. Biol Conserv 96:105–112
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Ingvarsson PK (2001) Restoration of genetic variation lost—the genetic rescue hypothesis. Trends Ecol Evol 16:62–63
IUCN (2010) IUCN Red List of Threatened Species. Version 2010.1. Available from URL: http://www.iucnredlist.org
Joseph L, Wilke T, Ten Have J, Chesser RT (2006) Implications of mitochondrial DNA polyphyly in two ecologically undifferentiated but morphologically distinct migratory birds, the masked and white-browed woodswallows Artamus spp. of inland Australia. J Avian Biol 37:625–636
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Keller LF, Arcese P, Smith JNM, Hochachka WM, Stearns SC (1994) Selection against inbred song sparrows during a natural population bottleneck. Nature 372:356–357
Larson S, Jameson R, Bodkin J, Staedler M, Bentzen P (2002) Microsatellite DNA and mitochondrial DNA variation in remnant and translocated sea otter (Enhydra lutris) populations. J Mammal 83:893–906
Laurance SGW, Gomez MS (2005) Clearing width and movements of understory rainforest birds. Biotropica 37:149–152
Laurance WF, Lovejoy TE, Vasconcelos HL, Bruna EM, Didham RK, Stouffer PC, Gascon C, Bierregaard RO, Laurance SG, Sampaio E (2002) Ecosystem decay of Amazonian forest fragments: a 22-year investigation. Conserv Biol 16:605–618
Laurance SGW, Stouffer PC, Laurance WE (2004) Effects of road clearings on movement patterns of understory rainforest birds in central Amazonia. Conserv Biol 18:1099–1109
Leberg PL, Firmin BD (2008) Role of inbreeding depression and purging in captive breeding and restoration programmes. Mol Ecol 17:334–343
Lee JY, Edwards SV (2008) Divergence across Australia’s Carpentarian Barrier: Statistical Phylogeography of the Red-Backed Fairy Wren (Malurus melanocephalus). Evolution 62:3117–3134
Lindenmayer DB, MacGregor C, Wood JT, Cunningham RB, Crane M, Michael D, Montague-Drake R, Brown D, Fortescue M, Dexter N, Hudson M, Gill AM (2009) What factors influence rapid post-fire site re-occupancy? A case study of the endangered Eastern Bristlebird in eastern Australia. Int J Wildland Fire 18:84–95
MacDougall-Shackleton EA, Derryberry EP, Foufopoulos J, Dobson AP, Hahn TP (2005) Parasite-mediated heterozygote advantage in an outbred songbird population. Biol Lett 1:105–107
Moritz C (1994) Defining evolutionarily-significant-units for conservation. Trends Ecol Evol 9:373–375
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in natural populations. Evolution 29:1–10
Nylander JAA (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Perrin C, Roberts DG (2010) Ten polymorphic microsatellite markers for the endangered Australian Eastern Bristlebird Dasyornis brachypterus. Conserv Genetic Resour 2:357–359
Phillimore AB, Owens PF (2006) Are subspecies useful in evolutionary and conservation biology? Proc R Soc B Biol Sci, 273:1049–1053
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ralls K, Ballou JD, Rideout BA, Frankham R (2000) Genetic management of chondrodystrophy in California condors. Anim Conserv 3:145–153
Raymond M, Rousset F (1995) GENEPOP version 3.3: a population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Reding DM, Freed LA, Cann RL, Fleischer RC (2010) Spatial and temporal patterns of genetic diversity in an endangered Hawaiian honeycreeper, the Hawaii Akepa (Loxops coccineus coccineus). Conserv Genet 11:225–240
Reed DH, Frankham R (2001) How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55:1095–1103
Roberts DG, Ayre DJ, Whelan RJ (2007) Urban plants as genetic reservoirs or threats to the integrity of bushland plant populations. Conserv Biol 21:842–852
Rodriguez A, Andren H, Jansson G (2001) Habitat-mediated predation risk and decision making of small birds at forest edges. Oikos 95:383–396
Schodde R, Mason IJ (1999) The directory of Australian birds. Passerines. CSIRO Publishing, Melbourne
Smith GT (1977) The effects of environmental change on six rare birds. Emu 77:173–179
Stouffer PC, Bierregaard RO, Strong C, Lovejoy TE (2006) Long-term landscape change and bird abundance in Amazonian rainforest fragments. Conserv Biol 20:1212–1223
Taylor SS, Jamieson IG, Wallis GP (2007) Historic and contemporary levels of genetic variation in two New Zealand passerines with different histories of decline. J Evol Biol 20:2035–2047
Templeton AR (1986) Coadaptation and outbreeding depression. In: Soulé ME (ed) Conservation biology: the science of scarcity, diversity. Sinauer Associates, Sunderland, MA, pp 105–116
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Uezu A, Metzger JP, Vielliard JME (2005) Effects of structural and functional connectivity and patch size on the abundance of seven Atlantic Forest bird species. Biol Conserv 123:507–519
Watson JEM, Whittaker RJ, Freudenberger D (2005) Bird community responses to habitat fragmentation: how consistent are they across landscapes? J Biogeogr 32:1353–1370
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Willi Y, Van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37:433–458
Wolf CM, Griffith B, Reed C, Temple SA (1996) Avian and mammalian translocations: update and reanalysis of 1987 survey data. Conserv Biol 10:1142–1154
Wright S (1969) Evolution and the genetics of populations. The theory of gene frequencies, vol 2. University of Chicago Press, Chicago
Yeh FC, Yang R, Boyle T (1999) POPGENE version 1.31: Microsoft Window-based freeware for population genetic analysis. http://www.ualberta.ca/~fyeh/
Acknowledgments
The authors are indebted to the following people who provided D. brachypterus and D. broadbenti samples: D. Bain, University of Wollongong; R. Booth & David Fleay, Wildlife Park; M. Bramwell, Victorian Department of Sustainability and Environment; M. Elphinstone, Southern Cross University; D. Oliver, Department of Environment and Climate Change; J. Sumner, Museum Victoria; R Palmer, CSIRO Sustainable Ecosystems. We thank M. Bramwell, D. Oliver, A. Chalklen, P. Latch & the Eastern Bristlebird Recovery Team for constructive discussions. We also thank M. Bramwell, D. Rohweder and D. Stewart for personal communications on recent population data and D. Bain and M. Elphinstone for technical assistance. This work was funded by the Australian Department of Environment, Water, Heritage and the Arts and the Victorian Department of Sustainability and Environment. This is contribution number 299 of the Ecological Genetics Group at the University of Wollongong.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roberts, D.G., Baker, J. & Perrin, C. Population genetic structure of the endangered Eastern Bristlebird, Dasyornis brachypterus; implications for conservation. Conserv Genet 12, 1075–1085 (2011). https://doi.org/10.1007/s10592-011-0210-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-011-0210-4