Abstract
We used demographic, spatial, and microsatellite data to assess fine-scale genetic structure in Ethiopian wolves found in the Bale Mountains and evaluated the impact of historical versus recent demographic processes on genetic variation. We applied several analytical methods, assuming equilibrium and nonequilibrium conditions, to assess demography and genetic structure. Genetic variation (H E = 0.584–0.607, allelic richness = 4.2–4.3) was higher than previously reported for this species and genetic structure was influenced by geography and social structure. Statistically significant F ST values (0.06–0.08) implied differentiation among subpopulations. STRUCTURE analyses showed that neighbouring packs often have shared co-ancestry and spatial autocorrelation showed higher genetic similarity between individuals within packs and between individuals in neighbouring packs compared to random pairs of individuals. Recent effective population sizes were lower than 2n (where n is the number of packs) and lower than the number of breeding individuals with N e /N ratios near 0.20. All subpopulations have experienced bottlenecks, one occurring due to a rabies outbreak in 2003. Nevertheless, differentiation among these subpopulations is consistent with long-term migration rates and fragmentation at the end of the Pleistocene. Enhanced drift due to population bottlenecks may be countered by higher migration into disease-affected subpopulations. Contemporary factors such as social structure and population bottlenecks are clearly influencing the level and distribution of genetic variation in this population, which has implications for its conservation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Knowledge of population genetic structure can provide information on both historical events and contemporary processes relevant to the conservation and management of endangered species. Since genetic structure is often the consequence of multiple factors and their interactions, the mechanisms underlying the observed level and distribution of genetic variation are not always obvious. For instance, the impact of historical demographic changes (e.g. range contraction or expansion) on population genetic structure or divergence will be influenced by current levels of gene flow resulting from dispersal or mating systems (McRae et al. 2005; Wayne and Koepfli 1996). Specifically, breeding structure can have important effects on fine-scale genetic structure by imposing social barriers to gene flow (Scribner and Chesser 1993) or producing non-random genetic structure consistent with sex-biased dispersal (Peakall et al. 2003). Similarly, habitat fragmentation (Keller and Largiadèr 2002; Riley et al. 2006) and population bottlenecks (Clegg et al. 2002) can have significant impacts on genetic structure. Given this complexity, studies of population genetic structure and the mechanisms by which it is shaped should interrogate different spatial and temporal scales, and demographic processes.
The Ethiopian wolf (Canis simensis) is a rare canid endemic to the Ethiopian highlands (Sillero-Zubiri et al. 2004). The species is highly social, forming cohesive packs of related kin (Randall et al. 2007) that share and defend small, exclusive territories (Sillero-Zubiri and Gottelli 1995b). Their distribution is restricted by a specialized diet on Afroalpine rodents which are confined to altitudes above 3000 m a.s.l. (Sillero-Zubiri and Gottelli 1995a). Using mitochondrial DNA and phylogeographic data, Gottelli et al. (2004) concluded that the species’ current geographic and genetic structure resulted from habitat reduction and fragmentation approximately 10,000–15,000 years ago when late Pleistocene global climatic changes reduced the extent of Afroalpine habitat to which Ethiopian wolves are adapted to a limited number of small, isolated mountain ranges. This coincided with a reduction in population size to probably a few thousand individuals (Gottelli et al. 2004). More recent habitat loss due to the expansion of human agriculture caused a further decline in numbers, resulting in a current global population estimate of approximately 500 individuals in seven small populations in Ethiopia (Marino 2003b).
In this study, we investigate the impact of contemporary demographic processes on the level and distribution of genetic variation among Ethiopian wolves in the Bale Mountains. In particular, we examine the impact of social structure, dispersal and human-induced disease outbreaks on genetic diversity and genetic structure. Social structure and breeding behaviour are important determinants of genetic diversity (Storz 1999) and where population substructure exists, the ‘social’ models introduced by Chesser (1991a, b) may be better predictors of genetic structure than isolation by distance (Wright 1943). Historic or recent population bottlenecks may also promote the loss of genetic variation by reducing effective population sizes (N e) and enhancing genetic drift (Nei et al. 1975; Wright 1931). Possible bottlenecks among Ethiopian wolves in the Bale Mountains caused by outbreaks of disease include at least two, possibly three, rabies outbreaks in the last 20 years (Randall et al. 2006; Randall et al. 2004; Sillero-Zubiri et al. 1996b) as well as at least one, possibly two, recent canine distemper outbreaks (Laurenson et al. 1998, EWCP unpublished data). Typically, these disease outbreaks result in 50–70% mortality in affected subpopulations (Randall et al. 2006). Thus, understanding the interacting effects of social structure and population bottlenecks on the amount and distribution of genetic variation is important for assessing whether conservation actions are necessary to maintain genetic viability and, thus, ensure population persistence (Gilpin and Soulé 1986; Lacy 1997).
To examine the relative impact of historic versus recent events on genetic structure, we assess genetic variation in three Ethiopian wolf subpopulations using 17 microsatellite markers. We apply a Bayesian clustering method to evaluate genetic structure without any a priori assumptions about the spatial distribution of individuals, followed by spatial-autocorrelation analysis to illustrate the relationship between genetic structure and the known geographic distribution of individuals, packs and subpopulations. We also measure levels of differentiation between subpopulations and test for the genetic signatures of bottlenecks. Differentiation between subpopulations may reflect the gradual accumulation of genetic differences due to drift since habitat fragmentation at the end of the Pleistocene. Alternatively, social structure and/or recent localized population bottlenecks may be enhancing genetic differentiation above that predicted from migration–drift equilibrium. We expect both long-term and contemporary processes are interacting to enhance fine scale genetic structure at both the pack and subpopulation level, and test these alternative hypotheses using a range of analytical approaches and tools.
Methods
Study population and sampling
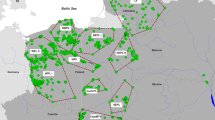
We studied three subpopulations of Ethiopian wolves (Web Valley, WV; Morebawa, MB, and Sanetti, SN) within the Afroalpine massif of the Bale Mountains (Fig. 1). Each subpopulation contained between eight and 16 focal packs whose size, composition and territory boundaries were known from field observations. Pack sizes ranged from two to 11 individuals (>1 year old). Pups were excluded from our analyses to limit bias due to the sampling of close relatives. Territory boundaries were constructed using 95% minimum convex polygons in the Animal Movement extension (Hooge and Eichenlaub 2000) of ARCVIEW 3.2 (Environmental Systems Research Institute, Inc., Redlands, CA, USA). Following Sillero-Zubiri and Gottelli (1995b), we included all breeding season (October to March) locations obtained during focal follows of packs on boundary patrols. Between 40 and 200 locations were obtained for each pack, except three packs that had between 25 and 40 locations, and five packs that had <25 locations. Subpopulation and territory centroids were determined from the average x and y coordinates among GPS locations.
The distribution of pack territories in the Bale Mountains in three subpopulations (WV, MB, SN). Pack ID and cluster number correspond to those in Table 2. Colours indicate one of 10 clusters to which packs were assigned based on the highest average probability of assignment (q) of genotyped individuals. Unfilled polygons indicate focal packs in which no individuals were genotyped in this study. Packs are present throughout wolf habitat in the Bale massif, but we only show the territories of focal packs included in this study
DNA was obtained from both faecal and tissue samples according to Randall et al. (2007). Wolves from two subpopulations (WV and SN) were genotyped for a previous study on the species’ mating system (Randall et al. 2007). For this study on fine-scale population genetic structure, we additionally genotyped wolves from a third subpopulation (MB) and wolves in the Web Valley immediately after a rabies outbreak in 2003 (Randall et al. 2004). The latter samples are collectively designated WV2 to distinguish them from samples collected from the Web Valley before the rabies outbreak (WV). Overall, we carried out analyses using genotypes from 140 wolves present in three subpopulations (43 wolves in WV, 44 wolves in MB, and 53 wolves in SN) and 16 individuals in WV2 (Table 1).
Genotyping
We genotyped individuals for 17 domestic dog microsatellite loci, including 16 tetranucleotide repeat loci (FH2001, FH2054, FH2119, FH2137, FH2138, FH2140, FH2159, FH2174, FH2226, FH2293, FH2320, FH2422, FH2472, FH2537, PEZ17, and PEZ19, in Breen et al. 2001; Neff et al. 1999) and one dinucleotide repeat locus (C05.377, Ostrander et al. 1993). DNA was extracted from faeces using QIAamp® DNA Stool Mini Kits (QIAGEN) and from tissue using QIAamp® DNA Mini Kits (QIAGEN) according to the manufacturer’s protocol. Genotypes were obtained by PCR amplification using the QIAGEN Multiplex PCR Kit with either (i) a fluorescent dye-labelled forward primer, or (ii) a hybrid forward primer consisting of the published forward primer with a M13F (-20) sequence (16 bp) added to the 5’ end and a fluorescent dye-labelled M13F (-20) primer (Boutin-Ganache et al. 2001). The reverse primer was unlabelled. Reactions were performed in 10 μl volumes containing 1.5 μl DNA, 1.0 μl primer mix, 0.4 μl 10 mg/ml BSA, 5.0 μl QIAGEN 10x master-mix, and the remainder ddH2O. Amplifications were performed on a programmable Peltier Thermal Cycler (MJ Research PTC-200) using multiplex cycling profiles for standard dye labelled primers and M13 hybrid primers as follows. Standard dye-labelled primers: 95°C for 15 min; 12 cycles at 94°C for 30 s, 60°C for 90 s dropping by 0.5°C per cycle, and 72°C for 60 s; then 33 cycles at 89°C for 30 s, 55°C for 90 s, and 72°C for 60 s plus a final extension at 60°C for 30 min. M13-labelled primers: 95°C for 15 min; 25 cycles at 94°C for 30 s, 59°C for 90 s, and 72°C for 60 s; then 20 cycles at 94°C for 30 s, 53°C for 90 s, and 72°C for 60 s plus a final extension at 60°C for 30 min. Genotyping reactions were run on a BeckmanCoulter CEQ™ 2000XL automated sequencer. Fragment sizes were scored automatically using CEQ™ 2000 software version 3.0 and checked manually with reference to a size standard. To account for genotyping errors when using non-invasive samples (Pompanon et al. 2005), faecal samples were genotyped using a multiple-tubes approach (Navidi et al. 1992; Taberlet et al. 1996) in which both alleles at a heterozygous locus were amplified at least twice and the single allele of a homozygous locus was amplified at least three times. Single-locus genotypes were treated as missing data if consistent results could not be achieved with the above approach, and samples that produced unreliable genotypes or no PCR product at four or more loci were discarded. Out of 680 faecal samples analysed, 119 yielded usable genotypes. In addition 121 tissue samples all yielded usable genotypes.
Genetic diversity
Genetic diversity was measured as the observed (H O) and expected (H E) heterozygosity (Nei 1987) using CERVUS (Marshall et al. 1998), and per locus allelic richness (El Mousadik and Petit 1996) using FSTAT version 2.9.3 (Goudet 1995). We checked whether our sampling protocol captured total allelic richness within subpopulations with EstimateS version 7.5 (Colwell 2005) using (1) a non-parametric estimator based on the distribution of rare alleles (Chao 1984), and (2) an asymptotic estimate based on extrapolation of moment-based rarefaction curves (Colwell and Coddington 1994). The asymptote (α) of the rarefaction curve was determined using the Michaelis–Menten function y = (αx)/(β + x), where y is the cumulative number of alleles, x is the number of individuals sampled, and β is a constant representing the slope or rate at which the curve becomes asymptotic (Colwell and Coddington 1994). After checking the data for normality, we applied an analysis of variance (ANOVA) to compare H O, H E, and A R between subpopulations. Significant differences in H O, HE and A R before and after the rabies outbreak (WV versus WV2) were tested using paired t-tests. Statistical tests were performed in SPSS 13.0 (SPSS Inc.).
Deviations from Hardy–Weinberg equilibrium (HWE) and linkage disequilibrium (LD) were assessed using GENEPOP version 3.2 (Raymond and Rousset 1995) with an adjusted P-value corresponding to alpha = 0.05 (α = 0.0004) after Bonferroni correction (Rice 1989). The level of inbreeding was measured using the inbreeding coefficient F IS. Global tests of heterozygote deficit were conducted for each subpopulation with critical values corrected for four comparisons (α = 0.0125), each locus within subpopulations with critical values corrected for 17 comparisons (α = 0.0029), and for each locus overall subpopulations with critical values corrected for 68 comparisons (α = 0.0007).
Genetic differentiation and structure
We measured genetic differentiation between subpopulations using Weir and Cockerham’s (1984) estimator of F ST in FSTAT version 2.9.3 (Goudet 1995). Statistical significance of pairwise F ST values were tested by 1,000 randomizations of genotypes among subpopulations (Goudet et al. 1996) using adjusted P-values corresponding to α = 0.05 after Bonferroni correction (Rice 1989). We also applied a hierarchical analysis of molecular variance (AMOVA, Excoffier et al. 1992) in GENALEX (Peakall and Smouse 2006) using F ST values to partition total variance at different levels of population structure and testing for significance by 1,000 permutations. To assess the influence of poorly sampled packs on these statistics, we ran separate analyses restricted to (i) packs with more than one genotyped individual, and (ii) packs with four or more genotyped individuals.
We implemented a Bayesian clustering method using STRUCTURE 2.1 (Pritchard et al. 2000) to infer the number of genetic clusters (K) without any a priori assumptions about sample location. The stability of the inferred clusters was evaluated using three independent runs at K = 1–20 with a burn-in period of 50,000 steps followed by 500,000 Markov chain Monte Carlo (MCMC) cycles. Summary statistics (log likelihood and alpha) were monitored to verify convergence during burn-in. We ran STRUCTURE assuming correlated allele frequencies and admixture to account for gene flow and mixed ancestry within the population. STRUCTURE calculates the log likelihood of the data (Ln P(X∣K)), for each K value and the probability of membership (q) in a cluster for each individual. We examined individual cluster assignments as a function of K (progressive hierarchical clustering pattern analysis) in comparison with the value for Ln P(X∣K) for each K value and our biological knowledge of the Ethiopian wolf system (i.e. social structure, packs and subpopulations) to determine the most true number of clusters to use for subsequent analyses (e.g. migrant identification).
Gene flow and migration
We assessed gene flow by spatial-autocorrelation using GENALEX version 6 (Peakall and Smouse 2006). The autocorrelation coefficient, r, estimates the genetic similarity between individuals within each geographic distance class. Geographic distances were calculated as the Euclidian distance between pack centroids (i.e. average x and y coordinates) for each comparison pair with distance classes allocated (0, 3, 6, 9, 12, 15, 20, 30 and 40 km) so that sample sizes were similar. The significance of the correlation was determined using 95% confidence intervals (CIs) around the null hypothesis of ‘no spatial genetic structure’ based on 1,000 randomization of individuals among distance classes. Bootstrapping was used to estimate the 95% CIs for estimates of r. Adult females and adult males were also tested separately to assess gene flow that may result from sex-biased dispersal.
Long-term migration between subpopulations was estimated in MIGRATE 2.4 (Beerli and Felsenstein 1999). The program estimates θ (4N e μ, where N e is the effective population size and μ is the mutation rate) and M (m/μ, where m is the unscaled migration rate independent of mutation) assuming equilibrium conditions and constant θ and M values. We used 10 Markov chains of 10,000 steps and three chains of 100,000 steps and an adaptive heating scheme (temperatures 1.0, 1.2, 1.5, 3.0). Runs were repeated until the confidence intervals for the posterior probabilities of θ and M overlapped and using prior estimates of θ and M as starting parameters for subsequent runs (four runs were sufficient and we report θ and M estimates for the final run). Long-term migration rates (m MIG) were estimated from M (Palstra et al. 2007; Turner et al. 2002).
Recent migration (over the last several generations) was estimated using a Bayesian MCMC analysis in BayesAss 1.3 (Wilson and Rannala 2003). Individuals were pre-assigned to subpopulations based on sampling location and the program returned means of the posterior probabilities for migration rates (m BAY) into each subpopulation. BayesAss uses a non-equilibrium method and does not assume HWE within subpopulations. Values were averaged from four runs using default parameters on the first run and different random seed and delta values for subsequent runs to check for consistency in the results (Austin et al. 2004). The confidence intervals derived by BAYESASS were compared to those that occur when there is no information in the data to determine if m BAY results are reliable.
First generation immigrants were identified using posterior probability assignment analysis in STRUCTURE with individuals pre-assigned to a cluster based on the highest average q value for all members of the same pack. Individuals were considered long-distance immigrants if their highest posterior probability assignment was >0.5 in a cluster from a different subpopulation and local immigrants if their highest posterior probability was >0.5 in a cluster from the same subpopulation.
Effective population size
We obtained estimates of long-term effective population size (N e ) for each subpopulation based on θ = 4N e μ, using estimates of θ and the average mutation rate of 10−2 for canid tetranucleotide microsatellites (Francisco et al. 1996). Francisco et al. (1996) evaluated over 3,000 meioses to quantify the mutation rate for the microsatellite loci used in this study and thus we feel that this mutation rate is robust. Long-term N e derived from a constant θ value may not reflect recent demography and breeding patterns. Therefore, estimates of current N e were made with Hill’s (1981) linkage disequilibrium (LD) method and Pudovkin et al.’s (1996) heterozygote excess method using N e Estimator v 1.3 (Peel et al. 2004). Hill’s method estimates N e by assessing LD and the correlation among alleles at different loci (r) in a sample. Pudovkin’s method is based on the heterozygote-excess principle where the allele frequency in males and females will be different due to binomial sampling error for small numbers of breeders. This difference generates an excess of heterozygotes in progeny compared to the proportion of heterozygotes expected under Hardy–Weinberg equilibrium. Thus, Pudovkin et al. (1996) showed that heterozygote excess can be used to estimate the number of breeding individuals. Both methods were selected because they do not require temporal data. Furthermore, the LD method delivers reliable estimates when the sample is the same or larger than the true N e (England et al. 2006), which is the case in this study.
Demographic history
To assess whether migration rates are consistent with the level of differentiation, we used the coalescent-based model of genetic diversity implemented in SIMCOAL 1.0 (Excoffier et al. 2000) to simulate genetic differentiation of three interconnected subpopulations under different demographic scenarios assuming equilibrium conditions. The program uses a stepwise mutation model (SMM) to predict pairwise F ST values based on the effective population size (N e ), sample size, migration rate (m), mutation rate (μ), and the time since fragmentation of our subpopulations. Information on the long-term effective population sizes and sample sizes allowed us to limit the number of parameters in our model. We varied N e at the time of divergence from 25 to 8,000 based on maximum population sizes (N) estimated by Gottelli et al. (Gottelli et al. 2004) for the last glacial age and maximum N e /N ratios estimated in this study (see below). Varying N e and sample size had little effect on simulated F ST values (data not shown). We used a μ of 10−2 (Francisco et al. 1996) and migration rates from 0 to 20%. The number of generations was varied from 350 to 3,500 to reflect separation times equivalent to 1,000–1,500 and 10,000–15,000 years ago. This broadly estimates the time since habitat fragmentation may have given rise to the current spatial and genetic structure in the Bale Mountains, assuming a 3–4 year generation time. We ran 500 simulations for each set of parameters and calculated average F ST using ARLEQUIN 2.000 (Schneider et al. 2000).
We tested each subpopulation and WV2 for evidence of a recent bottleneck using BOTTLENECK 1.1 (Piry et al. 1999) and the M ratio test (Garza and Williamson 2001). Both methods can be used to detect a recent bottleneck with a single representative sample of genetic diversity. BOTTLENECK tests for a significant heterozygosity excess compared to equilibrium expectations for a stable population based on the assumption that population reductions cause rare alleles to be lost faster than genetic diversity, resulting in a transient heterozygosity excess compared to the observed number of alleles (Cornuet and Luikart 1996). We applied the two-phase mutation model (TPM), which is considered appropriate for microsatellites (Di Rienzo et al. 1994), with a variance of 12 (Piry et al. 1999) and different percentages of the step-wise mutation model (SMM: 70, 80, 90%). We ran 1,000 iterations and tested significance with the Wilcoxon signed-rank test recommended by Maudet et al. (2002).
Garza and Williamson (2001) showed that the mean ratio (M ratio) of the number of alleles to the range in allele size is indicative of the demographic history of a population. In bottlenecked populations, the loss of rare alleles at intermediate allele sizes results in gaps in the frequency distribution of alleles and M ratios smaller than 1.0. We used the values suggested by the authors for the proportion of one-step mutations (p s = 90%) and average size of multi-step mutations (Δg = 3.5). Critical values were determined for θ (4N eμ) corresponding to pre-bottleneck effective population sizes (N e) of 250, 500 and 1,000. The program uses a fixed mutation rate (μ) of 5 × 10−4 per locus per generation, which is lower than the mutation rate for canid tetranucleotide loci (Francisco et al. 1996). Since bottlenecks would typically be under-supported by a lower mutation rate, the method is considered conservative for detecting significant deviations from equilibrium expectations in our data. Statistical significance of the observed ratio was tested by 10,000 simulations.
Finally, we assessed whether the subpopulations in the Bale Mountains are in equilibrium with the program 2mod (Ciofi et al. 1999) to test the likelihood of two models (immigration–drift–equilibrium versus non-equilibrium drift only). In the immigration–drift–equilibrium model, the degree of differentiation is a measure of the relative strength of drift versus immigration (Rannala and Hartigan 1996), while in the non-equilibrium drift model it is related to the time since fragmentation (scaled by population size) (O’Ryan et al. 1998). 2mod first estimates the probability of obtaining each dataset assuming that the mutation rate is much smaller than the immigration rate in the gene flow model, and that the reciprocal of the mutation rate is much longer than the divergence time in the drift model. 2mod then applies a Markov Chain Monte Carlo (MCMC) method to estimate the relative posterior probabilities of the two models.
Results
Genetic diversity
Twenty pairs of loci showed significant linkage disequilibrium (LD) after Bonferroni correction (P < 0.05), but only one pair was significant in more than one subpopulation. Since LD was not consistent across subpopulations, significant values are probably due to population structure or genetic drift. Only two pairs (FH2422/FH2537 in one subpopulation and FH2472/Pez17 in three subpopulations) were located on the same chromosome but are 56.9 and 8.4 Mb apart, respectively (Breen et al. 2001), thus LD is unlikely to be due to physical linkage.
All 17 loci were polymorphic across subpopulations with 3–10 alleles observed per locus. Expected heterozygosity (H E) ranged from 0.584 to 0.609 and mean per locus allelic richness (A R) ranged from 4.79 to 4.81 (Table 1). Observed heterozygosity (H O), H E, and A R were not significantly different among subpopulations, nor were they significantly different in the Web Valley before and after the rabies outbreak. Despite this result, four of the six alleles unique to WV were not present in WV2 individuals sampled after the outbreak. No heterozygote deficiencies were detected for any locus or subpopulation.
A total of 100 different alleles were detected in 140 individuals across subpopulations, of which 16 were alleles unique to a single subpopulation. WV had 6 of 16 (37.5%) unique alleles, MB had 4 of 16 (25.0%), and SN had 6 of 16 (37.5%). Unique alleles had an average frequency of 0.016. Our moment-based rarefaction curves show the same cumulative number of alleles in each subpopulation when corrected to the smallest sample size suggesting our samples are a reliable representation of total allelic diversity (Fig. S1). The asymptote and rare allele methods of estimating the total number of alleles in each subpopulation and WV2 also gave estimates roughly similar to the observed number of alleles in our samples (Table 1).
Genetic structure
All pairwise comparisons of F ST showed statistically significant genetic differentiation between subpopulations (WV–MB, F ST = 0.06; WV–SN, F ST = 0.08; MB–SN, F ST = 0.06; all P < 0.001). On the other hand, WV and WV2 were not significantly differentiated (F ST = 0.00). All variance components tested by AMOVA were significantly different from zero indicating considerable partitioning of genetic variation among packs and subpopulations. The majority of genetic variation existed within packs (83%, P < 0.001), however, the variation components among packs within subpopulations (12%) and among subpopulations (5%) were also statistically significant (P < 0.001; Table S1). These results changed little when the analysis was restricted to packs with more than one genotyped individual (n = 27) or packs with four or more genotyped individuals (n = 20, Table S1).
Hierarchical clustering pattern analysis of STRUCTURE results resolved the three subpopulations at K = 3 and revealed distinct substructure for the 29 packs, with the ultimate number of discernable genetic clusters (ten) matching the highest mean likelihood value of K = 10 (Ln = −4900.7, Table S2). In some cases, individual assignment probabilities (q) grouped all individuals in a given pack to the same cluster (n = 50 individuals in 9 of 27 packs with >1 individual genotyped, Table 2) and average pack assignments typically grouped one or more neighbouring packs to the same cluster (n = 18 packs). While many packs showed genetic admixture indicative of mixed ancestry, our results also imply stable lineages in some packs and, in many cases, shared co-ancestry between adjacent packs (Fig. 1).
Gene flow, migration, and effective population sizes
The spatial-autocorrelation (adults and yearlings) produced significant positive correlation coefficients in distance classes of 0 km (r = 0.349, P = 0.001), 3 km (r = 0.120, P = 0.001), and 6 km (r = 0.040, P = 0.001, Fig. 2a). Given the mean distance between centroids of adjoining territories (mean = 2.7 km, range = 1.0–5.9, for territories with >40 GPS locations) these results indicated higher genetic similarity between individuals within packs and, in some cases, between individuals in neighbouring packs compared to random pairs of individuals. A positive correlation was observed up to 8.5 km. This ‘patch width’ is less than the minimum distance between subpopulation centroids (WV–MB = 14 km, WV–SN = 27 km, MB–SN = 31 km), and could be used to define conservation units in the Bale Mountains Ethiopian wolf population (Aspi et al. 2006; Diniz-Filho and Telles 2002). Significant negative values were observed at distances equal to and greater than 12 km (r = −0.015, P < 0.05). The pattern is similar for males, indicating higher than average relatedness between adult males within packs (0 km, r = 0.364, P = 0.001) and significant positive values up to 6 km (r = 0.046, P = 0.001) but significantly negative values at distances equal to and greater than 9 km (r = −0.068, P < 0.001, Fig. 2b). Genetic similarity was significantly correlated between females in the same pack (0 km, r = 0.437, P = 0.001) and neighbouring packs (3 km, r = 0.122, P = 0.05). However, genetic structure was not detected among females at distances greater than ca. 6 km (Fig. 2c). These results suggest that gene flow is mediated by both males and females over short distances (i.e. between 6 and 8 km within subpopulations), but gene flow over larger distances (i.e. >9 km and, thus, between subpopulations) is attributed to females.
Spatial autocorrelation of genetic distance and geographic distance for a adults and yearlings, b adult males, and c adults females. Zero km identifies comparisons between individuals in the same pack. Upper (U) and lower (L) 95% CI for the null hypothesis of “no structure” are shown as are error bars for bootstrap values for 95% CIs of r
Recent estimates of N e were WV = 11.9–13.2, SN = 10.2–11.4, and MB = 22.6–25.2, implying a ratio of 0.17–0.22 for recent N e /N within subpopulations (Table 3). These N e values are lower than 2n (where n is number of packs) and number of breeding pairs in WV (16–18) and SN (14–15) when extra-pair copulations and multiple paternity are included (Randall et al. 2007). MIGRATE estimates of θ compatible with the observed level of genetic diversity (as measured by H E) were 1.52 for WV, 1.84 for MB, and 1.75 for SN. Using the equation θ = 4N e μ and an average mutation rate of 10−2, N e was 38 for WV, 46 for MB, and 44 for SN (Table 3). Estimates of θ were robust to the mutation model used (Table S3) and imply long-term N e /N ratios between 0.46 and 0.72 (Table 3).
The mean long-term migration rate (m MIG) estimated using M values returned by MIGRATE and the relation M = m/μ was 0.029. We observed asymmetrical migration between subpopulations, as evidenced by non-overlapping 95% CIs (Fig. 3). Migration rates from MB to both other subpopulations were higher than vice versa and migration rate from SN to WV was higher than vice versa (Fig. 3). Migration rates estimated in BayesAss were not different from those derived when there was no information in the data, implying that recent migration rates derived with BayesAss are not reliable for comparison with long-term migration rates. Posterior probability assignments in STRUCTURE identified eleven local (7 female, 4 male) and four (1 female, 3 male) long-distance first generation immigrants using the >0.5 posterior probability assignment criteria. All immigrants were adults. Five of the females and all four of the males identified as local migrants had within-subpopulation cluster assignments to other than their sampled pack of ≥0.93, while two females had assignments of 0.72 and 0.75. The female identified as a long distance migrant had a different subpopulation cluster assignment of 0.97, while the three males identified as long distance migrants had different subpopulation assignments of 0.92, 0.83 and 0.80.
Demographic history
Coalescent simulations suggested that a long-term effective migration rate of 0.029 is consistent with the observed F ST values between subpopulations (0.06–0.08), irrespective of the time since fragmentation (Fig. 4). Effective migration rates below 0.029 increase the simulated F ST value to >0.08. Thus, the observed level of drift is predicted by a mutation–migration–drift model.
Predicted F ST values for different number of years since fragmentation using coalescent simulations in SIMCOAL. Error bars are standard errors for the mean of pairwise values. Our empirical long-term migration rate of 0.029 produces average simulated F ST values consistent with our observed F ST values (0.06–0.08)
None of the tests for significant heterozygote excess in subpopulations were significant (P > 0.05, Table 4), but WV2 showed a significant heterozygote excess (P = 0.006–0.039 for three different percentages of SMM, Table 4). In contrast, M ratios in all subpopulations and WV2 were significantly lower than expected under mutation–drift equilibrium for pre-bottleneck effective population sizes even as large as 500 (P < 0.05, Table 4). WV2 had the smallest M ratios. A nonequilibrium drift-only model was supported in 97% of the tests run in 2mod.
Discussion
Our results reveal strong genetic structure in a population that has been in decline for thousands of years (Gottelli et al. 2004) as well as having suffered more recent bottlenecks due to disease outbreaks. Not surprisingly then, the Ethiopian wolf population in the Bale Mountains is not in equilibrium. Nonetheless, our application of different analytical methods provides varied perspectives through which we have assessed genetic signals and genetic structure (Austin et al. 2004; Hickerson and Cunningham 2005; Pearse et al. 2006). The consistency in the patterns observed using these complementary approaches suggests that our results are not affected by violations of equilibrium assumptions (Hickerson and Cunningham 2005; Jones et al. 2004).
Genetic diversity, genetic structure, and sex-biased dispersal
Levels of genetic diversity were similar in each subpopulation (H E 0.58–0.69, A 4.2–4.3), and higher than those previously published for Ethiopian wolves in the Bale Mountains (H E 0.24–0.38, A 2.0–2.1, Gottelli et al. 1994). However, such comparisons are of uncertain significance when different markers are analysed since polymorphism will vary among loci. For example, mean estimated H E in Mediterranean monk seals varied from 0.16 to 0.41 depending on whether monomorphic loci were included (Pastor et al. 2004). We used tetranucleotide repeat loci preferentially because they are typically more polymorphic than dinucleotide repeat markers (Mellersh et al. 2000).
Our study population showed high levels of genetic structuring with significant genetic partitioning at both pack and subpopulation levels. The largest amount of variation is found within packs (83%) in our AMOVA analysis. In nine of 29 packs, all genotyped individuals in a given pack were assigned to the same cluster by STRUCTURE, while genotyped individuals from 18 packs were assigned to the same cluster of neighbouring packs (Table 2, Fig. 1).
Assignment tests identified both female and male dispersers which questions prior notions about complete male philopatry in this species (Sillero-Zubiri et al. 1996a). Although males exhibit long-distance migration, significantly negative spatial autocorrelations at distances greater than 9 km imply that post-dispersal breeding is rare or absent. Female-biased gene flow was evident over short and long distances, although short-range dispersal to neighbouring packs appears to be more common. There was also high genetic similarity among males within the same or neighbouring packs, supporting habitual male philopatry but some male-mediated gene flow within subpopulations probably due to extra-pack paternity and occasional pack fission (Marino et al. 2006; Randall et al. 2007; Sillero-Zubiri et al. 2004).
Effective population sizes, bottlenecks and migration
Recent N e estimates produced N e /N ratios between 0.17 and 0.25 and in all cases, N e was lower than the number of breeding pairs determined from parentage analyses (Randall et al. 2007). In this population, limited dispersal, inbreeding, and differential breeding success among individuals (or breeding units) might reduce N e below that predicted from the number of breeding individuals (Chesser 1991b; Sugg and Chesser 1994). Migrate returned unexpectedly high N e /N ratios given the breeding behaviour of the species. These higher historical values likely result from a transient heterozygote excess above that predicted from the number of alleles found in a constant-sized subpopulation (Cornuet and Luikart 1996). Such a heterozygote excess can be explained by a loss of rare alleles during population bottlenecks (Nei et al. 1975; Cornuet and Luikart 1996) and has been used to explain unexpectedly high N e estimates in other studies (Aspi et al. 2006; Goossens et al. 2005; Storz et al. 2002). M-ratios indicative of one or more recent bottlenecks were evident in all three subpopulations. All subpopulations have been declining over several thousands of years (Gottelli et al. 2004), thus the differences between long-term N e /N ratios for each of the three subpopulations (WV = 0.62, MB = 0.46, SN = 0.72) is likely a discrepancy in the relative frequency, severity and or timing of more recent bottlenecks due to disease outbreaks in each of the three subpopulations. This is supported by field observations that recorded a 70% decline in wolf numbers in WV in 1992 and a 50% decline in wolf numbers in SN in 1990 due to rabies outbreaks (Sillero-Zubiri et al. 1996b). Regular monitoring of the MB subpopulation began only in 2002, but no disease outbreaks have been recorded through 2008, whereas further outbreaks have occurred in WV (rabies outbreaks in 2003 and 2008, Randall et al. 2004, EWCP unpublished data) and SN (CDV outbreak in 2005, EWCP unpublished data) suggesting Ethiopian wolves in these subpopulations are more susceptible to disease outbreaks.
Only WV2 showed significant heterozygote excess consistent with a reduction in effective population size and loss of rare alleles during the 2003 rabies disease outbreak (Randall et al. 2004). However, M ratios in all subpopulations and WV2 were significantly lower than predicted under equilibrium (0.75–0.81, P < 0.05). It is not uncommon for population bottlenecks to go undetected with certain genetic methods (Busch et al. 2007), and the power of the BOTTLENECK method may be limited to only very recent or especially prolonged reductions in N e (Cornuet and Luikart 1996). Our M ratios imply localized bottlenecks in all subpopulations within the last 500 years or less (Garza and Williamson 2001). This is in agreement with two contemporary processes in the Bale Mountains: (i) a loss of habitat to agricultural expansion in the last few decades, implying proportional reductions in population size (Marino 2003b), and (ii) dramatic, localized subpopulation declines associated with outbreaks in the last 20 years (Marino et al. 2006; Randall et al. 2006; Randall et al. 2004; Sillero-Zubiri et al. 1996b). Two lines of evidence suggest that disease outbreaks are principally responsible for the observed genetic bottlenecks: (i) the 2003 rabies outbreak resulted in a loss of rare alleles and an excess of heterozygosity that was not evident in the Web Valley subpopulation before the outbreak, and (ii) long-term N e /N estimates suggest the highest heterozygosity excesses occur in WV and SN, two subpopulations that appear most affected by disease according to field observations.
The observed F ST values (0.06–0.08) are consistent with simulated F ST values derived from a simple mutation–migration–drift model in SIMCOAL based on the mean long-term migration rate of 0.029. This is surprising given that the population is not in equilibrium. All subpopulations have suffered recent bottlenecks that, theoretically, reduce effective population sizes for several generations such that differentiation is enhanced (Garza and Williamson 2001). However, asymmetric migration rates between subpopulations also suggest relatively higher migration rates from more stable subpopulations into subpopulations more adversely affected by disease (MB → WV, MB → SN, and SN → WV). This pattern of migration may have been critical for maintaining Nm at a level that minimized drift due to bottlenecks.
Conservation implications
Ethiopian wolf subpopulations in the Bale Mountains are sufficiently distinct genetically to be classified as separate management units (Moritz 1995). It is therefore worrying that all subpopulations have suffered recent bottlenecks due to disease outbreaks. Much effort has been devoted to understanding the dynamics of disease in Ethiopian wolf populations and devising effective disease control measures (Haydon et al. 2002; Haydon et al. 2006; Knobel et al. 2008; Laurenson et al. 1997; Randall et al. 2006). In this study, we demonstrated that disease outbreaks may also be having an impact on the genetic viability of populations through the clear loss of rare alleles.
Following disease outbreaks, the erosion of genetic diversity will be determined by the number and genetic composition of breeding individuals within packs (Crow and Kimura 1970) and the number of generations required for population recovery (Nei et al. 1975). Since only the dominant female in each pack reproduces (Sillero-Zubiri et al. 1996a; Randall et al. 2007), Marino et al. (2006) showed that population recovery is determined by the number of breeding units (i.e. packs). Chesser’s (1991a) social models further suggest that maintaining the stability of genetic lineages within social groups (by protecting the incumbent breeding pair) will also maintain genetic diversity. Therefore, vaccination campaigns that target the dominant breeding female should assist demographic recovery (Haydon et al. 2002; Knobel et al. 2008) as well as preserve genetic diversity.
Furthermore, other studies have demonstrated that single bottlenecks (or ‘founder events’) may have minimal impact on genetic diversity or differentiation, whereas successive bottlenecks can substantially increase genetic differentiation and reduce genetic diversity (Clegg et al. 2002; Pruett and Winker 2005). Thus, the loss of genetic diversity may be exacerbated by the cumulative effect of recurring outbreaks. A detailed exploration of the future genetic viability of Ethiopian wolf populations is currently being undertaken using simulation modelling; particularly assessing the impact of recurring disease outbreaks and the need, if any, for overt genetic management.
Given the likelihood of future disease outbreaks, a metapopulation structure with suitable dispersal corridors appears to be vital for counteracting drift (this study) and maintaining population persistence (Haydon et al. 2006). However, habitat loss and disturbance due to increasing human, livestock and domestic dog density in the Bale Mountains massif (Marino 2003a) threatens the integrity of the Ethiopian wolf metapopulation. Consequently, conservation and management policies are urgently needed that exclude or strictly limit human activity and domestics dogs in the Bale Mountains and elsewhere where Ethiopian wolves persist in Ethiopia.
References
Aspi J, Roininen E, Ruokonen M, Kojola I, Vilà C (2006) Genetic diversity, population structure, effective population size and demographic history of the Finnish wolf population. Mol Ecol 15:1561–1576
Austin JD, Lougheed SC, Boag PT (2004) Controlling for the effects of history and nonequilibrium conditions in gene flow estimates in northern bullfrog (Rana catesbeiana) populations. Genetics 168:1491–1506
Beerli P, Felsenstein J (1999) Maximum likelihood estimation of migration rates and effective population numbers in two populations using a coalescent approach. Genetics 152:763–773
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31:24–28
Breen M, Jouquand S, Renier C et al (2001) Chromosome-specific single-locus FISH probes allow anchorage of an 1800-marker integrated radiation-hybrid/linkage map of the domestic dog genome to all chromosomes. Genome Res 11:1784–1795
Busch JD, Waser PM, DeWoody A (2007) Recent demographic bottlenecks are not accompanied by a genetic signature in banner-tailed kangaroo rats (Dipodomys spectabilis). Mol Ecol 16:2450–2462
Chao A (1984) Non-parametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Chesser RK (1991a) Gene diversity and female philopatry. Genetics 127:437–447
Chesser RK (1991b) Influence of gene flow and breeding tactics on gene diversity within populations. Genetics 129:573–583
Ciofi C, Beaumont MA, Swingland IR, Bruford MW (1999) Genetic divergence and units for conservation in the Komodo dragon Veranus komodoensis. Proc R Soc Lond B 266:2269–2274
Clegg SM, Degnan SM, Kikkawa J et al (2002) Genetic consequences of sequential founder events by an island-colonizing bird. Proc Natl Acad Sci USA 99:8127–8132
Colwell RK (2005) EstimateS: statistical estimation of species richness and shared species from samples. Version 7.5. User’s Guide and application published at: http://purl.oclc.org/estimates
Colwell RK, Coddington JA (1994) Estimating terrestrial biodiversity through extrapolation. Philos Trans R Soc Lond B 345:101–118
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent populations bottlenecks from allele frequency data. Genetics 144:2001–2014
Crow JF, Kimura M (1970) An introduction to population genetics theory. Harper and Row, New York
Di Rienzo A, Peterson AC, Garza JC et al (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Diniz-Filho J, Telles M (2002) Spatial autocorrelation analysis and the identification of operational units for conservation in continuous populations. Conserv Biol 16:924–935
El Mousadik A, Petit E (1996) High level of genetic differentiation for allelic richness among populations of the argan tree (Argania spinosa (L.) Skeels) endemic to Morroco. Theor Appl Genet 92:832–839
England P, Cornuet J, Berthier P, Tallmon D, Luikart G (2006) Estimating effective population size from linkage disequilibrium: severe bias in small samples. Conserv Genet 7:303–308
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Excoffier L, Novembre J, Schneider S (2000) SIMCOAL: a general coalescent program for the simulation of molecular data in interconnected populations with arbitrary demography. J Hered 91:506–509
Francisco LV, Langston AA, Mellersh CS, Neal CL, Ostrander EA (1996) A class of highly polymorphic tetranucleotide repeats for canine genetic mapping. Mamm Genome 7:359–362
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Gilpin ME, Soulé ME (1986) Minimum viable populations: processes of species extinction. In: Soulé ME (ed) Conservation biology: the science of scarcity and diversity. Sinauer Associates, Sunderland, MA, pp 13–34
Goossens B, Chikhi L, Jalil MF et al (2005) Patterns of genetic diversity and migration in increasingly fragmented and declining orang-utan (Pongo pygmaeus) populations from Sabah, Malaysia. Mol Ecol 14:441–456
Gottelli D, Sillero-Zubiri C, Applebaum GD et al (1994) Molecular genetics of the most endangered canid: the Ethiopian wolf Canis simensis. Mol Ecol 3:301–312
Gottelli D, Marino J, Sillero-Zubiri C, Funk SM (2004) The effect of the last glacial age on speciation and population genetic structure of the endangered Ethiopian wolf (Canis simensis). Mol Ecol 13:2275–2286
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Goudet J, Raymond M, DeMeeus T, Rousset F (1996) Testing differentiation in diploid populations. Genetics 144:1933–1940
Haydon DT, Laurenson MK, Sillero-Zubiri C (2002) Integrating epidemiology into population viability analysis: managing the risk posed by rabies and canine distemper to the Ethiopian wolf. Conserv Biol 16:1372–1383
Haydon DT, Randall DA, Mathews L et al (2006) Low-coverage vaccination strategies for the conservation of endangered species. Nature 443:692–695
Hickerson MJ, Cunningham CW (2005) Contrasting Quaternary histories in an ecologically divergent sister pair of low-dispersing intertidal fish (Xiphister) revealed by multilocus DNA analysis. Evolution 59:344–360
Hill WG (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res 38:209–216
Hooge PN, Eichenlaub B (2000) Animal movement extension to Arcview version 2.0. In Alaska Science Center—Biological Science Office, U.S. Geological Survey, Anchorage, AK, USA
Jones ME, Paetkau D, Geffen E, Moritz C (2004) Genetic diversity and population structure of Tansmanian devils, the largest marsupial carnivore. Mol Ecol 13:2197–2209
Keller I, Largiadèr CR (2002) Recent habitat fragmentation caused by major roads leads to reduction of gene flow and loss of genetic variability in ground beetles. Proc R Soc Lond B 270:417–423
Knobel D, Fooks AR, Brookes S et al (2008) Trapping and vaccination of endangered Ethiopian wolves to control an outbreak of rabies. J Appl Ecol 45:109–116
Lacy RC (1997) Importance of genetic variation to the viability of mammalian populations. J Mammal 78:320–335
Laurenson K, Shiferaw F, Sillero-Zubiri C (1997) Disease, domestic dogs and the Ethiopian wolf: the current situation. In: Sillero-Zubiri C, Macdonald DW (eds) The Ethiopian wolf: status survey and conservation action plan. IUCN, Gland, Switzerland and Cambridge, UK, pp 32–40
Laurenson MK, Sillero-Zubiri C, Thompson H et al (1998) Disease threats to endangered species: Ethiopian wolves, domestic dogs, and canine pathogens. Anim Conserv 1:273–280
Marino J (2003a) The spatial ecology of the Ethiopian wolf, Canis simensis. DPhil thesis, University of Oxford
Marino J (2003b) Threatened Ethiopian wolves persist in small isolated Afroalpine enclaves. Oryx 37:62–71
Marino J, Sillero-Zubiri C, Macdonald DW (2006) Trends, dynamics and resilience of an Ethiopian wolf population. Anim Conserv 9:49–58
Marshall T, Slate J, Kruuk L, Pemberton J (1998) Statistical confidence for likelihood-based paternity inference in natural populations. Mol Ecol 7:639–655
Maudet C, Miller C, Bassano B et al (2002) Microsatellite DNA and recent statistical methods in wildlife conservation management: applications in Alpine ibex [Capra ibex (ibex)]. Mol Ecol 11:421–436
McRae BH, Beier P, Dewald LE, Huynh LY, Keim P (2005) Habitat barriers limit gene flow and illuminate historical events in a wide-ranging carnivore, the American puma. Mol Ecol 14:1965–1977
Mellersh CS, Hitte C, Richman M et al (2000) An integrated linkage-radiation hybrid map of the canine genome. Mamm Genome 11:120–130
Moritz C (1995) Defining ‘evolutionary significant units’ for conservation. Trends Ecol Evol 9:373–375
Navidi W, Arnheim N, Waterman MS (1992) A multiple-tube approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am J Hum Genet 50:347–359
Neff MW, Broman KW, Mellersh CS et al (1999) A second-generation genetic linkage map of the domestic dog, Canis familiaris. Genetics 151:803–820
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
O’Ryan C, Harley EH, Bruford MW et al (1998) Microsatellite analysis of genetic diversity in fragmented South African buffalo populations. Anim Conserv 1:85–95
Ostrander EA, Sprague GF, Rine J (1993) Identification and characterization of dinucleotide repeat (CA)n markers for genetic mapping in dog. Genomics 16:207–213
Palstra FP, O’Connell F, Ruzzante DE (2007) Population structure and gene flow reversals in Atlantic salmon (Salmo salar) over contemporary and long-term temporal scales: effects of population size and life history. Mol Ecol 16:4504–4522
Pastor T, Garza JC, Allen P, Amos W, Aguilar A (2004) Low genetic variability in the highly endangered Mediterranean monk seal. J Hered 95:291–300
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peakall R, Ruibal M, Lindenmayer DB (2003) Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution 57:1182–1195
Pearse DE, Arndt AD, Velenzuela N et al (2006) Estimating population structure under nonequilibrium conditions in a conservation context: continent-wide population genetics of the giant Amazon river turtle, Podocnemis expansa (Chelonia; Podocnemididae). Mol Ecol 15:985–1006
Peel D, Ovenden JR, Peel SL (2004) NeEstimator: software for estimating effective population size, Version 1.3. In Queensland Government, Department of Primary Industries and Fisheries
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pompanon F, Bonin A, Bellemain E, Taberlet P (2005) Genotyping errors: causes, consequences and solutions. Nat Rev Genet 6:847–859
Pritchard JK, Stephens M, Donnelly PJ (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Pruett CL, Winker K (2005) Northwestern song sparrow populations show genetic effects of sequential colonization. Mol Ecol 14:1421–1434
Pudovkin AI, Zaykin DV, Hedgecock D (1996) On the potential for estimating the effective number of breeders from heterozygote-excess in progeny. Genetics 144:383–387
Randall DA, Williams SD, Kuzmin IV et al (2004) Rabies in endangered Ethiopian wolves. Emerg Infect Dis 10:2214–2217
Randall DA, Marino J, Haydon DT et al (2006) An integrated disease management strategy for the control of rabies in Ethiopian wolves. Biol Conserv 131:151–162
Randall DA, Pollinger JP, Wayne RK et al (2007) Inbreeding is reduced by female-biased dispersal and mating behavior in Ethiopian wolves. Behav Ecol 18:579–589
Rannala B, Hartigan JA (1996) Estimating gene flow in island populations. Genet Res 67:147–158
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Riley SPD, Pollinger JP, Sauvajot RM et al (2006) A southern California freeway is a physical and social barrier to gene flow in carnivores. Mol Ecol 15:1733–1741
Schneider S, Roessli D, Excoffier L (2000) ARLEQUIN, version 2.000: a software for population genetics analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Scribner KT, Chesser RK (1993) Environmental and demographic correlates of spatial and seasonal genetic structure in the eastern cottontail (Sylvilagus floridanus). J Mammal 74:1026–1045
Sillero-Zubiri C, Gottelli D (1995a) Diet and feeding behavior of Ethiopian wolves (Canis simensis). J Mammal 76:531–541
Sillero-Zubiri C, Gottelli D (1995b) Spatial organization in the Ethiopian wolf Canis simensis: large packs and small stable home ranges. J Zool 237:65–81
Sillero-Zubiri C, Gottelli D, Macdonald DW (1996a) Male philopatry, extra-pack copulations and inbreeding avoidance in Ethiopian wolves (Canis simensis). Behav Ecol Sociobiol 38:331–340
Sillero-Zubiri C, King AA, Macdonald DW (1996b) Rabies and mortality in Ethiopian wolves (Canis simensis). J Wildl Dis 32:80–86
Sillero-Zubiri C, Marino J, Gottelli D, Macdonald DW (2004) Ethiopian wolves. In: Macdonald DW, Sillero-Zubiri C (eds) Biology and conservation of wild canids. Oxford University Press, Oxford, pp 311–322
Storz JF (1999) Genetic consequences of mammalian social structure. J Mammal 80:553–569
Storz JF, Beaumont MA, Alberts SC (2002) Genetic evidence for a long-term population decline in a savannah-dwelling primate: inferences from a hierarchical Bayesian model. Mol Biol Evol 19:1981–1990
Sugg DW, Chesser RK (1994) Effective population sizes with multiple paternity. Genetics 137:1147–1155
Taberlet P, Griffin AS, Goossens B et al (1996) Reliable genotyping of samples with very low DNA quantities using PCR. Nucleic Acids Res 26:3189–3194
Turner TF, Wares JP, Gold JR (2002) Genetic effective size is three orders of magnitude smaller than adult census size in an abundant, estuarine-dependent marine fish (Sciaenops ocellatus). Genetics 162:1329–1339
Wayne RK, Koepfli KP (1996) Demographic and historical effects on genetic variation in carnivores. In: Gittleman JL (ed) Carnivore behavior, ecology, and evolution. Cornell University Press, Ithaca, pp 453–484
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Wright S (1931) Evolution of Mendelian populations. Genetics 16:97–159
Wright S (1943) Isolation by distance. Genetics 28:114–118
Acknowledgements
We thank the Ethiopian Wildlife Conservation Authority, the Oromia Rural Land and Natural Resources Administration Authority, and the Bale Mountains National Park for permission to undertake this research. We are grateful for the support of the Ethiopian Wolf Conservation Programme (EWCP) and, in particular, Stuart Williams, Lucy Tallents, Edriss Ebu, Hussein Adam, Mustefa Dule, and Gedlu Tessera for their assistance in the field. The field work was funded by the Iris Darnton Trust, Jesus College, and the Sophie Danforth Conservation Biology Fund (Rhode Island Park Zoo). The genetic work was funded by a National Science Foundation (US) grant to RKW and supported by the Conservation Genetics Resource Center at UCLA. DAR was supported by an Overseas Student Award (UK) and a Clarendon Fund Award from the University of Oxford.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Randall, D.A., Pollinger, J.P., Argaw, K. et al. Fine-scale genetic structure in Ethiopian wolves imposed by sociality, migration, and population bottlenecks. Conserv Genet 11, 89–101 (2010). https://doi.org/10.1007/s10592-009-0005-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-009-0005-z