Abstract
Due to societal changes and altered demands for firewood, the traditional forest management of coppicing has been largely abandoned. As a result, many forest herbs that are specifically adapted to regular opening of the canopy, have suffered significant declines in abundance, and the remaining populations of these species often tend to be small and isolated. Reduced population sizes and pronounced spatial isolation may cause loss of within-population genetic diversity and increased between-population differentiation through random genetic drift and inbreeding. In this study, we investigated genetic diversity and genetic structure of 15 populations of the food-deceptive orchid Orchis mascula using AFLP markers. Within-population genetic diversity significantly increased with increasing population size, indicating genetic impoverishment in small populations. Genetic differentiation, on the other hand, was rather low (ΦST = 0.083) and there was no significant relationship between genetic and geographic distances, suggesting substantial gene flow within the study area. However, strong differences in levels of within-population diversity and among-population differentiation were found for populations located in forests that have been regularly coppiced and populations found in forests that were neglected for more than 50 years and that were totally overgrown by shrubs. Our data thus indicate that a lack of coppicing leads to decreased genetic diversity and increased differentiation in this orchid species, most likely as a result of genetic drift following demographic bottlenecks. From a conservation point of view, this study combined with previous results on the demography of O. mascula in relation to forest management illustrates the importance of coppicing in maintaining viable populations of forest herbs in the long-term.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In much of Western Europe, coppicing used to be the main method of forest management since medieval times (Rackham 1980). During the twentieth century, however, this kind of forest management went into rapid decline mainly due to altered demands for firewood and societal changes. In Southern Belgium, for example, where this research was conducted, the area coppiced woodland decreased from 105,000 ha in 1899 to only 15,000 ha in 1999 (85% decrease). The remaining coppices are not regularly cut anymore, or have been converted to timber plantations. As a result, open spaces within woods have become very small, are often not connected with open spaces in the wider landscape, and are no longer bordered by a dynamic mixture of young growth and temporary open space (Peterken 1991). These changes may have reduced population sizes, extirpated local populations, and increased distances among remnant populations, all which may affect population genetics of plant species. Increased spatial isolation and decreased population sizes can lead to reduced gene flow, genetic erosion and increased genetic divergence, through random genetic drift and increased levels of inbreeding (Barrett and Kohn 1991; Ellstrand and Elam 1993; Young et al. 1996).
Besides alterations in spatial population characteristics, changes in forest management also influence local environmental conditions, which, in turn, can have an effect on the distribution of genetic variation among populations, through several evolutionary processes, including selection, differential gene exchange and chance associations caused by genetic drift and founder effects (Antonovics 1971; Gram and Sork 2001). First, natural selection can cause populations to adapt to the variable levels of light penetrating to the forest soil, resulting in micro-geographical variation of genetic diversity that is associated with the different types of forest management.
Secondly, genetic heterogeneity can also be the result of differential gene exchange brought about by variation in flowering probabilities among forest management types. Since many forest herbs are specifically adapted to regular opening of the canopy, alterations in forest management can result in pronounced reductions in flowering probabilities once a certain threshold value of light penetrating to the soil has been exceeded (Mitchell and Woodward 1988). This may be particularly true for orchids. Although most orchid species tolerate moderate levels of shade, recent shading and leaf removal experiments have indeed demonstrated strong responses to light availability. In Spiranthes spiralis, for example, reducing daylight to 1% photosynthetic active radiation resulted in a dramatic decline (93%) in the number of plants surviving to the next year (Willems et al. 2001). In the woodland orchids Cephalanthera rubra and Cypripedium calceolus, shading did not affect survival, but it resulted in prolonged dormancy and thus decreased flowering (Shefferson et al. 2005). Within a region, differences in flowering probabilities related to different types of forest management may thus create genetic barriers that, in turn, may exacerbate local differences.
In this study, we investigated the population genetics of the tuberous orchid Orchis mascula in the Viroin region (Belgium). Previous research on demographic structure and fruit and seed set (Jacquemyn et al. 2008) has shown that this species benefits substantially from coppicing. The increased light intensity due to coppicing positively affected the rate of flowering and fruit set (Jacquemyn et al. 2008), potentially affecting gene flow by both pollen and seeds. However, the genetic consequences of forest management have not been investigated yet. Therefore, 15 populations were selected within the study area. Seven populations were located in forests that were not coppiced for more than 50 years and were severely overgrown by shrubs such as Buxus sempervirens and Prunus spinosa, before they were coppiced in 2000 (in the context of a European Life project that aimed to restore these sites to their original state). Eight populations were located in forests that have been coppiced on a regular basis. More specifically, we wanted to answer the following questions:
-
1.
Is within-population genetic diversity related to the size of the investigated populations?
-
2.
How does forest management affect the within-population genetic diversity and genetic structure of the investigated populations?
-
3.
Are the extant populations genetically differentiated from each other and how are genetic distances related to geographic distances?
Materials and methods
Study species
Orchis mascula (Early Purple Orchid) is a relatively small, perennial orchid that is widely distributed across Europe, western Asia and northern Africa. It usually occurs in forests or grasslands on limestone. In many parts of Europe, this species has suffered dramatic declines in abundance (Jacquemyn et al. 2005; Kull and Hutchings 2006), most likely as a result of the abandonment of traditional coppicing practices. O. mascula flowers in Spring, mostly from mid-May till the second week of June (Nilsson 1983). The red–violet flowers are borne in many-flowered spikes that vary in height between 15 and 40 cm and carry up to 40 flowers. The flowers are nectarless, but produce a strong terpene blend, mainly consisting of monoterpene hydrocarbons (Nilsson 1983). O. mascula is allogamous and pollinators are required for successful fruit set. Most frequent pollinators of O. mascula are bumblebees (Bombus), cuckoo bumblebees (Psithyrus), and solitary bees of the genera Eucera, Nomada, Andrena and Osmia (Nilsson 1983). Pollination occurs mainly during the first days after anthesis before pollinators have learned to avoid the flowers. Percentage fruit set is mostly low (between 3% and 20%) and ca. 50% of individual plants do not set fruit in a given year (Nilsson 1983; Johnson and Nilsson 1999). After seeds have been released and germinate, the first tuber develops from the protocorm before any other organs have been formed and merely functions as a storage organ for the products of mycophagy that are translocated from the protocorm (Rasmussen 1995). As a result, the underground stage of O. mascula lasts for at least 2 years.
Study area
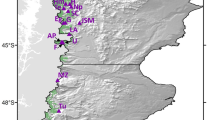
This study was conducted in the Calestienne region (Belgium), which covers a narrow (5 km wide, 100 km long), SW–NE oriented geological belt of Devonian limestone hills, interspersed with river plains and plateaus. The actual study area is situated in the southwestern part of this region, roughly coinciding with the valley of the river Viroin 75 km south of Brussels, and covers about 80 km2 (ca. 50° N, 4.5° E) (Fig. 1). This area consists of a mixture of calcareous grasslands, woodlands on limestone, agricultural land and small villages (for more details: see Adriaens et al. (2006)). Ash (Fraxinus excelsior), field maple (Acer campestre), hornbeam (Carpinus betulus), beech (Fagus sylvatica) and oak (Quercus robur) make up the dominant tree species of most woodlands, which are characterized by a very rich understory.
Map of the study area and the studied populations. Numbers refer to populations described in Table 1. Forests within the study area are depicted in grey
Fifteen populations of O. mascula, randomly chosen from all populations occurring in the study area (Fig. 1), were sampled in May 2006. Seven populations were located in forests that were left undisturbed for more than 50 years. Most of these populations were coppiced again in 2000 in the context of a European Life project that aimed to restore these sites to their original state. Before restoration had begun, forests were heavily invaded by shrubs such as Buxus sempervirens and Prunus spinosa reducing population sizes of O. mascula and other forest herbs to a very few individuals. However, at the time of sampling (2006), most O. mascula populations had recovered and attained reasonable population sizes. The other eight populations were located in forests that have been managed on a regular basis, had a very thin shrub layer and a rich understory of spring forest herbs.
Sampling procedure and AFLP protocol
In each population, the total number of individuals was recorded and young leaf material was collected from 20 randomly chosen plants and immediately frozen in liquid nitrogen. Before DNA extraction, leaf material was freeze dried for 48 h and homogenized with a mill (Retsch MM 200) to fine powder. Total DNA was extracted from 30 mg of freeze dried leaf material using the Nucleospin® 96 Plant Kit (Machery-Nagel). DNA integrity and concentration were estimated on 1.5% (w/v) agarose gels.
AFLP analysis was carried out according to Vos et al. (1995) and following the protocol of Roldán-Ruiz et al. (2000). The enzymes EcoRI and MseI were used for DNA digestion. Each individual plant was fingerprinted with four primer combinations. The primer extensions used were EcoRI-ACGT/MseI-CGA, EcoRI-ACGT/MseI-CGC, EcoRI-ACAG/MseI-CGT and EcoRI-ACAG/MseI-CTA. Fragment separation and detection took place on an ABI Prism 3130xl capillary sequencer. GeneScan 500 Rox labelled size standard (Perkin Elmer) was loaded in each run. In all cases a peak amplitude threshold of 50 was set for the analysis. The fluorescent AFLP fingerprints were scored using GeneMapper v. 3.7 (Applied Biosystems). We scored the presence or absence of each marker in each plant using the standard peak detection parameters recommended by the manufacturer for AFLP-runs. All polymorphic AFLP-bands in the range 50–475 bp were scored automatically using GeneMapper, and then visually inspected to ensure the accurateness of the results. To assess reproducibility of the markers retained, three independent DNA extractions were carried out for 10 individuals and AFLP fingerprints were generated for each replicate and primer combination (a total of 120 AFLP fingerprints). The replicated samples used to estimate the error rate were fully randomized with all other samples in order to avoid biases in this estimation. Only once the scoring had been completed, the replications were compared to estimate the reproducibility of our results. In total, 64 differences were observed among a total of 5,880 comparisons, yielding an error rate of 1.1% (Bonin et al. 2004).
Data analysis
Total genetic diversity was partitioned among groups of populations (populations located in recently managed forests versus populations in undisturbed forests), among populations within groups, and within populations by carrying out a hierarchical analysis of molecular variance (AMOVA) on Euclidean pairwise distances between individuals calculated according to Huff et al. (1993) and using GenAlEx v. 6.0 (Peakall and Smouse 2006). Significances were determined using a permutation test (n = 9,999). The Φst analogue for G st was derived from the Euclidean genetic distances and its significance was determined using the Monte Carlo procedure available in GenAlEx (9,999 simulations).
Pairwise genetic distances among the 15 investigated populations and their level of significance were also obtained from the AMOVA. Again 9,999 permutations were performed. A principal coordinates analysis was performed on this matrix using GenAlEx and the first two components were plotted graphically. Evidence of isolation by distance among populations was obtained by examining correlations between the matrices of genetic distances and geographical distances (Slatkin 1993). Significance of the observed relationships was obtained using a Mantel test (Mantel 1967). A total of 9,999 random permutations were performed.
Three measures of within-population genetic diversity were estimated: the proportion of polymorphic loci (P), Nei’s gene diversity (H j) and band richness (Br). The proportion of polymorphic loci and Nei’s gene diversity were estimated using AFLPsurv V.1.0 (Vekemans et al. 2002). Estimates of allelic frequencies at AFLP loci were calculated using the Bayesian method with a non-uniform prior distribution of allele frequencies following Zhivotovsky (1999), assuming either no, or some deviation (F IS = 0.1) from Hardy–Weinberg genotypic proportions according to the outcrossing nature of the species (Nilsson 1983, Jacquemyn, unpublished results). However, the results for different F IS values did not differ much and only results for F IS = 0 are presented. After estimating allele frequencies, statistics of gene diversity and population genetic structure were computed according to Lynch and Milligan (1994). For each population, we calculated the proportion of polymorphic loci (P) at the 5% level and Nei’s gene diversity (H j). Finally, band richness was calculated for a standardized sample size according to the rarefaction method of Petit et al. (1998). This measure of genetic diversity represents the number of phenotypes expected at each locus (i.e. each scored AFLP fragment) and can be interpreted as an analogue of allelic richness (Coart et al. 2005).
To investigate whether population size affected the proportion of polymorphic loci, Nei’s gene diversity and band richness, ordinary least squares (OLS) regressions were used. Population size was log10 transformed prior to analysis. To compare levels of within-population genetic diversity between populations that were recently restored and those that were managed regularly, simple t-tests were used. Finally, to investigate whether possible differences in genetic diversity among groups of populations were mediated by differences in population size, an analysis of covariance (ANCOVA) was used.
Results
The size of the 15 populations studied ranged from 33 to more than 1,000 individuals (Table 1). The four AFLP primer combinations resulted in 196 unambiguously scorable, polymorphic bands. Each individual displayed a unique banding pattern and the differences among populations were thus attributable to frequency differences in variable markers, rather than to private markers.
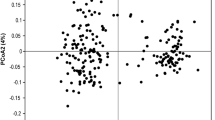
The AMOVA indicated significant (P = 0.001), but low genetic differentiation between populations (ΦST = 0.083) (Table 2). In contrast, no differentiation was found between groups of populations. Pairwise ΦST values ranged from 0.021 to 0.164 (Table 3). All pairwise genetic distances between populations were also highly significant (P < 0.001) (Table 3). The first two principal coordinates, which explained 16% and 15% of the total variance, showed a central cluster of strongly related populations and a suite of populations scattered widely around this cluster (Fig. 2). This grouping coincided with the geographical position of the populations in the landscape (Fig. 1) and the way they were managed in the past (Table 1). All populations contained in the central cluster occurred in forests that were managed regularly, whereas all other populations were found in sites that were previously completely overgrown, but were recently restored to their original state. There was no significant relationship between pairwise genetic distances and geographic distances when all populations were considered (r M = 0.086, P > 0.05) (Fig. 3a), nor when recently restored (r M = 0.283, P > 0.05) and regularly managed (r M = −0.174, P > 0.05) populations were analysed separately (Fig. 3b). When restricting comparisons to those within each management type, pairwise genetic distances were substantially larger for the recently restored populations than for the regularly managed populations (Fig. 3b), with the overall ΦST = 0.125 and 0.048, respectively.
Relationship between genetic differentiation and geographical distance for (a) all 15 populations of Orchis mascula and (b) populations that were regularly managed during the last 100 years (open circles) and populations that were recently cut after they were left undisturbed for more than 50 years and were severely overgrown by shrubs (full circles)
The proportion of polymorphic loci ranged from 42.3 to 52.6 (Table 1). Average expected heterozygosity was 0.160 (range: 0.137–0.173). Both measures of gene diversity were significantly and positively related (F 1,13 = 15.88 and 12.93, P < 0.01) to population size. Band richness ranged from 1.39 to 1.53 (mean: 1.48; SD: 0.04) (Table 1) and was also significantly related to population size (F 1,13 = 17.08, P = 0.001) (Fig. 4). The proportion of polymorphic loci and band richness were significantly different between populations that were recently restored and those that were managed regularly (t 13 = 3.27, P = 0.006 and t 13 = 3.37, P = 0.005, respectively). Nei’s gene diversity, on the other hand, was not significantly different between both types of populations (t 13 = 1.86, P = 0.086). When analysing the combined effects of population size and forest management type, only population size showed a significant effect on the proportion of polymorphic loci (F 1,11 = 10.44, P = 0.008) and band richness (F 1,11 = 13.09, P = 0.004), indicating that the effects of forest management were primarily mediated by variation in population size. When Nei’s gene diversity was considered, both population size (F 1,11 = 14.29, P = 0.003) and forest management (F 1,11 = 5.33, P = 0.041) showed a significant effect, but the effect of population size was far more pronounced than that of forest management. The interaction between population size and forest management was not significant (F 1,11 = 4.51, P = 0.057).
Discussion
Genetic diversity and forest management
The observed levels of gene diversity (H j) in the studied populations ranged from 0.137 to 0.173 and are well within the range reported for species with a mixed breeding system (Nybom and Bartish 2000). Our results further showed lower genetic diversity in populations located in forests that were left undisturbed for more than 50 years, indicating that a lack of coppicing might lead to decreased genetic diversity in this orchid species and providing evidence that forest management not only affects demographic properties of this species (Jacquemyn et al. 2008), but also influences its genetic properties. However, when population size was included in the analysis, it was shown that the effects of forest management on gene diversity were mainly the result of variation in population size among forest types (Fig. 4), implying that decreases in genetic diversity are for the most part mediated by differences in population size rather than by direct effects of forest management.
There are several lines of evidence that suggest that the observed low genetic diversity in populations located in unmanaged forests can be best explained by genetic drift associated with demographic bottlenecks. First, theory predicts that populations that have experienced a recent bottleneck will show low allelic diversity and heterozygosity (Nei et al. 1975). However, the loss of allelic diversity occurs faster than the reduction of expected heterozygosity (Widmer and Lexer 2001). We found significant differences in the proportion of polymorphic loci and band richness (an analogue of allelic richness), but not in expected heterozygosity between populations that were managed regularly and occurred in species-rich forests and populations that were recently restored and occurred in places that were previously overgrown by thickets of Buxus sempervirens and Prunus spinosa. Before restoration of these sites began, populations were much smaller than the population sizes observed in this study. Second, a population bottleneck is expected to cause a random loss of allelic variation as a consequence of random genetic drift. Inspection of Fig. 2 indeed suggests random loss of alleles as populations that recently went through a demographic bottleneck are scattered widely around the regularly managed populations that clearly cluster together in the central part of the figure.
Population genetic structure
These results are corroborated by the patterns of population genetic structure. The AMOVA indicated significant genetic differentiation among populations within groups, but not between groups. In addition, pronounced differences in the level of genetic differentiation were found between regularly managed populations and populations that were left undisturbed for many years (ΦST = 0.048 and 0.125, respectively). The low genetic differentiation among the regularly managed populations may be explained by high levels of gene flow and low levels of drift, whereas the higher levels of genetic differentiation in unmanaged populations may indicate that genetic drift is prevalent (Hutchison and Templeton 1999; Koizumi et al. 2006). Because the AMOVA analysis showed no between-group differentiation, the possibility that selection for shade tolerance in undisturbed populations affected genetic differentiation, seems unlikely, because in that case genetic variation should to some extent be related to the type of forest management. If, on the other hand, geographical location would have influenced the patterns of genetic variation, we should have expected isolation-by-distance, which was also not the case. These results point to genetic drift as the major factor affecting genetic diversity in the unmanaged populations, and suggest that a lack of forest management, and the reduction in population size that follows, may substantially decrease genetic diversity of this species.
Nevertheless, the overall level of genetic differentiation among populations was low (ΦST = 0.083), and even lower when only the managed populations were considered (ΦST = 0.048). Low genetic differentiation might indicate high levels of gene flow. This can be explained by the fact the O. mascula is still quite common within the study area with numerous populations scattered around. A second explanation can be found in the pollination system of the species. Within the orchid family, two broad categories of pollination systems can be distinguished: species that offer substantial rewards to their pollinators and species that are pollinated by deceit. It is expected that in the absence of any reward flower visits will be brief and few. As deceived pollinators quickly loose interest, after a few attempts they will leave the patch, increasing long-distance gene flow (Peakall and Beattie 1996; Ferdy et al. 1998) and thus decreasing the probability of geitonogamous pollination (Smithson 2002; Johnson et al. 2004). Lower levels of genetic differentiation among populations have indeed been observed in deceptive species than in rewarding species (Tremblay and Ackerman 2003; Cozzolino and Widmer 2005).
Implications for conservation
O. mascula has suffered significant declines during the last five decades in many parts of Europe (Jacquemyn et al. 2005; Kull and Hutchings 2006). In the Netherlands, for example, being one of the most abundant orchids in the Zuid-Limburg region several populations of more than thousand individuals have been decimated or have completely disappeared (Kreutz and Dekker 2000). Although this species is still relatively abundant in the study area, our data show that small populations show signs of genetic erosion. These results add to a growing body of studies that have shown that population size may strongly affect plant population genetic diversity and that common species may be as susceptible to the genetic consequences of fragmentation as rare species (reviewed in Honnay and Jacquemyn 2007). Furthermore, this research also suggests that populations that have recently gone through a demographic bottleneck may harbour lower genetic diversity. Since many orchid populations are currently more or less recovering from extremely small population sizes as a result of recently established nature management practices, it is very likely that other species went through severe genetic bottlenecks as well, negatively affecting their long-term survival chances. Cozzolino et al. (2003), for example, also showed decreased genetic diversity in the orchid Anacamptis palustris, which they explained as being the result of recent genetic bottlenecks. Although the variation surveyed is most likely selectively neutral, its loss may indicate the correlated loss of potentially adaptive genetic variation (Young et al. 1996). Low genetic diversity may also be associated with decreased fitness through decreasing heterozygosity and the expression of deleterious alleles (Fischer and Matthies 1998; Keller and Waller 2002). With regards to O. mascula, prevention of population decline therefore seems to be the most important conservation measure at this stage, to ensure long-term viability of this species in the study. This can be best achieved by regular coppicing of forests as it increases the probability of flowering, fruit and seed set and, as a result, seedling recruitment (Jacquemyn et al. 2007, 2008).
References
Adriaens D, Honnay O, Hermy M (2006) No evidence of a plant extinction debt in highly fragmented calcareous grasslands in Belgium. Biol Conserv 133:212–224
Antonovics J (1971) The effects of a heterogeneous environment on the genetics of natural populations. Am Sci 59:593–599
Barrett SCH, Kohn J (1991) The genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, Oxford, pp 3–30
Bonin A, Bellemain E, Eidesen B, Pompanon F, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetics studies. Mol Ecol 13:3261–3273
Coart E, Van Glabeke S, Petit RJ, Van Bockstaele E, Roldán-Ruiz I (2005) Range wide versus local patterns of genetic diversity in hornbeam (Carpinus betulus L.) Conserv Genet 6:259–273
Cozzolino S, Widmer A (2005) Orchid diversity: an evolutionary consequence of deception? Trends Ecol Evol 20:487–494
Cozzolino S, Noce ME, Musacchio A, Widmer A (2003) Variation at chloroplast minisatellite locus reveals the signature of habitat fragmentation and genetic bottlenecks in the rare orchid Anacamptis palustris (Orchidaceae). Am J Bot 90:1681–1687
Ellstrand NC, Elam DR (1993) Population genetic consequences of small population size: implications for plant conservation. Annu Rev Ecol Syst 24:217–242
Ferdy JB, Gouyon PH, Moret J, Godelle B (1998) Pollinator behavior and deceptive pollination: learning process and floral evolution. Am Nat 152:696–705
Fischer M, Matthies D (1998) RAPD variation in relation to population size and plant fitness in the rare Gentianella germanica (Gentianaceae). Am J Bot 85:811–819
Gram WK, Sork VL (2001) Association between environmental and genetic heterogeneity in forest tree populations. Ecology 82:2012–2021
Honnay O, Jacquemyn H (2007) Susceptibility of common and rare plants to the genetic consequences of habitat fragmentation. Conserv Biol 21:823–831
Huff DR, Peakall R, Smouse PE (1993) RAPD variation within and among populations of outcrossing buffalograss (Buchloë dactyloides (Nutt.) Engel). Theor Appl Genet 86:927–934
Hutchison DW, Templeton AR (1999) Correlation of pairwise genetic and geographic distance measures: inferring the relative influences of gene flow and drift on the genetic distribution of genetic variability. Evolution 53:1898–1914
Jacquemyn H, Brys R, Hermy M, Willems JH (2005) Does nectar reward affect rarity and extinction probabilities of orchid species? An assessment using historical records from Belgium and the Netherlands. Biol Conserv 121:257–263
Jacquemyn H, Vandepitte K, Brys R, Honnay O, Roldán-Ruiz I (2007) Fitness variation and genetic diversity of small, remnant populations of the food deceptive orchid Orchis purpurea. Biol Conserv 139:203–210
Jacquemyn H, Brys R, Honnay O, Hermy M (2008) Effects of coppicing on demographic structure, fruit and seed set in Orchis mascula. Basic Appl Ecol (in press). doi:10.1016/j.baae.2007.05.002
Johnson SD, Nilsson LA (1999) Pollen carryover, geitonogamy, and the evolution of deceptive pollination systems. Ecology 80:2607–2619
Johnson SD, Craig PI, Ågren J (2004) The effects of nectar addition on pollen removal and geitonogamy in the non-rewarding orchid Anacamptis morio. Proc Roy Soc London B 271:803–809
Keller LF, Waller DM (2002) Inbreeding effects in wild populations. Trends Ecol Evol 17:230–241
Koizumi I, Yamamoto S, Maekawa K (2006) Decomposed pairwise regression analysis of genetic and geographic distances reveals a metapopulation structure of stream-dwelling Dolly Varden charr. Mol Ecol 15:3175–3189
Kreutz CAJ, Dekker H (2000) De orchideeën van Nederland – ecologie, verspreiding, bedreiging, beheer. Uitgave Kreutz and Seckel, Landgraaf & Raalte
Kull T, Hutchings MJ (2006) A comparative analysis of decline in the distribution ranges of orchid species in Estonia and the United Kingdom. Biol Conserv 129:31–39
Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3:91–99
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mitchell PL, Woodward FI (1988) Responses of three woodland herbs to reduced photosynthetically active radiation and low red to far-red ratio in shade. J Ecol 76:807–825
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nilsson LA (1983) Anthecology of Orchis mascula (Orchidaceae). Nord J Bot 3:157–179
Nybom H, Bartish IV (2000) Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect Plant Ecol Evol Syst 3:93–114
Peakall R, Beattie AJ (1996) Ecological and genetic consequences of pollination by sexual deception in the orchid Caladenia tentaculata. Evolution 50:2207–2220
Peakall R, Smouse PE (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Peterken GF (1991) Managing semi-natural woods: a suitable case for coppice. Quart J Forest 85:21–29
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conserv Biol 12:844–855
Rackham O (1980) Ancient woodland, its history, vegetation and uses in England. Arnold, London
Rasmussen HN (1995) Terrestrial orchids: from seed to mycotrophic plant. Cambridge University Press, Cambridge
Roldán-Ruiz I, Dendauw J, Van Bockstaele E, Depicker A, De Loose M (2000) AFLP markers reveal high polymorphism rates in ryegrasses (Lolium spp.). Mol Breed 6:125–134
Shefferson RP, Kull T, Tali K (2005) Adult whole-plant dormancy induced by stress in long-lived orchids. Ecology 86:3099–3104
Slatkin M (1993) Isolation by distance in equilibrium and non-equilibrium populations. Evolution 47:264–279
Smithson A (2002) The consequences of rewardlessness in orchids: reward-supplementation experiments with Anacamptis morio (Orchidaceae). Am J Bot 89:1579–1587
Tremblay RL, Ackerman JD (2003) The genetic structure of orchid populations and its evolutionary consequences. Lanksteriana 7:87–92
Vekemans X, Beauwens T, Lemaire M, Roldán-Ruiz I (2002) Data from amplified fragment length polymorphism (AFLP) markers show indication of size homoplasy and of a relationship between degree of homoplasy and fragment size. Mol Ecol 11:139–151
Vos P, Hogers R, Bleeker M, Reijans M, Vandelee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Widmer A, Lexer C (2001) Glacial refugia: sanctuaries for allelic richness, but not for gene diversity. Trends Ecol Evol 16:267–269
Willems JH, Balounová Z, Kindlmann P (2001) The effect of shading on seed production and plant survival in the threatened species Spiranthes spiralis (Orchidaceae). Lindleyana 16:31–37
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418
Zhivotovsky LA (1999) Estimating population structure in diploids with multilocus dominant DNA markers. Mol Ecol 8:907–913
Acknowledgements
We would like to thank Nancy Mergan for providing excellent technical assistance for the generation of AFLP data. Two anonymous reviewers provided very useful comments that improved significantly the quality of this ms. This research was funded by the Flemish Fund Scientific Research (FWO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jacquemyn, H., Brys, R., Adriaens, D. et al. Effects of population size and forest management on genetic diversity and structure of the tuberous orchid Orchis mascula . Conserv Genet 10, 161–168 (2009). https://doi.org/10.1007/s10592-008-9543-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-008-9543-z