Abstract
In this paper, we analyze the genetic variability in four Tunisian natural populations of Medicago ciliaris using 19 quantitative traits and six polymorphic microsatellite loci. We investigated the amplification transferability of 30 microsatellites developed in the model legume M. truncatula to M. ciliaris. Results revealed that about 56.66% of analyzed markers are valuable genetic markers for M. ciliaris. The most genetic diversity at quantitative traits and microsatellite loci was found to occur within populations (>80%). Low differentiations among populations at quantitative traits Q ST = 0.146 and molecular markers F ST = 0.18 were found. The majority of measured traits exhibited no significant difference in the level of Q ST and F ST . Furthermore, significant correlations established between these traits and eco-geographical factors suggested that natural selection should be invoked to explain the level of phenotypic divergence among populations rather than drift. There was no significant correlation between population differentiation at quantitative traits and molecular markers. Significant spatial genetic structure consistent with models of isolation by distance was detected within all studied populations. The site-of-origin environmental factors explain about 9.07% of total phenotypic genetic variation among populations. The eco-geographical factors that influence more the variation of measured traits among populations are the soil texture and altitude. Nevertheless, there were no consistent pattern of associations between gene diversity (He) and environmental factors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Maintenance of genetic diversity within populations is currently regarded as a primary goal in conservation efforts, as it will enhance the threatened species ability to adapt to future environmental changes (Shrestha et al. 2002; Jump and Penuelas 2005). A better preservation of natural populations in the face of environmental changes requires a good understanding of their genetic variation pattern (Palo et al. 2003; Pressoir and Berthaud 2004). Spatial genetic structure can result from two major evolutionary mechanisms; random genetic drift and natural selection. The relative importance of these two forces in shaping the pattern of spatial genetic structure is a central question in population genetics (Merilä and Crnokrak 2001). One way to distinguish between neutral and selective forces in natural populations is to examine variation from locus to locus, expecting that genetic drift and migration should affect all loci similarly, whereas natural selection should affect some loci differently (Jaramillo-Correa et al. 2001). Q ST estimates genetic divergence among populations at quantitative traits (Spitze 1993), which is analogous to F ST measured using molecular markers (Podolsky and Holtsford 1995). There are three possible outcomes from the comparison of Q ST and F ST . If neutral differentiation among populations (F ST ) is in the same magnitude of that for quantitative traits (Q ST ) or is significantly more than Q ST , the hypothesis that among populations variance is due to random drift cannot be rejected or convergent selection may be involved as a cause for the reduced differentiation. The third case where Q ST > F ST , suggesting a prominent role for natural selection in accounting for patterns of quantitative traits differentiation among populations (Merilä and Crnokrak 2001).
Local adaptation of populations to climate has been demonstrated often since the pioneering work of Clausen et al. (1940) and Turesson (1925). In natural environments, the spatial distribution of individual plants within populations often depends on environmental factors that affect seedling establishment, such as temperature, precipitation, growing season length and changes in day length during latitudinal range changes (Berlow et al. 2002; Cruse-Sanders and Hamrick 2004). The genus Medicago is one of the most widespread genera of the Fabaceae, including approximately 83 different species and 18 infraspecies taxa (Bena et al. 1998). Two thirds are annuals and one third perennial (Lesins and Lesins 1979). In Tunisia, this genus represents an important proportion of the native flora through all bioclimatic stages (Pottier-Alapetite 1979). However, information on genetic diversity available within and among natural populations of Medicago annual species is meager and poorly documented. Medicago ciliaris is an annual, diploid (2n = 16) and self-pollinating forage species. In Tunisia, it grows spontaneously in northern areas; restricted to humid, sub-humid and to superior and inferior semi-arid stages (Abdelkefi et al. 1996). M. ciliaris is nodulated efficiently in its natural habitat by Sinorhizobium medicae (Zribi et al. 2007). In the world, it is distributed from the Canary Islands to Israel on both sides of the Mediterranean basin and it could be intercrossed readily with M. intertexta and also with M. muricoleptis (Lesins and Lesins 1979).
M. ciliaris has the advantage of belonging to a genus with a model legume, M. truncatula (Barker et al. 1990). Microsatellites, also referred to as simple sequence repeats (SSRs), are tandem repeats of short oligonucleotides that are ubiquitously interspersed in eukaryotic genomes (Kashi et al. 1997). They are highly polymorphic and efficient markers for genetic mapping and the characterization of plant germplasm (Saal et al. 2001). Transferability of EST-SSRs from M. truncatula have been recently evaluated for amplification and polymorphism within the genus (Julier et al. 2003; Eujayl et al. 2004; Gutierrez et al. 2005), and in all of these studies, high levels of both cross-amplification and polymorphism were detected in several Medicago spp. M. ciliaris has not been included in these investigations. In the present study, we analyzed the genetic variation within and among natural populations of M. ciliaris using 19 quantitative traits and six polymorphic SSR markers. The aims of this paper were: (i) to assess the degree of genetic diversity available within and among natural populations of M. ciliaris, (ii) to compare genetic differentiation among natural populations at quantitative traits (Q ST ) and SSR markers (F ST ), and (iii) to estimate associations between quantitative traits, molecular markers and environmental factors (texture, climate and altitude).
Materials and methods
Plant material and sampling
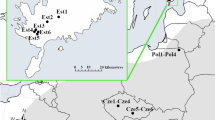
The plant material used in this study consisted of four Tunisian natural populations of M. ciliaris, covering different regions of plant distribution, sampled in July 2003 in inferior semi-arid (Enfidha), superior semi-arid (Soliman), sub-humid (Rhayet) and humid (Mateur) bioclimatic stages (Fig. 1). A collection site of mature pods, as defined here, is an ecologically homogenous area of about 500 m2 which contains one population. To minimize the chance of sampling the same individual more than once, the minimum distance between sampling pods was 4 m. These populations were selected based on their belonging to some different edaphic and climatic areas and they are accessible and abundant under natural conditions. Lines were created by one generation of spontaneous selfing in the greenhouse. Each population was constituted by 12 lines. We assumed that offspring should be genetically identical in each presumable line. Consequently, the within line variance can be considered as environmental however among lines variance component is assumed to be solely genetic (Falconer and MacKay 1996; Bonnin et al. 1997).
Map of Tunisia with the location of M. ciliaris sampling sites. 1 = Enfidha (latitude (N) = 36°07′; longitude (E) = 10°22′), 8 = Soliman (latitude (N) = 36°41′; longitude (E) = 10°32′), 10 = Rhayet (latitude (N) = 36°39′; longitude (E) = 09°27′) and 11 = Mateur (latitude (N) = 37°01′; longitude (E) = 09°40′)
Quantitative traits
Forty-eight (48) genotypes (original plants) of M. ciliaris were analyzed in this trial. In order to estimate environmental variance within a genotype (i.e. the within line variance) (Falconer and MacKay 1996), five seeds per genotype were used. A total of 240 plants were analyzed. Lines seeds were surface-sterilized and scarified with concentrated H2SO4 for 12 min and rinsed 10 times with sterile distilled water. The soaked seeds have been sown in Petri dishes on agar agar 0.9% medium before being vernalized at 4°C for 96 h. Seedlings were transplanted in plastic hampers (30/50 cm) filled with a mixture of sand/peat (1:3/2:3) with five plants, representative of one genotype, per hamper. Hampers were placed in a greenhouse at the Centre de Biotechnologie à la Technopole de Borj-Cédria (CBBC) in October 2004. A completely randomized design was used. Nineteen (19) morphological traits related to vegetative and reproductive development of plants were measured to assess quantitative genetic differentiation among studied populations of M. ciliaris (Table 1). Some of these traits were used by Bonnin et al. (1997) and they were also used by Badri et al. (2004a, b). All traits showed high heritability levels. For weight determination, plant organs were dried at 70°C for 48 h.
Molecular markers
Extraction of DNA from leaves was carried out according to Rogers and Bendish (1988) modified protocol by Gherardi et al. (1998). Molecular characterization of studied lines was performed using a total of 17 SSR loci (Table 2). PCR was carried out in a volume of 20 μl with 30 ng of genomic DNA, 11.4 μl of distilled water, 50 ng of each primer, 0.2 mM dNTP’s and 0.16 U of Taq polymerase (Uptima) in 2× 10 Taq buffer and 1.5 mM MgCl2. Each reaction was overlaid with 25 μl of mineral oil to prevent evaporation. Amplifications were carried out in a thermal cycler (Biometra) under the following conditions: 4 min at 94°C, followed by 40 cycles of 30 s at 94°C, 30 s at the annealing temperature, 30 s at 72°C and a final 6 min extension step at 72°C. Depending on the primer pair, annealing temperatures were 55°C (at most) and 50°C. Samples were stored at 4°C before use. Amplified fragments were resolved by gel electrophoresis on 3.5% agarose run in 0.5× TBE (45 mmol/l Tris, 45 mmol/l boric acid, 0.02 mmol/l EDTA pH8) using 110 V for about 3 h, stained with ethidium bromide (EtB) and photographed under UV light (312 nm). The Marcel ladder (Bio-Rad) was used as size standard.
Statistical analyses
Quantitative genetic variation
Genetic variance in quantitative traits was partitioned into within and among populations components using SAS Proc GLM (type III) (SAS 7.02 Institute, Inc. 1998). The model included population and genotype within population. To estimate variance components, population and genotype were considered as random effects. Phenotypic mean values of each population were compared for all quantitative characters with Duncan multiple range test. Total phenotypic variance \((\sigma_T^2)\) was the sum of three components of variance: (i) arising among populations \((\sigma_p^2),\) (ii) between genotypes within populations \((\sigma_g^2)\) and (iii) due to residual error arising between individuals within original genotype \((\sigma_e^2).\) Broad-sense heritability (H 2) was estimated for each trait within each population as the ratio of the variance arising between genotypes \((\sigma_g^2)\) on the sum of \((\sigma_g^2)\) and \((\sigma_e^2)\) (Bonnin et al. 1997).
The level of population differentiation at quantitative traits (Q ST ) was estimated as described in Bonnin et al. (1996) as the following:
For our design, using samples of lines from a selfing species, the genetic variance within populations, \(\sigma_g^2,\) is estimated by the among line nested within population component of variance. However, this genetic variance includes both additive and non-additive components, thus leading to an underestimation of Q ST values. Therefore, our results should be considered with caution. The calculation of 95% confidence intervals (CIs) for Q ST was performed by 5000 bootstrap replicates in SAS Interactive Matrix Language (IML). Association between Q ST and geographical distance matrices was examined with Mantel (1967) test. Coefficient of genetic variation (CV g ) was obtained as shown below (Bonnin et al. 1997).
where m is the population phenotypic mean.
Molecular genetic variation
Molecular genetic diversity was analyzed within each population using four indices: (i) the number of multilocus genotypes (Geno), (ii) the mean observed heterozygosity (Ho), (iii) the gene diversity (He), expected heterozygosity, Nei 1978), and (iv) the linkage disequilibria (LD%) under Hardy–Weinberg equilibrium using Genetix software version 4.02 (Belkhir 2001). Linkage disequilibrium was estimated as described in Volis et al. (2001). F ST was used as a measure of genetic differentiation among populations for the polymorphic loci. It was defined as a standardized genetic variance among populations and it was estimated according to Weir and Cockerham’s (1984) method. Overall F ST among populations and pairwise estimates for all pairs of populations were computed, running the FSTAT program (Goudet 1995). Confidence intervals (CIs), based on re-sampling techniques, are provided with the software and involve jackknifing over all loci to obtain the mean and the standard error of the overall F ST , and bootstrapping over loci to obtain 95% CIs. Isolation by distance was analyzed as described in Rousset (1997) through Mantel tests (Mantel 1967) carried out between matrices of log-transformed geographic distances and odds-transformed genetic distances F ST /(1−F ST ). Relationships among populations were studied by the constructed Neighbor-joining (NJ) tree. Support for clustering was determined by a bootstrap procedure applied on SSR alleles (5000 replications). The NJ analysis and bootstrapping were performed with Darwin software (Perrier and Jacquemoud-Collet 2006). In addition, we assessed spatial genetic structure within population using spatial autocorrelation analyses. These were performed with kinship coefficients (Loiselle et al. 1995) using SPAGeDi software (Hardy and Vekemans 2002). As described in Van Rossum et al. (2004), to test for isolation-by-distance, the multilocus kinship coefficient for each pair of individuals was plotted against the logarithm of the geographical distance separating them.
Quantitative traits versus SSR markers
Sampling variances on estimates of variance components can be obtained using bootstrap or Jackknife procedures. As suggested by Jaramillo-Correa et al. (2001), we used bootstrapping over loci and lines (5000 bootstraps) to estimate 95% CIs, respectively, of F ST and Q ST from the observed distribution. The degree of population divergence in neutral quantitative traits and single locus markers (as measured by Q ST and F ST , respectively) are expected to be similar and independent of mutation rates, and depend only on effective population size and migration rates which affect all traits equally (Stenoien et al. 2005). Consequently, any difference between Q ST and F ST indices estimated for the same set of populations should be attributed to the effect of natural selection (Merilä and Crnokrak 2001). The correlation between genetic variation at quantitative traits and SSR markers was estimated in two ways. We first estimated the Pearson correlation coefficient (r) between CV g , at both the trait-specific and multi-trait levels, and gene diversity of each locus (He) and over all loci (He all). Secondly, Mantel (1967) test was used to estimate correlations between matrices of pairwise Q ST and F ST (Gomez-Mestre and Tejedo 2004).
Associations of quantitative traits and SSRs with environmental factors
Thirteen eco-geographical factors of sampling sites of natural populations of M. ciliaris were scored: texture, pH, saturation (ml/100g), electro-conductivity (mmho/cm), total calcareous (%), active calcareous (%), organic matter (%), carbon (%), assimilated \({\rm P}_{2}\rm{O}_{5}\) and K2O, climate, annual rainfall (mm) and altitude (m). Only three factors (texture, climate and altitude) out of these 13 parameters, showing significant differences, between studied sites were retained. To estimate the influence of the eco-geographical factors on the behavior of natural populations at quantitative traits two analyses were performed. Variance components estimates within and among populations were computed from quantitative data using SAS Proc Mixed (SAS Institute 1998) with the following mixed model:
where y ijklmn are the observations for all quantitative traits, μ the population mean, P i the random effect of population, L j(i) the random effect of genotype nested within population, T k the fixed effect of texture, A l the fixed effect of altitude, R m the effect of climate considered as co-variable, and e ijklmn a random residual effect. Then, correlations between quantitative traits and environmental factors were computed using Pearson correlation coefficients (r). Significance level was set to 0.05, and adjusted for multiple comparisons by Bonferroni corrections. On the other hand, associations of gene diversity of each locus (He) and mean gene diversity over all loci (He all) with eco-geographical factors were estimated using the Spearman rank correlation analysis (Turpeinen et al. 2001).
Results
Quantitative traits
Variance analysis of the population effect showed that 15 out of the 19 measured traits exhibited significant means differences between populations (Table 3). The population sampled from Rhayet site showed the lowest total weight (PTOT). Furthermore, populations collected in superior and inferior semi-arid stages, the case of Soliman and Enfidha respectively, revealed higher production of pods (NGOU) and they allocated greater effort to reproduction (REP) while lower values were found for Rhayet population.
The most percentage of genetic variance was found within populations (85.44%). Table 3 gives the coefficients of genetic variation (CV g ) of each trait and for each population in M. ciliaris. The average CV g was 31.92% in Enfidha (SD = ±11.56%), 37.90% in Soliman (SD = ±14.39%), 39.86% in Rhayet (SD = ±17.60%) and 33.77% in Mateur (SD = ±13.35%). We will arbitrarily consider as large those CV g above 25%. Twelve (12) traits out of 19 measured traits in Mateur, 13 out of 19 in Enfidha, 14 out of 19 in Soliman, and 15 out of 19 in Rhayet, showed large CV g . Therefore, the higher within-population genetic variation (CV g ) (15/19) was found in Rhayet.
Heritability (H 2) of measured traits ranges from 0.20 for P100G to 0.66 for D1F with an average of 0.50 (Table 3). Quantitative differentiation among populations (Q ST ) varies between Q ST = 0 and Q ST = 0.47 with a mean of Q ST = 0.146 (Table 3). There was a strong positive correlation (r = 0.84; P = 0.034) between Q ST and geographic distances matrices. For 171 possible correlations between measured traits, 126 were significant and 96 out of them are positive (Table 4). The flowering time trait (FLOR) was positively correlated with emergence date of first and sixth leaves (D1F and D6F) and stem weight (PTIG); however it was negatively correlated with pods production (NGOU) and with the effort allocated to reproduction (REP).
Molecular markers
In a preliminary study, the analysis of transferability amplification of 30 SSR loci from the model legume M. truncatula to M. ciliaris showed that about 56.66% of them amplified with M. ciliaris. This finding suggests that SSR markers developed from M. truncatula genome could be considered as valuable genetic markers for M. ciliaris. Seventeen (17) SSR loci out of these 30 tested markers were used to analyze molecular genetic differentiation among natural populations of M. ciliaris.
Among the 17 used SSR loci, only 6 (TP36B, MTIC451, MTIC452, JF20, TPG20C and MTIC86) out of them were polymorphic. Higher number of alleles per locus was observed for Expressed Sequence Tags (EST) SSR loci, followed by those from Microsatellite-Enriched Genomic Library (MEGL) and BAC (Bacterial Artificial Chromosome) SSR markers. Using the 3.5% agarose gels, we were able to detect a minimal size difference of 2 pb between alleles of each SSR marker. A total of 28 alleles were detected over loci. Among the 48 studied lines of M. ciliaris, 36 multilocus genotypes were distinguished. The number of multilocus genotypes ranged from 7 out of 12 in Enfidha to 11 in Mateur. The within populations molecular variability parameters are summarized in the Table 5. Low rate of observed heterozygosity (Ho), with a mean of about 1.3%, reflects that the studied species is preferentially self-pollinating. The percentage of linkage disequilibrium (LD%) between SSR markers ranged from 60% to 100% with an average of 73%.
High percentage of variance was found to occur within populations (80.94%). Overall, similar levels of within populations’ genetic variation (He) were found for analyzed populations. Molecular differentiation among populations (F ST ) varies from 0.08 between Enfidha and Soliman, and 0.30 between Soliman and Rhayet with a mean of F ST = 0.18 (Table 6). It is obvious that F ST does not support the isolation by distance model as an evolutionary mechanism for natural populations differentiation of M. ciliaris (r = 0.31; P = 0.47). Similarly, NJ analysis (Fig. 2) showed that studied lines of M. ciliaris from the same region group not all together and there seems to be no association between geographical and genetic distances among populations. However, there was a significant relationship (P < 0.05) between within-population genetic variation and geographical distance. We found a significant linear relationship between decreasing pairwise kinship coefficients and the logarithm of increasing geographical distance in Enfidha (r = −0.53; P < 0.001), Soliman (r = −0.52; P < 0.001), Rhayet (r = −0.49; P < 0.001) and Mateur (r = −0.31; P = 0.006) (Fig. 3).
Unrooted Neighbor-joining tree of the 48 lines of M. ciliaris sampled from the four studied populations. Each tip represents a single line. 1, 8, 10 and 11 indicate the populations of origin (Enfidha, Soliman, Rhayet and Mateur, respectively). The numbers on the tips indicate bootstrap values (expressed in percentages) and are shown for all clusters with >50% bootstrap support
Quantitative traits versus SSR markers
We found three different patterns of population differentiation across these 19 quantitative traits for M. ciliaris (Table 3). First, we found that 7 out of the 19 measured traits displayed significantly smaller Q ST than F ST , while only one out of these traits exhibited significantly larger Q ST than F ST . Second, we found that 11 out of these 19 traits displayed no significant difference in the level of Q ST and F ST . There were no consistent patterns of associations between CV g at multi-trait level and He over all loci (r = −0.55; P = 0.45), and between CV g at specific-trait level and He of each locus (P > 0.00044) (data not shown). Furthermore, no significant correlation (r = 0.095; P = 0.33) was found between Q ST and F ST matrices.
Associations of quantitative traits and SSR markers with eco-geographical factors
The site-of-origin environmental factors explain about 9.07% of total phenotypic genetic variation among analyzed populations of M. ciliaris. For the 76 possible correlations between analyzed quantitative traits and eco-geographical variables, 9 are significant (P < 0.00088) and 5 out of them are positive (Table 7). The environmental factors that influence more the quantitative traits of natural populations of M. ciliaris are the soil texture and altitude (44.44%), followed by climate (11.11%). On the other hand, no significant associations were found between gene diversity at each locus (He) as well as mean gene diversity over all loci (Heall) with environmental factors (data not shown).
Discussion
Quantitative and molecular population differentiation levels in M. ciliaris
Low population differentiation at quantitative traits (Q ST ) and SSR markers (F ST ) was found in M. ciliaris, indicating that studied populations have not been isolated for long periods of time. This result may be explained by the restricted geographical distribution of this species in Tunisia. In contrast, Badri et al. (2004a) reported higher level of quantitative and molecular genetic differentiation among natural populations of M. truncatula growing spontaneously in the same areas as those of M. ciliaris. While M. truncatula is a widespread species in Tunisia, M. ciliaris is limited to the north of country extending from humid to inferior semi-arid stages. Accordingly, geographical range has been shown to be a good predictor of the levels of genetic variation in plants (Gitzendanner and Soltis 2000). Similar levels of within-population genetic variation at quantitative traits and SSR markers were found for studied populations of M. ciliaris. Furthermore, most of this genetic variability was found to occur within populations. This finding is discordant with that found for several studies based on molecular markers. Indeed, self-fertilizing plant species tend to have lower within-population and higher among-population genetic variation than out-crossing plant species (Charlesworth 2003). This is a direct consequence of the mating system. Inbreeding reduces the effective population size, and consequently enhances the effect of genetic drift. The majority of correlations established between the 19 measured quantitative traits are positive. These results suggest that earlier emerging seedlings were the earliest-flowering and they allocated the largest effort to reproduction. This finding was reported for Tunisian natural populations of M. truncatula and M. laciniata (Badri et al. 2007) and French populations of M. truncatula (Bonnin et al. 1997). Low mean heterozygosity, about 1.3%, found for this collection of M. ciliaris indicates that this species is preferentially self-pollinating. In contrast to Q ST , no significant correlation was found between F ST and geographical distances; rejecting the hypothesis of isolation by distance. Nevertheless, we detected a significant pattern of fine-scale spatial genetic structure consistent with the model of isolation-by-distance: pods separated by short distances are more likely to be genetically related than those that are farther apart.
Quantitative traits versus SSR markers
Comparison of differentiation at quantitative traits (Q ST ) and molecular markers (F ST ), supposed neutral, allows to determine the relative importance of natural selection and genetic drift in the differentiation process (Spitze 1993). We found three different patterns of population differentiation across the 19 measured traits for M. ciliaris. First, we found that the majority of measured traits (11/19) displayed no significant difference in the level of Q ST and F ST , indicating that the effects of drift and selection are indistinguishable to explain the level of divergence (Merilä and Crnokrak 2001). However, significant correlations were established between these traits and eco-geographical factors (7/9), consistent with selection for local adaptation rather than genetic drift. Second, we found that 7 out of these traits displayed significantly smaller Q ST than F ST . This finding and the observed lack of correlation between F ST values and geographical distance among populations support the hypothesis of convergent selection for these traits. Third, only one trait out of these measured traits exhibited significantly higher Q ST than F ST , consistent with divergent selection. Significant associations found between this trait (SFT6) with climate and altitude factors may suggest that this particular character is adapting in response to the regional differences detected in these eco-geographical factors. A synthetic review performed by Merilä and Crnokrak (2001) reported that more than 20 independent studies have compared F ST and Q ST values. The overall trend revealed by those studies is that natural selection (Q ST > F ST ) appears to be a major differentiating and orienting force of regional evolutionary change, maintaining genetic polymorphisms under conditions of environmental heterogeneity and stress (Bonnin et al. 1996; Kremer et al. 1997; Podolsky and Holtsford 1995). Other evolutionary forces, such as mutation, drift and migration, do interact with selection but appear subordinate to natural selection. Depending on markers type some differences in F ST values were found but they are also dependent on plant species as reported by Isabel et al. (1999).
We found no significant correlation between population differentiation at quantitative traits and SSR markers. Accordingly, numerous studies do not support such correlation and hence molecular markers become inaccurate predictors of variation at quantitative traits (Reed and Frankham 2001; Gomez-Mestre and Tejedo 2004; Volis et al. 2005). Absence of such association between genetic variation at quantitative traits and molecular markers might be explained by several genetic and environmental factors. These factors include a non-additive genetic component such as epistasis or dominance (Crnokrak and Roff 1995), the conversion of non-additive genetic variance components to additive through genetic drift (Lopez-Fanjul et al. 2003), variation in mutation rates across loci, and variation in selection pressures and eco-geographical effects (Reed and Frankham 2001). Nevertheless, in some cases quantitative and neutral molecular distances have been reported to show a significant positive correlation (Merilä and Crnokrak 2001; Stenoien et al. 2005; Zhan et al. 2005), suggesting that variation at molecular markers can be used as a predictor of the degree of quantitative differentiation. A review covering more than 70 studies performed on various species revealed that established correlations between the population differentiation at molecular markers and quantitative traits were not significant, with an overall correlation coefficient of −0.08 (Reed and Frankham 2001). As the population differentiation at quantitative traits was found to be significantly correlated with geographical distances, it is not surprising that we failed to detect a relationship between Q ST and F ST .
Associations of quantitative traits and SSRs with environmental factors
An additional factor that may cause genetic divergence of populations is the presence of local adaptation via genotype-by-environment interactions. In this study, established correlations between quantitative traits and eco-geographical factors (soil texture, altitude and climate) suggest that these particular traits are adapting in response to the regional differences detected in such factors. In agreement with our findings, significant correlations between quantitative traits and environmental factors were observed in populations of Triticum dicoccoides (wild emmer wheat) (Li et al. 2001) and in Hordeum spontaneum (wild barley) (Huang et al. 2002) showing a significant micro-geographical genetic differentiation in response to climate (solar radiation, temperature and aridity stress). This micro-geographical adaptive differentiation with respect to climate was also found in some tree species such as Pinus edulis (pinon pine) (Mitton and Duran 2004). Overall, the moderate number of correlations established between quantitative traits and eco-geographical factors and the low Q ST found for M. ciliaris suggest its weak phenotypic plasticity to adapt to local varying environmental conditions.
Several studies reported significant correlations between SSR markers and eco-geographical factors suggesting the operation of natural selection on these markers by creating SSR loci regional divergence. These results were observed in wild emmer wheat (Li et al. 2000), in wild barely (Ivandic et al. 2002) and in the selfing plant, Arabidopsis thaliana, (Innan et al. 1997). However, in agreement with Jaramillo-Correa et al. (2001) our findings do not support this hypothesis suggesting that used SSR markers were selectively neutral.
The high proportion of amplification transferability of SSR markers from M. truncatula to M. ciliaris suggests that genetic and genomic tools developed for the model legume M. truncatula will radically improve genetic characterizations and breeding programs of M. ciliaris. We found a moderate degree of differentiation among analyzed populations of M. ciliaris in both quantitative traits and SSR markers. There was no significant correlation between population differentiation at quantitative traits and molecular markers. In contrast to the among populations genetic structure, we detected a significant pattern of fine-scale spatial genetic structure consistent with the model of isolation-by-distance. Further study is needed to analyze the mechanisms underlying adaptive genetic differentiation within and among natural populations of M. ciliaris in relation to environmental factors. We call also for further research to focus on ecological genetics of salt stress tolerance in this species.
References
Abdelkefi A, Boussaid M, Biborchi A, Haddioui A, Salhi-Hannachi A, Marrakchi M (1996) Genetic diversity inventory and evaluation of spontaneous species belonging to Medicago L. genus in Tunisia. Cah Options Méditerr 18:143–149
Badri M, Zribi K, Soula S, Lazrek F, Badri Y, Khaldi M, Yousfi N, Djebali N, Huguet T, Aouani ME (2004a) Specificities and usefulness of medics-sinorhizobia symbioses in Tunisian areas: preliminary assessment of the statement. In: Abstracts of the 5th Tunisian-Japanese on Culture, Science and Technology, Sfax, Tunisia
Badri M, Huguet T, Aouani ME (2004b) Comparison of quantitative genetic traits among natural populations of the model legume Medicago truncatula in Tunisian arid areas. In: Abstracts of arid regions of aridoculture et cultures oasiennes, Djerba, Tunisia
Badri M, Ilahi H, Huguet T, Aouani ME (2007) Quantitative and molecular genetic variation in sympatric populations of Medicago laciniata and M. truncatula (Fabaceae): relationships with eco-geographical factors. Genet Res 89:107–122
Barker DG, Bianchi S, London F, Datee Y, Duc G, Essad S, Flament P, Gallusci P, Génier G, Guy P, Muel X, Tourneur J, Dénarié J, Huguet T (1990) Medicago truncatula, a model for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol Biol Rep 8:40–49
Belkhir K (2001) Genetix software, version 4.01. Laboratoire Génome, Populations, Interactions, CNRS UPR 9060, Montpellier, France (genetix@crit.univ-montp2.fr)
Bena G, Prosperi JM, Lejeune B, Olivieri I (1998) Evolution of annual species of the genus Mediacgo: a molecular phylogenetic approach. J Mol Evol 9(3):552–559
Berlow EL, D’Antonio CM, Reynolds SA (2002) Shrub expansion in montane meadows: the interaction of local-scale disturbance and site aridity. Ecol Appl 12:1103–1118
Bonnin I, Prosperi JM, Olivieri I (1996) Genetic markers and quantitative genetic variation in Medicago truncatula (Leguminoseae): a comparative analysis of population structure. Genetics 143:1795–1805
Bonnin I, Prosperi JM, Olivieri I (1997) Comparison of quantitative genetic parameters between two natural populations of a selfing plant species, Medicago truncatula Gaertn. Theor Appl Genet 94:641–651
Charlesworth D (2003) Effects of inbreeding on the genetic diversity of populations. Philos Trans Roy Soc B 358(1434):1051–1570
Clausen JD, Keck D, Hiesey WM (1940) Experimental studies on the nature of species. In: Effect of varied environments on western North American plants. Carnegie Institute of Washington, Washington, DC
Crnokrak P, Roff DA (1995) Dominance variance-associations with selection and fitness. Heredity 75:530–540
Cruse-Sanders JM, Hamrick JL (2004) Spatial and genetic structure within populations of wild American ginseng (Panax quinquefolius L., Araliaceae). J Hered 95(4):309–321
Eujayl I, Sledge MK, Wang L, May GD, Chekhovskiy K, Zwonitzer JC, Mian MAR (2004) Medicago truncatula EST-SSRs reveal cross-species genetic markers for Medicago spp. Theor Appl Genet 108:414–422
Falconer DS, MacKay TFC (1996) Introduction to quantitative genetics, 4th edn. Longman Group Ltd, London
Gherardi M, Mangin B, Bonnet D, Goffinet B, Huguet T (1998) A method to measure genetic distance between allogamous populations of alfalfa (Medicago sativa) using RAPD molecular markers. Theor Appl Genet 96:406–412
Gitzendanner MA, Soltis PS (2000) Patterns of genetic variation in rare and widespread plant congeners. Am J Bot 87:783–792
Gomez-Mestre I, Tejedo M (2004) Contrasting patterns of quantitative and neutral genetic variation in locally adapted populations of the natterjack toad, Bufo calamita. Evolution 58(10):2343–2352
Goudet J (1995) FSTAT, version 1.2. A computer program to calculate F-statistics. J Hered 86:485–486
Gutierrez MV, Vaz Patto MC, Huguet T, Cubero JI, Moreno MT, Torres AM (2005) Cross-species amplification of Medicago truncatula microsatellites across three major pulse crops. Theor Appl Genet 110:1210–1217
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Huang QY, Beharav A, Youchun UC, Kirzhner V, Nevo E (2002) Mosaic microecological differential stress causes adaptive microsatellite divergence in wild barley, Hordeum spontaneum, at Neve Yaar, Israel. Genome 45:1216–1229
Innan H, Terauchi R, Miyashita NT (1997) Microsatellite polymorphism in natural populations of wild plant Arabidopsis thaliana. Genetics 146:1441–1452
Isabel N, Beaulieu J, Thériault P, Bousquet J (1999) Direct evidence for biased gene diversity estimates from dominant random amplified polymorphic DNA (RAPD) fingerprints. Mol Ecol 8:477–483
Ivandic V, Hackett CA, Nevo E, Keith R, Thomas WTB, Forster BP (2002) Analysis of simple sequence repeats (SSRs) in wild barley from the Fertile Crescent: associations with ecology, geography and flowering time. Plant Mol Biol 48:511–527
Jaramillo-Correa JP, Beaulieu J, Bousquet J (2001) Contrasting evolutionary forces driving population structure at expressed sequence tag polymorphisms, allozymes and quantitative traits in white spruce. Mol Ecol 10:2729–2740
Julier B, Flajoulot S, Barre P, Cardinet G, Santoni S, Huguet T, Huyghe C (2003) Construction of two genetic linkage maps in cultivated tetraploid alfalfa (Medicago sativa) using microsatellite and AFLP markers. BMC Plant Biol 3:9
Jump AS, Penuelas J (2005) Running to stand still: adaptation and the response of plants to rapid climate change. Ecol Lett 8:1010–1020
Kashi Y, King D, Soller M (1997) Simple sequence repeats as a source of quantitative variation. Trends Genet 13:74–78
Kremer A, Zanetto A, Ducousso A (1997) Multilocus and multitrait measures of differentiation for gene markers and phenotypic traits. Genetics 145:1229–1241
Lesins KA, Lesins I (1979) Genus Medicago (Leguminosae). A Taxogenetic Study. The Hague, The Netherlands
Li Y, Röder MS, Fahima T, Beiles A, Korol A, Nevo E (2000) Natural selection causing microsatellite divergence in wild emmer wheat at the ecologically variable microsite at Ammiad, Israel. Theor Appl Genet 100:985–999
Li YC, Krugman T, Fahima T, Beiles A, Korol AB, Nevo E (2001) Spatiotemporal allozyme divergence caused by aridity stress in a natural population of wild wheat, Triticum dicoccoides, at the Ammiad microsite, Israel. Theor Appl Genet 102:853–864
Loiselle BA, Sork VL, Nason J, Graham C (1995) Spatial genetic structure of a tropical understory shrub, Psychotria officinalis (Rubiaceae). Am J Bot 82:1420–1425
Lopez-Fanjul C, Fernandez A, Toro MA (2003) The effect of neutral nonadditive gene action on the quantitative index of population divergence. Genetics 164:1627–1633
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:1055–1067
Merilä J, Crnokrak P (2001) Comparison of differentiation at marker loci and quantitative traits. J Evol Biol 14:892–903
Mitton JB, Duran KL (2004) Genetic variation in pinon pine, Pinus edulis, associated with summer precipitation. Mol Ecol 13:1259–1264
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Palo JU, O’Hara RB, Laugen AT, Laurila A, Primmer CR, Merilä J (2003) Latitudinal divergence of common frog (Rana temporaria) life history traits by natural selection: evidence from a comparison of molecular and quantitative genetic data. Mol Ecol 13:2865–2869
Perrier X, Jacquemoud-Collet JP (2006) DARwin software version 5.0.148 (http://darwin.cirad.fr/darwin)
Podolsky RH, Holtsford TP (1995) Population structure of morphological traits in Clarkia dudleyana. I. Comparison of F ST between allozymes and morphological traits. Genetics 140:733–744
Pottier-Alapetite G (1979) Flore de la Tunisie, Angiospermes, dicotylédones, Apétales-Dialypétales. Publications scientifiques tunisiennes, Tunis
Pressoir G, Berthaud J (2004) Population structure and strong divergent selection shape phenotypic diversification in maize landraces. Heredity 92:95–101
Reed DH, Frankham R (2001) How closely correlated are molecular and quantitative measures of genetic variation? A meta-analysis. Evolution 55:1095–1103
Rogers SO, Bendish AJ (1988) Extraction of DNA from plant tissues. In: Gelvin SB, Schiperoot RA (eds) Plant molecular biology manual. Kluwer, Academic publishers, Dordrecht Boston London
Rousset F (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Saal B, Plieske J, Hu J, Quiros CF, Struss D (2001) Microsatellite markers for genome analysis in Brassica. II. Assignment of rapeseed microsatellites to the A and C genomes and genetic mapping in Brassica oleracea L. Theor Appl Genet 102:695–699
SAS Institute (1998) SAS/STAT User’ Guide, version 70. SAS Institute Inc, Cary NC
Shrestha MK, Golan-Goldhirsh A, Ward D (2002) Population genetic structure and the conservation of isolated populations of Acacia raddiana in the Negev Desert. Biol Conserv 108:119–127
Spitze K (1993) Population structure in Daphnia obtusa: quantitative genetic and allozymic variation. Genetics 135:367–374
Stenoien HK, Fenster CB, Tonteri A, Savolainen O (2005) Genetic variability in natural populations of Arabidopsis thaliana in northern Europe. Mol Ecol 14:137–148
Turesson G (1925) The plant species in relation to habitat and climate. Hereditas 6:147–236
Turpeinen T, Tenhola T, Manninen O, Nevo E, Nissilä E (2001) Microsatellite diversity associated with ecological factors in Hordeum spontaneum populations in Israel. Mol Ecol 10:1577–1591
Van Rosum F, Bonnin I, Fénart S, Pauwels M, Petit D, Saumitou-Laprade P (2004) Spatial genetic structure within a metallicolous population of Arabidopsis halleri, a clonal, self-incompatible and heavy-metal-tolerant species. Mol Ecol 13:2959–2967
Volis S, Yakubov B, Shulgina I, Ward D, Mendlinger S (2005) Distinguishing adaptive from nonadaptive genetic differentiation: comparison of Q ST and F ST at two spatial scales. Heredity 95:466–475
Volis S, Yakubov B, Shulgina I, Ward D, Zur V, Mendlinger S (2001) Tests for adaptive RAPD variation in population genetic structure of wild barley, Hordeum spontaneum Koch. Biol J Linn Soc 74:289–303
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Zhan J, Linde CC, Jürgens T, Merz U, Steinebrunner F, Mc Donald BA (2005) Variation for neutral markers is correlated with variation for quantitative traits in the plant pathogenic fungus Mycosphaerella graminicola. Mol Ecol 14:2683–2693
Zribi K, Badri Y, Saidi S, van Berkum P, Aouani ME (2007) Medicago ciliaris growing in Tunisian soils is preferentially nodulated by Sinorhizobium medicae. Aust J Soil Res 45:473–477
Acknowledgements
We would like to thank Soumaya Arraouadi for molecular technical assistance and two anonymous referees for helpful comments on the manuscript. This work was funded in part by Tunisian-French collaborative programs (CMCU 00F0909 and PICS 712).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Badri, M., Zitoun, A., Soula, S. et al. Low levels of quantitative and molecular genetic differentiation among natural populations of Medicago ciliaris Kroch. (Fabaceae) of different Tunisian eco-geographical origin. Conserv Genet 9, 1509–1520 (2008). https://doi.org/10.1007/s10592-007-9483-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-007-9483-z