Abstract
In order to establish a strategy for conservation, the distribution of genetic diversity in four natural populations of Ocotea catharinensis in the Brazilian Atlantic rain forest was investigated using 18 allozyme loci. Estimates of the average number of alleles per loci (2.2), percentage of polymorphic loci (83.3%) and expected genetic diversity (0.427) in adult individuals were high; suggesting that all populations have genetic potential for conservation. The inbreeding within populations (\( \hat{f} = - 0.0 1 1 \)) and the total inbreeding (\( \hat{F} = 0. 1 3 3 \)) suggest population structure, since a high level of divergence among populations (\( \hat{\theta }_{\text{P}} = 0. 1 4 3 \)) was also detected. Significant values of spatial genetic structure were found inside the four populations. This study demonstrates that the realized gene flow among the remaining populations of O. catharinensis is not sufficient to stop population divergence due to genetic drift and local selection, which threatens the future viability of this species.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Forest fragments are small and separate remnants of reduced continuous forest areas, which probably have limited ecosystem functionality compared to the original forest (Young and Boyle 2000). The first consequence of forest fragmentation is demographic (Lande 1993; Hanski and Ovaskainen 2000; Jaquiéry et al. 2008), but fragmentation has also genetic consequences, which contributes to risks of extinction (Frankham 1995; Lande 1995; Reed 2005). Forest fragmentation has shown to produce an immediate loss of alleles that is associated with a reduction in population size (White et al. 1999; Cascante et al. 2002). Theoretical studies (Barrett and Kohn 1991) predict that severe fluctuations in the number of individuals or continually small population sizes result in genetic drift, which in turn leads to the fixation of alleles and consequently to a loss of genetic variation. Another consequence of small population size is susceptibility to inbreeding, which reduces heterozygosity and the performance of various fitness-related traits and thereby substantially increases the probability of extinction (Reed and Frankham 2003; Matthies et al. 2004; Reed 2005).
Moreover, forest fragmentation may disrupt the mating system and modify gene flow, resulting in significantly elevated levels of inbreeding, population divergence and a reduction in genetic diversity within populations (Hanson et al. 2008). Inbreeding is a concern to outcrossing populations where highly deleterious mutations and polymorphisms for overdominant alleles may contribute to inbreeding depression (Cheptou and Schoen 2007). However, the negative effects predicted in the genetic theory of small populations may be absent in forest fragments if they are experiencing an effective seed and pollen dispersal over long distances (Hamrick 2004; Kramer et al. 2008).
The study of genetic structure and diversity permits knowledge of the organization and distribution of the genetic variability among and within natural populations (Hamrick et al. 1992; Young et al. 1996; Dick et al. 2008). These understandings have helped to provide important assumptions regarding conservation and the elaboration of management strategies (Bawa 1992; Álvarez-Buylla et al. 1996; Reis et al. 2000; Sebbenn et al. 2008). In addition, the knowledge of genetic variation in conservation strategies secures populations a high adaptive potential to withstand environmental stress such as climatic change, pollution and diseases (Geburek 2000).
Studies have established the fact that most of the genetic variation is distributed within populations (Auler et al. 2002; Gaiotto et al. 2003; Conte et al. 2008), and in many cases the distribution within population is not random, due to the limited dispersal of pollen and seeds or selection pressures or historical events (Heywood 1991; Vekemans and Hardy 2004; Hardy et al. 2006; Dick 2008). This therefore promotes spatial genetic structure (SGS).
Many studies on plants have been devoted to characterizing SGS (Heywood 1991; Epperson et al. 1999; Mantovani et al. 2006; Hardy et al. 2006; Born et al. 2008), because the presence of SGS within populations has important implications in conservation programs as it can affect estimates of genetic parameters (Young and Boyle 2000; Jones and Hubbell 2006), such as the outcrossing rate in natural populations (Ennos and Clegg 1982; Bittencourt and Sebbenn 2007), influencing the sampling strategies for improvement of conservation projects ex situ and it aids management of populations, where exploitation patterns can affect the levels of genetic diversity (Doligez and Joly 1997; Sebbenn et al. 2008).
The species Ocotea catharinensis Mez. (Lauraceae) is a native tree of the Atlantic rain forest (ARF), with natural occurrence in the south and southeast of Brazil. O. catharinensis was the most common and typical tree in the ARF of Santa Catarina State in southern Brazil. It accounted for a third of all tree volume in a hectare (Reitz et al. 1978). Because of its economic value due to its high quality wood and essential oils, this species has intensively been exploited since 1940 (Carvalho 1994). Intensive exploitation along with expansion of agricultural frontiers have caused forest fragmentation and losses of the numbers of individuals and populations. The remaining ARF fragments in Santa Catarina State that may house O. catharinensis populations represent 14.46% (13,693.34 km2) of its original area (IESB 2007). Today, O. catharinensis is threatened of extinction in its natural habitat and also appears in the International Union of Conservation of Nature and Natural Resources Red List of endangered species as vulnerable to extinction (Varty and Guadagnin 1999).
To conserve natural populations of O. catharinensis in the ARF more effectively, demographic and genetic information about this species needs to be obtained. However, there are no studies yet that evaluate the demography, genetic diversity and fine-scale spatial genetic structure in natural populations of O. catharinensis. Thus, the objective of this study was to analyze the demography, genetic diversity and structure and intrapopulation spatial genetic structure of four natural populations of O. catharinensis using allozymic markers. Since we studied two populations inside conservation units in continuous forest and other two inside small, isolated and fragmented forests in private proprieties, the following questions were raised: (1) Do the populations in small, isolated and fragmented forests have equivalent levels of genetic diversity as the ones in the continuous forest? (2) How is it possible to enhance reproductive success and survival of O. catharinensis in the remaining forest fragments? (3) Since seeds of O. catharinensis are animal- and gravity-dispersed (Moraes and Paoli 1995, 1999), should spatial genetic structure be expected in all the studied populations?
Materials and methods
Species biology
Ocotea catharinensis Mez. (Lauraceae) is a long-living, evergreen and dominant canopy species, reaching a height of 45 m and 150 cm in diameter at breast height. The species is indigenous to south and southeast of Brazil. The natural distribution of the species in Brazil ranges from 21°20′ S (Minas Gerais State) to 30°15′ S (Rio Grande do Sul State) and it is found exclusively in the Atlantic rain forest, between altitudes of 30 and 1,400 m (Carvalho 1994). O. catharinensis’ small and white hermaphrodite flowers are pollinated by diverse insects such as bees, wasps, flies and beetles (Reitz et al. 1978). Its seeds are animal- and gravity-dispersed. Animal seed dispersal is carried out by primates such as Cebus apella, Alouatta fusca and Brachyteles arachnoids (Moraes and Paoli 1995, 1999) and birds, e.g., Pipile jacutinga (Galetti et al. 1997). Seedlings of O. catharinensis establish in shade, regardless of the presence of gaps for their development (Moraes and Paoli 1999).
Field collections
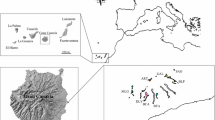
Studies were carried out in four natural populations of O. catharinensis in the cities of Ilhota [conservation unit—Parque Botânico do Morro Baú (PMB); 26°47′56″ S, 48°55′49″ W], Santo Amaro da Imperatriz [conservation unit—Parque Estadual da Serra do Tabuleiro (PEST); 27°44′43″ S, 48°49′13″ W], Corupá [Private property of MOBASA/BATISTTELLA (CP); 26°25′06″ S, 49°22′02″ W] and Grão Pará [Private property of Mr. Antônio Alberton (GP) 28°14′17″ S, 49°17′40″ W], all located in the state of Santa Catarina (SC) in the Brazilian ARF (Fig. 1). The distance between populations ranged from 60.7 (CP to MB) to 202.1 km (GP to CP), with an average of 122.6 km. These populations were separated from each other as a result of forest fragmentation.
In all study areas, we georeferenced and numbered all the reproductive individuals of O. catharinensis with aluminum plates (Fig. 2). We collected demographic data [density (D), diameter at breast height (DBH), Clark and Evans (1954) aggregation index (R), mean distance to the nearest neighbor (MD) and local declivities (LD)] and leaf samples from all the reproductive trees in the study areas: PMB (n = 51; 12.5 ha; 500 m × 250 m; LD = 13%), PEST (n = 35; 7.5 ha; 500 m × 150 m; LD = 17%), CP (n = 49; 3.00 ha; 200 m × 150 m; LD = 12%) and GP (n = 53; 14.0 ha; 350 m × 400 m; LD = 13%). Samples were put into plastic bags in ice containers and taken to the laboratory, where they were stored at 5°C until needed.
Laboratory procedures
The populations were analyzed by horizontal starch-gel electrophoresis (13%). The enzymes glutamate dehydrogenase (locus Gtdh-1; EC 1.1.1.2), diaphorase (Dia-1 and Dia-2; EC 1.8.1.4), glutamate oxaloacetate transaminase (Got-1 and Got-2; EC 2.6.1.1), isocitrate dehydrogenase (Idh-1; EC 1.1.1.42), phosphoglucomutase (Pgm-1; EC 5.4.2.2), superoxide dismutase (Sod-1; EC 1.15.1.1), acid phosphatase (Acp-1; EC 3.1.3.2), shikimate dehydrogenase (Skdh-1; EC 1.1.1.25), esterase (Est-1,Est-2 and Est-3; EC 3.1.1.1), peroxidase (Po-1 and Po-2; EC 1.11.1.7), malate dehydrogenase (Mdh-1 and Mdh-2; EC 1.1.1.37) and malic enzyme (Me-1; EC 1.1.1.40) were assayed using a continuous, number 10, Tris–citrate buffer system (pH 7.5) (Alfenas and Brune 1998).
Data analysis
For demographic characterization we analyzed Clark and Evans (1954) mean distances between neighbors (MD) and aggregation index (R). In addition, a Z-test was applied to determine the statistical level (P-value) at which the observed pattern deviates from an expected random pattern, when R = 1 the spatial pattern is random, R < 1 clumping occurs (0 = minimum) and R > 1 when a uniform distribution pattern exists (2.149 = maximum). All calculations were accomplished through SGS program (Degen et al. 2001).
For genetic characterization we identified the genotypes and estimated the allelic frequencies for the studied populations from the interpretation of the zymograms. We used the GDA computer program (Lewis and Zaykin 2001) to estimate the percentage of polymorphic loci (P), the mean number of alleles per locus (A), the mean observed (H o) and expected (H e) heterozygosities, and the fixation indices for each population (f i). P was obtained considering the number of loci that presented the most frequent allele with an occurrence of less than 95% in relation to the total number of loci. A was estimated by dividing the total number of alleles by the number of loci. H o was obtained by counting the heterozygote. H e was obtained by Nei’s (1978) formula \( \hat{H}_{\text{e}} = 1 - \sum {p_{i}^{2} } \), where p i is the mean frequency of the allele i in the loci. 95% bootstrap confidence intervals were estimated on f i by resampling loci 10,000 times using the GDA program (Lewis and Zaykin 2001).
Wright’s (1951, 1965) F-statistics (F IS, F IT and F ST) were estimated for the populations with BIOSYS-2 program (Swofford and Selander 1997), using Nei’s (1987) corrections for small sample size. In addition, genetic structure was quantified using Weir and Cockerham’s (1984) F-statistics (f = F IS, F = F IT and θ P = F ST) implemented in GDA program (Lewis and Zaykin 2001). 95% bootstrap confidence intervals (CI) were estimated from F, f and θ P by re-sampling loci 10,000 times using the GDA program (Lewis and Zaykin 2001).
The apparent outcrossing rate \( \left(\hat{t}_{\text{a}} = \left( {1 - \hat{f}} \right)/\left( {1 + \hat{f}} \right)\right) \) was estimated from f as suggested by Carlini-Garcia et al. (2001). And the apparent gene flow \( \left(\hat{N}m = \left( {1/4\alpha } \right)[(1/\hat{F}_{\text{ST}} ) - 1]\right) \) among populations was estimated indirectly, using the island model proposed by Crow and Aoki (1984), which corrects the analyses for a small number of populations. In this expression the genetic divergence among populations (F ST) was substituted by θ P, on account of it is less biased as suggested by Cockerham and Weir (1993), and the correction of the population number (n) was calculated as α = [n/(n-1)]2.
Spatial genetic structure in the four populations was assessed using Moran’s I coefficient (Moran 1950; Sokal and Oden 1978) for each locus. Moran’s I was calculated using alleles with a frequency higher than 5% and for each of seven consecutive distance classes from 0–20 m (class 1) to 120–140 m (class 7) with 20 m distance intervals. 20 m was the minimum distance class size that could be tested in order to have 30 pairs of data points in each distance class, the minimum statistical sample size requirement for Moran’s I (Degen et al. 2001). An average Moran’s I coefficient was also calculated for all loci as a summary statistic. A permutation procedure using Monte-Carlo simulations was applied to test the significance of deviations from randomness of the spatial distribution for each calculated measure (Manly 1997). Each permutation consisted of a random redistribution of multilocus genotypes over the spatial coordinate of the sampled trees. For each distance class, the observed values were compared with 95% confidence intervals constructed for each of the parameters from the distributions obtained after 1,000 permutations. The expected value for the absence of spatial autocorrelation was −1/(n−1) (Sokal and Wartenberg 1983), n was the population sample size. These calculations were performed through the SGS program (Degen et al. 2001). Additionally, Wright’s (1943) neighborhood sizes were estimated for the populations by using the observed average Moran’s I values for the first distance class, and comparing these to theoretical values in Table 4 of Epperson et al. (1999).
Results
Demographic characterization
Population CP presented the highest tree density with an average distance among trees of 8.22 m (Table 1). PEST, PMB and GP presented an equally moderate tree density with an average distance among trees of 17.06, 16.98 and 18.98 m, respectively (Table 1). Based on Clark and Evans (1954) aggregation index (R), this value indicates a rejection of null hypothesis of a strictly random pattern of distribution for the trees within the four populations, yet it suggests a much greater tendency toward randomness than clumping (Table 1). Pooling the four populations, the DBH average of 188 trees was 45.51 ± 23.06 cm, ranging from 6.85 to 148.30 cm. Population PEST presented the lowest DBH, and PMB, CP and GP presented significantly higher values of DBH compared to PEST (Table 1). DBH values for CP and GP did not differ significantly.
Genetic diversity
Among the eighteen loci resolved into the 12 enzymatic systems, two loci were monomorphic (Got-1and Gtdh-1), and the other sixteen were polymorphic. About 38 alleles were found in population GP, 37 alleles were found in populations CP and PMB, and 36 were found in PEST. The maximum number of alleles per locus was three and the mean number of alleles per locus for the species was 2.2 for all loci (Table 2). The observed and expected heterozygosity values obtained for PEST and PMB deviated from Hardy–Weinberg equilibrium, indicating excess of heterozygote. CP also deviated from Hardy–Weinberg equilibrium, endogamy was detected. Unlike GP that was in Hardy–Weinberg equilibrium (Table 2).
The estimate of the fixation index (\( \hat{F}_{\text{IT}} = 0.0 9 7 ;\,\hat{F} = 0. 1 3 3 \), CI = 0.128–0.136) indicated a significant deficit of heterozygosity for the total sample, but the subpopulation fixation index indicated excess of heterozygosity (\( \hat{F}_{\text{IS}} = - 0.0 2 3;\,\hat{f} = - 0.0 1 1 \), CI = −0.016 to −0.009). The genetic divergence among the four populations (\( \hat{F}_{\text{ST}} = 0. 1 1 8;\,\hat{\theta }_{P} = 0. 1 4 3 \), CI = 0.138–0.143) was significant. In agreement with this, the number of migrants per generation (\( \hat{N}m \)) using \( \hat{\theta }_{P} \) was 0.842. The \( \hat{\theta }_{P} \) values calculated among the closest geographical populations (CP-PMB = 0.206; PMB-PEST = 0.126; PEST-GP = 0.102) were significant. In this study, the populations showed a discontinuous distribution, like in the case of the stepping stone model.
Spatial genetic structure
There was significant fine-scale spatial genetic structure within the four populations of O. catharinensis, indicating that the trees share common alleles with a frequency greater in comparison with that expected at random (Fig. 3). Significant Moran’s I values were found among trees separated by less than 40 m in CP and less than 60 m in PEST and GP (Fig. 3). For PMB, significant Moran’s I values were found among trees separated by less than 80 m, except for trees found in the distance class from 20 to 40 m (Fig. 3). The average Moran’s I values for all pairwise tree comparisons for CP, PMB, PEST and GP, within the first distance class (0–20 m) were estimated to be 0.072, 0.166, 0.118 and 0.191, respectively (Fig. 3). In concordance, neighborhood size was approximately 83 individuals in the case of CP and about 25 individual for PMB, PEST and GP.
Discussion
Demographic patterns
The deviation from an expected random spatial pattern found in the four populations of O. catharinensis is a trend of most tropical tree species (Hubbell 1979; He et al. 1997; Condit et al. 2000; Plotkin et al. 2000, 2002). The tendency toward randomness could be related to the age of the sampled trees, where spatial pattern tends to random distribution as age increases (Itoh et al. 1997; Condit et al. 2000; Plotkin et al. 2000). Spatial aggregation of tree species in tropical forests results from many abiotic and biotic factors, such as edaphic factors, limited seed dispersal and habitat specialization (Ng et al. 2006; Gardner and Engelhardt 2008; Muller-Landau et al. 2008). As the four sampled areas are mature forest and have a similar declivity, differences in spatial distribution of individuals could be more probably related to biotic dispersion of seed. Therefore, O. catharinensis animal- and gravity-dispersed seeds are contributing to the aggregated spatial distribution of adult individuals and each local frugivore community that interacts with the species causes variation in the spatial pattern and dispersal distance.
Diameter at breast height values revealed a large amount of reproductive trees in all the study areas. These trees through seed dispersal can help the species reclaim its original area. Nevertheless, seedlings of O. catharinensis establish in shade, requiring mature or advanced successional stage forest for their development (Carvalho 1994; Moraes and Paoli 1999). To enhance reproductive success and survival of O. catharinensis the remaining mature forest fragments must be protected and landscape connectivity reestablished. To achieve this goal, Harvey et al. (2008) and Perfecto and Vandermeer (2008) provide conservation plans appropriate for Santa Catarina State by reconciling farming and biodiversity conservation. As Santa Catarina State is dominated by agricultural landscapes and conservation units respond for less than 10% of its areas, owners of rural properties must be treated as stakeholders in conserving biodiversity and incentives for sustainable development of the landscape must be provided.
Brazil has very strict laws for forest conservation, the national environment policy (Brazilian federal law no. 6938/1981), the forestry law (Brazilian federal law no. 4771/1965) both updated until the year 2006, and ARF conservation law (Brazilian federal law no. 11428/2006), already treat rural property owners as stakeholders in conserving biodiversity, but there is a lack of policies to encourage conservation. The law in Brazil requires that 20% of the rural property area be reserved to ARF conservation; however in most cases this law is not respected. With proper law enforcement and incentives, in situ conservation of the ARF in rural properties in Brazil may overcome conservation units in area and achieve the minimum requirement established in the Convention on Biological Diversity (CBD 2003) for ecosystem conservation.
As O. catharinensis seedlings need mature or advanced successional stage forest to establish and these forests account for less than 14.46% of ARF original area in Santa Catarina (IESB 2007), a viable solution for expanding O. catharinensis populations and reconnecting forest fragments is the use of agroforestry systems. On the one hand, agroforestry systems may offer shade for the establishment of O. catharinensis seedlings and on the other hand, it can be integrated into biodiversity corridors for a variety of uses, such as timber and non-timber forest products, thereby minimizing the exploitation of protected areas (Huang et al. 2002) and ameliorating the environmental characteristics that produce the edge effect (Perfecto et al. 2007). Therefore, with sustainable agriculture activity and proper law enforcements, the rural landscape could be integrated in the conservation of O. catharinensis and maintain the forest ecosystem functionality.
Genetic diversity
The heterozygosity found in this study (\( \hat{H}_{T} = 0. 4 40 \)) is above the mean heterozygosity found for woody perennial outcrossing species (\( \hat{H}_{T} = 0. 2 8 3 \); Hamrick and Godt 1996; Hamrick 2004). The expected genetic diversity (\( \hat{H}_{e} = 0. 4 2 7 \)), the mean number of alleles per locus (\( \hat{A} = 2. 2 \)) and the mean percentage of polymorphic loci (\( \hat{P} = 8 3\% \)) of O. catharinensis were above the mean values found in Lauraceae (Table 3). The \( \hat{A} \) and \( \hat{P} \) values found for O. catharinensis were closer to those found for other two Ocotea species from the ARF, O. porosa (\( \hat{A} = 2.0 7 \) and \( \hat{P} = 7 1. 4\% \); Silva et al. 2003) and O. odorifera (\( \hat{A} = 2. 3 6 \) and \( \hat{P} = 6 7. 8\% \); Kageyama et al. 2003) than to those for Cryptocarya aschersoniana (\( \hat{A} = 2. 9 4 ;\,\hat{P} = 9 5. 1\% \); Moraes and Derbyshires 2002) another Lauraceae species from the ARF, probably due to the phylogenetic relationships inside Lauraceae.
The high genetic diversity values detected for O. catharinensis demonstrate that all the studied populations have genetic potential for conservation. Moreover, the high genetic diversity observed might be related to the wide geographical distribution in the ARF and to the mating system, apparently outcross pollination (\( \hat{t}_{\text{a}} = 1.023 \)). Perennial species with a long life cycle, sexual reproduction, predominant outcrossing pollination and wide distribution, accumulate a great genetic variability within the populations (Hamrick et al. 1992; Leimu et al. 2006).
The studied populations occurring inside conservation units in continuous forest and inside small, isolated and fragmented forests in private proprieties presented equally moderate levels of genetic diversity, probably due to the fact that the sampled individuals were present before the forest fragmentation events initiated in the mid-1900. This can be inferred because O. catharinensis is a long-living tropical tree species that may live for more than 300 years, concerning the high dimensions that the trees can reach (DBH > 150 cm) and the very slow growth rate (Carvalho 1994).
Our F-statistics demonstrated population genetic structure and a low migration rate, approximately one migrant per generation. The first event as a determinant of gene flow among populations is an effective pollen dispersal (Hamrick and Nason 2000). A lower divergence among populations can be achieved if the pollen disperser travels over long distances (Kramer et al. 2008). As O. catharinensis is pollinated by various small insects such as wasps, bees, flies and beetles (Reitz et al. 1978) and reproductive trees are located in closed-canopy forest with an average distance among trees of 15 m, probably its pollen dispersers are traveling over short distances, promoting high genetic divergence among the populations. Although long-distance pollination mediated by small insects is common for tropical tree species (Kramer et al. 2008), short pollination distances appear when tree density is high and in closed-canopy forest (Dick et al. 2008; Hanson et al. 2008).
As gene flow is a two stage event and depends on seedling establishment (Hamrick and Nason 2000) we suggest that the dynamic of allele movement in O. catharinensis is strongly related to its restricted seed dispersal carried out by gravity and animals, promoting spatial genetic structure within the populations and high divergences among the populations. Dick et al. (2008) reviewed 42 tropical tree species and found high genetic differentiation among populations (\( \hat{F}_{ST} = 0. 1 7 7 \)) related to low tree density, restricted seed dispersion and biogeographic barriers. An evidence of a biogeographic barrier affecting genetic differentiation among populations in O. catharinensis is the Jaraguá Montain located between populations CP and PMB, where \( \theta_{P} \) presented the highest value (0.206) among the closest populations.
Our results showed that the realized gene flow (\( \hat{N}m = 0. 8 4 2 \)) is not sufficient to stop the increase in the genetic differentiation among population due to genetic drift and local selection, threatening the future viability of O. catharinensis populations. The rule of one migrant per generation (OMPG rule) has been proposed as the required minimum to protect populations from the accumulation of deleterious mutations (Mace and Lande 1991; Hedrick 1995; Mills and Allendorf 1996). Nevertheless, two conditions need to be fulfilled for the OMPG rule: large population sizes and high growth rates (Couvet 2002; Bouchy et al. 2005). Moreover, when the condition of having less than one migrant per generation associated with local adaptations could lead to an increase of the probability of population extinction (Couvet 2002). To reduce O. catharinensis probability of extinction, urgent efforts towards conservation must be intensified in these four different populations. The remaining long-living trees should be used as seed-trees to help the species reclaim its original area and as genetic connectors among fragmented populations.
Spatial genetic structure
The analysis of fine-scale spatial genetic structure within the populations detected SGS in adult trees separated by less than 80 m. In this context, genetic similarity is higher among neighboring individuals than among more distant ones, which is in agreement with the expected pattern of SGS at drift-dispersal equilibrium predicted by the theory of genetic isolation by distance (Vekemans and Hardy 2004). SGS within tropical tree populations has been found for several studies (e.g., Degen et al. 2001; Luna et al. 2005; Hardy et al. 2006; Born et al. 2008) and can be a result of selection or genetic drift, due to environmental heterogeneity or genetic isolation by distance, respectively (Heywood 1991; Epperson 1993).
The distances of SGS in the studied populations were similar to those found for others insect-pollinated and gravity- and animal- seed dispersed Brazilian Lauraceae tree species, for instance, Ocotea odorifera (Kageyama et al. 2003) with significant SGS found in trees separated by less than 74 m (Moran’s I = 0.05) and shorter than Cryptocarya moschata (Moraes et al. 2004) with significant SGS found in trees separated by less than 150 m (Moran’s I = 0.05). Moreover, when our SGS results are combined with the findings of Kageyama et al. (2003) and Moraes et al. (2004) and compared with other Asian Lauraceae species that are insect-pollinated, whose seeds are bird-dispersed and lack SGS (e.g., Neolitsea sericea, Chung et al. 2000; Cinnamomum insularimontanum: Chung et al. 2003), we demonstrate how biotic seed dispersal influences the SGS. Therefore, we should expect genotypes to be randomly distributed within populations with bird-seed dispersal (Chung et al. 2003) and should expect SGS within local populations caused by gravity- and mammal-seed dispersal (Moraes et al. 2004; Hardy et al. 2006).
The observed Moran’s I values for the first distance class corresponds approximately to a neighborhood size of 83 individuals in the case of CP and about 25 individuals for the other populations. Under different densities, the area of O. catharinensis demes corresponds approximately to 5 ha for CP and 6 ha for PMB, PEST and GP. The lowest SGS patterns found for population CP may probably reflect the sampling strategy that only surveyed 3 ha and 49 individuals, less than the expected deme area and size, respectively. These results also alert us that small forest fragments with at least 5 ha may contain exclusive alleles with high adaptive potential to withstand environmental stress. Therefore, the conservation of genetic diversity across an entire population and in small fragments must be considered in conserving the species evolutionary potential (Chung et al. 2001; Cruse-Sanders and Hamrick 2004).
Conservation implications
The high genetic diversity in the studied populations illustrates the importance of preserving them for the conservation of the species evolutionary potential. Our results also proved that the realized gene flow is not enough to reduce the genetic differentiation among populations due to genetic drift and local selection. As illegal deforestation continues in the Brazilian ARF, the removal of small forest patches, up to 6 ha, can eliminate an entire O. catharinensis deme and consequently, eliminate alleles within these demes. Moreover, forest fragmentation may increase, reducing the connectivity between populations of O. catharinensis and hence, the SGS and genetic differentiation could also increase. However, connectivity between fragments could be reestablished to maintain gene flow among populations if rural landscape is integrated to conservation planning through agroforestry systems.
The analyses of demographic patterns and SGS are powerful tools for developing conservation strategies and pointing out undisturbed populations (Chung and Nason 2007). As the four studied populations revealed spatial aggregation of trees and SGS, these features may represent a natural state in O. catharinensis undisturbed populations. Therefore, we can use our results as a pattern to alert conservation managers that increased SGS values may indicate forest fragmentation effects, or even the lack of a frugivore community that disperses O. catharinensis seeds. On the other hand, the absence of SGS may indicate anthropogenic disturbance, such as harvesting. Accordingly, an efficient restoration plan within forest fragments, which would provide a balanced family structure, should involve the entry of new alleles through seeds. These seeds could come from gene flow corridors or even by direct human intervention. Although, a special regard should be taken when using these seeds, they must come from nearby populations to avoid maladaptation problems (e.g., McKay et al. 2005; Bower and Aitken 2008). Concerning in situ and ex situ genetic conservation, the seeds must be collected from trees at least 80 m apart reducing genetic similarity among seed-trees to obtain a representative sample.
References
Alfenas AC, Brune W (1998) Identificações específicas de enzimas em géis. In: Alfenas AC (ed) Eletroforese de isoenzimas e proteínas afins: fundamentos e aplicações em plantas e microrganismos. Editora UFV, Viçosa, pp 201–328
Álvarez-Buylla ER, García-Barrios R, Lara-Moreno C, Martínez-Ramos M (1996) Demographic and genetic models in conservation biology: applications and perspectives for tropical rain forest tree species. Annu Rev Ecol Syst 27:387–421. doi:10.1146/annurev.ecolsys.27.1.387
Auler NMF, Reis MS, Guerra MP, Nodari RO (2002) The genetics and conservation of Araucaria angustifolia I. Genetic structure and diversity of natural populations by means of non-adaptative variation in the state of Santa Catarina, Brazil. Genet Mol Biol 25:323–327. doi:10.1590/S1415-47572002000300014
Barrett SC, Kohn JR (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 3–30
Bawa KS (1992) Mating systems, genetic differentiation and speciation in tropical rain forest plants. Biotropica 24:250–255. doi:10.2307/2388519
Bittencourt JVM, Sebbenn AM (2007) Patterns of pollen and seed dispersal in a small, fragmented population of the wind-pollinated tree Araucaria angustifolia in southern Brazil. Heredity 99:580–591. doi:10.1038/sj.hdy.6801019
Born C, Hardy OJ, Chevallier MH, Ossari S, Attéké C, Wickings J, Hossaert-McKey M (2008) Small-scale spatial genetic structure in the Central African rainforest tree species Aucoumea klaineana: a stepwise approach to infer impact of limited gene dispersal, population history and habitat fragmentation. Mol Ecol 17:2041–2050. doi:10.1111/j.1365-294X.2007.03685.x
Bouchy P, Theodorou K, Couvet D (2005) Metapopulation viability: influence of migration. Conserv Genet 6:75–85. doi:10.1007/s10592-004-7744-7
Bower AD, Aitken SN (2008) Ecological genetics and seed transfer guidelines for Pinus albicaulis (Pinaceae). Am J Bot 95:66–76. doi:10.3732/ajb.95.1.66
Carlini-Garcia LA, Vencovsky R, Coelho ASG (2001) Bootstrap method applied over resampling levels in the estimation of genetic parameters of populations. Scientia Agricola. doi: 10.1590/S0103-90162001000400021
Carvalho PER (1994) Espécies florestais brasileiras: recomendações silviculturais, potencialidades e uso da madeira. EMBRAPA-CNPF, Colombo
Cascante A, Quesada M, Lobo JJ, Fuchs EA (2002) Effects of dry tropical forest fragmentation on the reproductive success and genetic structure of the tree Samanea saman. Conserv Biol 16:137–147. doi:10.1046/j.1523-1739.2002.00317.x
CBD (Convention on Biological Diversity) (2003) Global strategy for plant conservation (UNEP/CBD/SBSTTA/9/14/2003) Montreal, Quebec. Available from http://www.cbd.int/doc/meetings/sbstta/sbstta-09/official/sbstta-09-14-en.pdf (accessed October 2008)
Cheptou PO, Schoen DJ (2007) Combining population genetics and demographical approaches in evolutionary studies of plant mating systems. Oikos 116:271–279. doi:10.1111/j.0030-1299.2007.14655.x
Chung MY, Nason JD (2007) Spatial demographic and genetic consequences of harvesting within populations of the terrestrial orchid Cymbidium goeringii. Biol Conserv 37:125–137. doi:10.1016/j.biocon.2007.01.021
Chung MG, Chung MY, Oh GS, Epperson BK (2000) Spatial genetic structure in a Neolitsea sericea population (Lauraceae). Heredity 85:490–497. doi:10.1046/j.1365-2540.2000.00781.x
Chung MG, Chung MY, Epperson BK (2001) Conservation genetics of an endangered herb, Hanabusaya asiatica (Campanulaceae). Plant Biol 3:42–49. doi:10.1055/s-2001-11744
Chung MY, Nason JD, Epperson BK, Chung MG (2003) Temporal aspects of the fine-scale genetic structure in a population of Cinnamomum insularimontanum (Lauraceae). Heredity 90:98–106. doi:10.1038/sj.hdy.6800187
Clark PJ, Evans FC (1954) Distance to nearest neighbor as a measure of spatial relationships in populations. Ecology 35:445–453. doi:10.2307/1931034
Cockerham CC, Weir BS (1993) Estimation of gene flow from F-Statistics. Evolution Int J Org Evolution 47:855–863. doi:10.2307/2410189
Condit R, Ashton PS, Baker P, Bunyavejchewin S, Gunatilleke S, Gunatilleke N, Hubbell SP, Foster RB, Itoh A, LaFrankie JV, Lee HS, Losos E, Manokaran N, Sukumar R, Yamakura T (2000) Spatial patterns in the distribution of tropical tree species. Science 288:1414–1418. doi:10.1126/science.288.5470.1414
Conte R, Reis MS, Mantovani A, Vencovsky R (2008) Genetic structure and mating system of Euterpe edulis Mart. populations: a comparative analysis using microsatellite and allozyme markers. J Hered. doi:10.1093/jhered/esn055
Couvet D (2002) Deleterious effects of restricted gene flow in fragmented populations. Conserv Biol 16:369–376. doi:10.1046/j.1523-1739.2002.99518.x
Crow JF, Aoki K (1984) Group selection for a polygenic behavioral trait: estimating the degree of population subdivision. Proc Natl Acad Sci USA 81:6073–6077. doi:10.1073/pnas.81.19.6073
Cruse-Sanders JM, Hamrick JL (2004) Spatial and genetic structure within populations of wild American ginseng (Panax quinquefolius L., Araliaceae). J Hered 95:309–321. doi:10.1093/jhered/esh046
Degen B, Petit R, Kremer A (2001) SGS—spatial genetic software: a computer program for analysis of spatial genetic and phenotypic structures of individuals and populations. J Hered 92:447–448. doi:10.1093/jhered/92.5.447
Dick CW (2008) New interpretations of fine-scale spatial genetic structure. Mol Ecol 17:1873–1876. doi:10.1111/j.1365-294X.2008.03728.x
Dick CW, Hardy OJ, Jones FA, Petit R (2008) Spatial scales of pollen and seed-mediated gene flow in tropical rain forest trees. Trop Plant Biol 1:20–33. doi:10.1007/s12042-007-9006-6
Doligez A, Joly HI (1997) Genetic diversity and spatial structure within a natural stand of a tropical forest tree species, Carapa procera (Meliaceae), in French Guiana. Heredity 79:72–82. doi:10.1038/hdy.1997.124
Ennos RA, Clegg MT (1982) Effect of population substructuring on estimates of outcrossing rate in plant population. Heredity 48:283–292. doi:10.1038/hdy.1982.33
Epperson BK (1993) Recent advances in correlation studies of spatial patterns of genetic variations. Evol Biol 27:95–155
Epperson BK, Huang Z, Li TQ (1999) Measures of spatial structure in samples of genotypes for multiallelic loci. Genet Res 73:251–261. doi:10.1017/S001667239900378X
Frankham R (1995) Inbreeding and extinction: a threshold effect. Conserv Biol 9:792–799. doi:10.1046/j.1523-1739.1995.09040792.x
Gaiotto FA, Grattapaglia D, Vencovsky R (2003) Genetic structure, mating system, and long-distance gene flow in heart of palm (Euterpe edulis Mart.). J Hered 94:399–406. doi:10.1093/jhered/esg087
Galetti M, Martuscelli P, Olmos F, Aleixo A (1997) Ecology and conservation of the jacutinga Pipile jacutinga in the Atlantic forest of Brazil. Biol Conserv 82:31–39. doi:10.1016/S0006-3207(97)00004-9
Gardner RH, Engelhardt KAM (2008) Spatial processes that maintain biodiversity in plant communities. Perspect Plant Ecol Evol Syst 9:211–228. doi:10.1016/j.ppees.2007.08.003
Geburek T (2000) Effects of environmental pollution on the genetics of forest trees. In: Young AG, Boshier D, Boyle TJ (eds) Forest conservation genetics: principles and practice. CSIRO Publishing, Melbourne, pp 135–158
Gibson JP, Wheelwright NT (1995) Genetic structure in a population of a tropical tree Ocotea tenera (Lauraceae): influence of avian seed dispersal. Oecologia 103:49–54
Hamrick JL (2004) Response of forest trees to global environmental changes. For Ecol Manage 197:323–335. doi:10.1016/j.foreco.2004.05.023
Hamrick JL, Godt MJW (1996) Effects of life history traits on genetic diversity in plant species. Philos Trans R Soc Lond, B 351:1291–1298. doi:10.1098/rstb.1996.0112
Hamrick JL, Loveless MD (1986) Isozyme variation in tropical trees: procedures and preliminary results. Biotropica 18:201–207. doi:10.2307/2388485
Hamrick JL, Nason JD (2000) Gene flow in forest trees. In: Young AG, Boshier D, Boyle TJ (eds) Forest conservation genetics: principles and practice. CSIRO Publishing, Melbourne, pp 81–90
Hamrick JL, Godt MJW, Sherman-Broyles (1992) Factors influencing levels of genetic diversity in woody plant species. New For 6:95–124. doi:10.1007/BF00120641
Hanski I, Ovaskainen O (2000) The metapopulation capacity of a fragmented landscape. Nature 404:755–758. doi:10.1038/35008063
Hanson TR, Brunsfeld SJ, Finegan B, Waits LP (2008) Pollen dispersal and genetic structure of the tropical tree Dipteryx panamensis in a fragmented Costa Rican landscape. Mol Ecol 17:2060–2073. doi:10.1111/j.1365-294X.2008.03726.x
Hardy OJ, Maggia L, Bandou E, Breyne P, Caron H, Chevallier M, Doliguez A, Dutech C, Kremer A, Latouche-Halle C, Troispoux V, Veron V, Degen B (2006) Fine-scale genetic structure and gene dispersal inferences in 10 Neotropical tree species. Mol Ecol 15:559–571. doi:10.1111/j.1365-294X.2005.02785.x
Harvey CA, Komar O, Chazdon R, Ferguson BG, Finegan B, Griffith DM, Martínez-Ramos M, Morales H, Nigh R, Soto-Pinto L, Breugel MV, Wishnie M (2008) Integrating agricultural landscapes with biodiversity conservation in the Mesoamerican hotspot. Conserv Biol 22:8–15. doi:10.1111/j.1523-1739.2007.00863.x
He FL, Legendre P, LaFrankie JV (1997) Distribution patterns of tree species in a Malaysian tropical rain forest. J Veg Sci 8:105–114. doi:10.2307/3237248
Hedrick PW (1995) Gene flow and genetic restoration: the Florida panther as a case study. Conserv Biol 9:996–1007. doi:10.1046/j.1523-1739.1995.9050996.x
Heywood JS (1991) Spatial analysis of genetic variation in plant populations. Annu Rev Ecol Syst 22:335–355. doi:10.1146/annurev.es.22.110191.002003
Huang W, Luukkanen O, Johanson S, Kaaraka V, Räisänen S, Vihemäki H (2002) Agroforestry for biodiversity conservation of nature reserves: functional group identification and analysis. Agrofor Syst 55:65–72. doi:10.1023/A:1020284225155
Hubbell SP (1979) Tree dispersion, abundance and diversity in a dry tropical forest. Science 203:1299–1309. doi:10.1126/science.203.4387.1299
IESB (2007) Projeto de conservação e utilização sustentável da diversidade biológica brasileira—levantamento da cobertura vegetal nativa do Bioma Mata Atlântico: relatório final PROBIO 03/2004. Available from: http://www.mma.gov.br
Itoh A, Yamakura T, Ogino K, Lee HS, Ashton PS (1997) Spatial distribution patterns of two predominant emergent trees in a tropical rainforest in Sarawak, Malaysia. Plant Ecol 132:121–136. doi:10.1023/A:1009779424279
Jaquiéry J, Guillaume F, Perrin N (2008) Predicting the deleterious effects of mutation load in fragmented populations. Conserv Biol. doi:10.1111/j.1523-1739.2008.01052.x
Jones FA, Hubbell SP (2006) Demographic spatial genetic structure of the Neotropical tree, Jacaranda copaia. Mol Ecol 15:3205–3217. doi:10.1111/j.1365-294X.2006.03023.x
Kageyama PY, Cunha GC, Barreto KD, Gandara FB, Camargo FRA, Sebbenn AM (2003) Diversidade e autocorrelação genética espacial em populações de Ocotea odorífera (Lauraceae). Scientia Forestalis 64:108–109
Kramer AT, Ison J, Ashley MV, Howe HF (2008) The paradox of forest fragmentation genetics. Conserv Biol. doi:10.1111/j.1523-1739.2008.00944.x
Lande R (1993) Risks of population extinction from demographic and environmental stochasticity and random catastrophes. Am Nat 142:911–927. doi:10.1086/285580
Lande R (1995) Mutation and conservation. Conserv Biol 9:782–791. doi:10.1046/j.1523-1739.1995.09040782.x
Leimu R, Mutikainen P, Koricheva J, Fisher M (2006) How general are positive relationships between plant population size, fitness and genetic variation? J Ecol 94:942–952. doi:10.1111/j.1365-2745.2006.01150.x
Lewis PO, Zaykin D (2001) Genetic data analysis: Computer program for the analysis of allelic data. Version 1.0 (d16c). Available from: http://lewis.eeb.uconn.edu/lewishome/software.html
Luna R, Epperson BK, Oyama K (2005) Spatial genetic structure of two sympatric Neotropical palms with contrasting life histories. Heredity 95:298–305. doi:10.1038/sj.hdy.6800655
Mace GM, Lande R (1991) Assessing extinction threats—toward a reevaluation of IUCN threatened species categories. Conserv Biol 5:148–157. doi:10.1111/j.1523-1739.1991.tb00119.x
Manly BFJ (1997) Randomization, bootstrap and Monte Carlo methods in Biology, 2nd edn. Chapman and Hall, London
Mantovani A, Morellato LPC, Reis MS (2006) Internal genetic structure and outcrossing rate in a natural population of Araucaria angustifolia (Bert.) O. Kuntze. J Hered 97:466–472. doi:10.1093/jhered/esl031
Matthies D, Bräuer I, Maibom W, Tscharntke (2004) Population size and the risk of local extinction: empirical evidence from rare plants. Oikos 105:481–488. doi:10.1111/j.0030-1299.2004.12800.x
McKay JK, Christian CE, Harrison S, Rice KJ (2005) “How local is local?” A review of practical and conceptual issues in the genetics of restoration. Restor Ecol 13:432–440. doi:10.1111/j.1526-100X.2005.00058.x
Mills LS, Allendorf FW (1996) The one-migrant-per-generation rule in conservation and management. Conserv Biol 10:1509–1518. doi:10.1046/j.1523-1739.1996.10061509.x
Moraes PLR, Derbyshires MTVC (2002) Estrutura genética de populações naturais de Cryptocarya aschersoniana Mez. (Lauraceae) através de marcadores isoenzimáticos. Biota Neotropica 2:1–19
Moraes PLR, Paoli AA (1995) Dispersão e germinação de sementes de Crptocarya moschata Nees & Martius ex Nees, Ocotea catharinensis Mez. e Endlicheria paniculata (Sprengel) Macbride (Lauraceae). Arq Biol Tecnol 38:1119–1129
Moraes PLR, Paoli AA (1999) Morfologia e estabelecimento de plântulas de Cryptocarya moschata Nees, Ocotea catharinensis Mez. e Endlicheria paniculata (Spreng.)—Lauraceae. Rev Bras Bot 22:287–295
Moraes PLR, Monteiro R, Vencovsky R (2004) Intrapopulation genetic structure in Cryptocarya moschata Nees (Lauraceae). Rev Bras Bot 27:475–487. doi:10.1590/S0100-84042004000300008
Moran PAP (1950) Notes on continuous stochastic phenomena. Biometrika 37:17–23
Muller-Landau HC, Wright SJ, Calderón O, Condit R, Hubbell SP (2008) Interspecific variation in primary seed dispersal in a tropical forest. J Ecol 96:653–667. doi:10.1111/j.1365-2745.2008.01399.x
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, NY
Ng KKS, Lee SL, Saw LG, Plotkin JB, Koh CL (2006) Spatial structure and genetic diversity of three tropical tree species with different habitat preferences within a natural forest. Tree Genet Genomes 2:121–131. doi:10.1007/s11295-006-0035-3
Perfecto I, Vandermeer J (2008) Biodiversity conservation in tropical agroecosystems: a new conservation paradigm. Ann N Y Acad Sci 1134:173–200. doi:10.1196/annals.1439.011
Perfecto I, Armbrecht I, Philpott SM, Soto-Pinto L, Dietsch TV (2007) Shaded coffee and the stability of rainforest margins in northern Latin America. In: Tscharntke T, Leuschner C, Zeller M, Guhardja E, Bidin A (eds) Stability of tropical rainforest margins, linking ecological, economical and social constraints of land use and conservation. Springer, Heidelberg, pp 227–263
Plotkin JB, Potts M, Leslie N, Manokaran N, LaFrankie J, Ashton PS (2000) Species-area curves, spatial aggregation, and habitat specialization in tropical forests. J Theor Biol 207:81–99. doi:10.1006/jtbi.2000.2158
Plotkin JB, Chave J, Ashton PS (2002) Cluster analysis of spatial patterns in Malaysian tree species. Am Nat 160:629–643. doi:10.1086/342823
Reed DH (2005) Relationship between population size and fitness. Conserv Biol 19:563–568. doi:10.1111/j.1523-1739.2005.00444.x
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237. doi:10.1046/j.1523-1739.2003.01236.x
Reis MS, Fantini AC, Nodari RO, Reis A, Guerra MP, Mantovani A (2000) Management and conservation of natural populations in Atlantic rain forest: the case study of palm heart (Euterpe edulis Martius). Biotropica 32:894–902. doi:10.1111/j.1744-7429.2000.tb00627.x
Reitz R, Klein RM, Reis A (1978) Projeto madeira de Santa Catarina. Sellowia 28–30:1–320
Sebbenn AM, Degen B, Azevedo VCR, Silva MB, Lacerda AEB, Ciampi AY, Kanashiro M, Carneiro FS, Thompson I, Loveless MD (2008) Modelling the long-term impacts of selective logging on genetic diversity and demographic structure of four tropical tree species in the Amazon forest. For Ecol Manage 254:335–349. doi:10.1016/j.foreco.2007.08.009
Silva JZ, Puchalski A, Mantovani M, Reis MS (2003) Caracterização genética Preliminar de uma população Natural de Imbuia (Ocotea porosa), uma espécie ameaçada de extinção. In: Anais do 49° Congresso de Genética, Sociedade Brasileira de Genética, Ribeirão Preto
Sokal RR, Oden NL (1978) Spatial autocorrelation in biology: I. Methodology. Biol J Linn Soc Lond 10:199–228. doi:10.1111/j.1095-8312.1978.tb00013.x
Sokal RR, Wartenberg DE (1983) A test of spatial autocorrelation analysis using an isolation-by-distance model. Genetics 105:219–247
Swofford DL, Selander RBA (1997) Biosys-2. A computer program for the analysis of allelic variation genetics. University of Illinois, Urbana
Varty N, Guadagnin DL (1999) Ocotea catharinensis. In: IUCN 2008. 2008 IUCN red list of threatened species, Available in http://www.redlist.org
Vekemans X, Hardy OJ (2004) New insights from fine-scale spatial genetic structure analyses in plant populations. Mol Ecol 13:921–935. doi:10.1046/j.1365-294X.2004.02076.x
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution Int J Org Evolution 38:1358–1370. doi:10.2307/2408641
White GM, Boshier DH, Powell W (1999) Genetic variation within a fragmented population of Swietenia humilis Zucc. Mol Ecol 8:1899–1909. doi:10.1046/j.1365-294x.1999.00790.x
Wright S (1943) Isolation by distance. Genetics 28:114–138
Wright S (1951) The genetical structure of populations. Ann Eugen 15:395–420
Wright S (1965) The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution Int J Org Evolution 19:395–420. doi:10.2307/2406450
Young AG, Boyle TJ (2000) Forest fragmentation. In: Young AG, Boshier D, Boyle TJ (eds) Forest conservation genetics: principles and practice. CSIRO Publishing, Melbourne, pp 123–134
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418. doi:10.1016/0169-5347(96)10045-8
Acknowledgments
We are grateful to the Núcleo de Pesquisas em Florestas Tropicais of Universidade Federal de Santa Catarina (UFSC) for help during the field work; to the Laboratório de Fisiologia do Desenvolvimento e Genética Vegetal of UFSC for providing the infrastructure for allozyme analysis; to Herbário Barbosa Rodrigues, FATMA, MOBASA/BATISTTELLA, and Mr. Antônio Alberton for providing research licence and help in their areas; to professor Dr. Ademir Reis for his botanical knowledge; to Dr. Alexandre M. Sebbenn for the discussion. The authors thank the anonymous reviewers for several important insights and for helpful comments on an earlier version of the manuscript. Roberto Tarazi received a master scholarship from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), Adelar Mantovani received a pos doc scholarship from CNPq (National Council of Technological and Scientific Development) and Maurício Sedrez dos Reis received research productivity scholarships from CNPq. This research article is part of Roberto Tarazi Master’s Dissertation in Plant Genetic Resources of UFSC (Programa de Pós-graduação em Recursos Genéticos Vegetais da UFSC).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tarazi, R., Mantovani, A. & dos Reis, M.S. Fine-scale spatial genetic structure and allozymic diversity in natural populations of Ocotea catharinensis Mez. (Lauraceae). Conserv Genet 11, 965–976 (2010). https://doi.org/10.1007/s10592-009-9939-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-9939-4