Abstract
The saltwater crocodile (Crocodylus porosus) is the largest and most broadly distributed crocodilian species, and thus is of special conservation and economic interest. Similar to other parts of its range throughout the Indo-Pacific, C. porosus distributed in the Republic of Palau have experienced a severe population decline over the past century primarily due to commercial hunting and eradication campaigns. In addition, several thousand crocodiles of undocumented species and origin were imported into Palau during the 1930’s for commercial farming purposes, potentially polluting the gene pool of the endemic saltwater crocodiles. Analysis of 39 individuals collected throughout the Republic of Palau revealed a single mitochondrial DNA control region haplotype shared by populations sampled in Sulawesi, Borneo and Australia. The mtDNA results, in combination with microsatellite genotypic data at six loci, detected no evidence for inter-specific hybridization between endemic Palauan C. porosus and potentially introduced Crocodylus species. There was no evidence for a genetic bottleneck in the Palauan population, however an excess of rare alleles was identified, indirectly suggesting a recent history of admixture potentially linked to introductions of non-native C. porosus. Following from these findings, Palauan C. porosus should be included in the single ESU previously established for all saltwater crocodiles given the recovery of a fixed, but geographically widespread haplotype. Although Palauan C. porosus exhibited significant genetic differentiation relative to all other sampled populations, it’s delineation as a distinct management unit is precluded at the present time by evidence that the genetic integrity of the population may have been compromised by the introduction of non-native saltwater crocodiles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The saltwater crocodile, Crocodylus porosus, is considered an endangered or threatened species throughout the majority of its range, and is of special conservation and economic interest (Ross 1998). It is the largest and broadest ranging of crocodilian species, occurring in coastal and estuarine habitats across the Indo-Pacific region, from Northern Australia, throughout Southeast Asia, to India and Sri Lanka. The distribution of C. porosus in the western Pacific Ocean ranges from the Solomon Islands to Vanuatu, and in the Republic of Palau, a 160 km long archipelago centered at approximately 7°30′ N latitude, 134°30′ E longitude, in the Caroline Islands group in Micronesia.
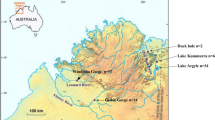
Palau is composed of approximately 350 coralline and volcanic islands, largely surrounded by a barrier reef (Fig. 1). Babeldaob is the largest island of about 333 km2, and comprises roughly 80% of Palau’s total landmass. Approximately 80% of Babeldaob’s 157 km of coastline are covered by mangrove forest, prime habitat for C. porosus. Ngeruktabel (20.2 km2) and Beleliu Island (12.4 km2) follow in size and include extensive mangrove swamps. A comprehensive review of Palauan herpetofauna and zoogeographic history is presented in Crombie and Pregill (1999).
The crocodiles of the Palauan islands are relatively isolated, separated from other populations of C. porosus, and additional crocodilian species by considerable expanses of open ocean (Fig. 1). The Philippine islands, located approximately 800 km to the west, are home to severely reduced populations of C. porosus and the endangered Philippine freshwater crocodile, C. mindorensis. Equally distant from Palau, the island of New Guinea continues to harbor significant populations of both C. porosus and the freshwater New Guinea crocodile, C. novaeguineae. Populations of C. porosus also persist on Sulawesi and Borneo. The only other Crocodylus species occurring in Southeast Asia is the endangered Siamese crocodile, C. siamensis. This species has been farmed extensively for many decades in Cambodia, Indonesia and Thailand, but remnant populations are thought to exist in eastern Borneo and Cambodia (Ross 1998).
The wild populations of C. porosus on Palau, historically estimated at approximately 1500 individuals (Messel and King 1991), were decimated in the decades following World War II, and large individuals were preferentially eradicated during a series of commercial hunting initiatives after two crocodile/human-related conflicts that occurred in 1964 and 1965. In the proceeding years, crocodile populations were noticeably depleted, both in number and adult size. Unofficial surveys subsequently reported the sightings of only 23 animals of unreported size at the close of 1967, and 1 year later, 85 animals were observed, six of which were considered large enough to be a danger to people (2–3 m long) (Brazaitis et al. 2003). Although Messel and King (1991) estimated that several hundred crocodiles of “commercial size” (1.4–3 m) may have existed in Palau in the early 1990’s, an official United States Fish and Wildlife Service survey conducted in 1993 reported sighting 45 crocodiles between 1.5 and 3.5 m in length (Brazaitis et al. 2003).
In addition to the severe population contraction following the hunting and eradication campaigns, several thousand crocodiles of undocumented species and origin were imported into Palau during the 1930’s for commercial farming purposes. Farming initiatives ceased, however, following the major naval and land battles occurring on or around Palau during World War II, leaving behind no records of the fate of the farmed crocodiles or their disposition. The historical legacy of these programs persist, however, as the original presence of imported, non-native crocodilians gave rise to the belief, still held by local inhabitants, that more than one dissimilar crocodilian species reside on Palau.
The potential hybridization between Palauan C. porosus and introduced Crocodylus species combined with the reported severity of the population contraction during the mid-20th century have significant implications for crocodilian conservation and management on Palau. The present study utilizes Crocodylus-specific mitochondrial and nuclear microsatellite genetic markers to assess the distinctiveness of the Palauan C. porosus relative to conspecific populations distributed throughout the Indo-Pacific, and to test for the genetic signatures of population contraction and/or recent hybridization and introgression with non-native crocodilians introduced over the course of the past century.
Materials and methods
Sampling and data collection
Blood samples from 39 C. porosus individuals were collected in the Republic of Palau between June 2003 and June 2005. Five C. porosus individuals were also sampled from Queensland, Australia in October 2005. All collection sites were geo-referenced with latitude and longitude coordinates via GPS (Table 1). Additionally, museum specimens (e.g. teeth) of exemplar individuals from throughout the range of C. porosus in the Indo-Pacific region were sampled as listed in Table 1. Sequences previously deposited in GenBank were also utilized (Table 1). Moreover, microsatellite genotypic datasets of population samplings on Papua New Guinea and Indonesia (Gratten 2003) were used as reference populations by which to compare the relative genetic composition, population structuring and evolutionary novelty of the Palauan population. Specifically, calibrated microsatellite genotypes (see below for calibration details) were utilized from the following distinct populations pursuant to the findings of Gratten (2003): northwestern Papua New Guinea (nwPNG; n = 27), northeastern Papua New Guinea (nePNG; n = 32), New Britain, Papua New Guinea (NB; n = 21), North Solomons Province, Solomon Islands (NSP; n = 12), southern Papua New Guinea (sPNG; n = 31), Sunda Shelf sampled in Borneo (SUN; n = 19), and Sulawesi (SUL; n = 11). Lastly, exemplar individuals of C. mindorensis (n = 1), C. novaeguineae (n = 3), and C. siamensis (n = 11) were sampled from the St. Augustine Alligator Farm for the purpose of conducting comparative analyses of inter-specific haplotypic and genotypic divergences relative to C. porosus.
Palauan crocodiles were captured at night, sampled by veni-puncture, and released unharmed at the capture site. Blood samples from zoological collection animals were taken during the course of routine medical treatments.
DNA was extracted from all blood samples using the DNeasy Tissue kit and manufacturer protocols (Qiagen, Inc.). Roots of teeth from museum specimens were powderized using mortar and pestle, and DNA was extracted in a dedicated facility for ancient DNA work according to a modified protocol (available from corresponding author) following all necessary precautions to prevent contamination by extant specimens.
A ∼620 basepair fragment of the mitochondrial control region encompassing Domains I and II was amplified in whole using external primers tPhe-L and CR2H (Ray and Densmore 2002) for the DNA extractions from blood samples or, in the case of the DNA extractions from museum specimens, as a set of four overlapping fragments not exceeding 180 basepairs in length each (tPhe-L/CporCR1B: AACTAATGGATGGGTGTTRGGG; CporCR2A: CAGCTATGTATTATAAGGCATTCATTT/CporCR2B: CAGGCAATAGCAAGATGRGTA; CporCR3A: ACCTCTGGTTATCACTCTC/CporCR3B: AGATATAAGCCCCCGGGTAG; CporCR4A: TGGGGAGATCTCATCCMCTAC/CR2H). All PCR reactions were carried out on an MJ Research DNA engine thermal cycler in 25 μl reactions containing: ∼20–50 ng of DNA, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 0.5 μM of each primer and 0.5 U of AmpliTaq DNA polymerase (PE Biosystems). Cycling conditions for all primer pairs consisted of 95°C for 2 min, 35 cycles of 95°C for 30 s, 50°C for 30 s, 72°C for 30 s, and a final extension of 72°C for 7 min. Double-stranded PCR products were sequenced using Big Dye 3.1 terminators on an ABI 3730 DNA sequencer (Applied Biosystems).
Genotypic data were obtained for the field-collected samples from Palau and Australia at six microsatellite loci (Cj16, Cj18, Cj104, Cj107, Cj127, Cj131; FitzSimmons et al. 2000). In addition, a representative sample of 30 individuals from the Gratten (2003) study was genotyped at the same six loci to calibrate allele calls collected in different laboratories. Allele calls at all but one locus were of different absolute sizes than in Gratten (2003), however, the discrepancies were consistent in all cases allowing for calibration to accurately combine the two datasets. All PCR reactions were carried out on an MJ Research DNA engine thermal cycler in 12.5 μl reactions containing: ∼20–50 ng of DNA, 10 mM Tris–HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 200 μM dNTPs, 7.5 μg bovine serum albumin (BSA), 0.5 μM of each primer and 0.5 U of AmpliTaq DNA polymerase (PE Biosystems). Reaction conditions for all primers were optimized using a ‘touchdown’ cycling program which consisted of: 95°C for 10 min; 35 cycles of 95°C for 30 s, annealing for 30 s, and 72°C for 45 s; and a final step of 72°C for 7 min (Russello et al. 2001). The annealing step in the ‘touchdown’ program decreased 2°C every other cycle from 59°C until it reached 51°C (the 9th cycle) at which point the remaining cycles continued with a 51°C annealing temperature.
Haplotypic variation and genealogical relationships
Haplotypic (h; Nei 1987) and nucleotide (π; Nei 1987) diversity estimates for the Palau population were calculated based on mitochondrial DNA (mtDNA) control region (CR) sequences as executed in ARLEQUIN (Schneider et al. 2000). Pairwise genetic distances were calculated in PAUP*4.0b10 (Swofford 2002) assuming the HKY + G model of nucleotide substitution as selected following a series of hierarchical likelihood ratio tests as implemented in Modeltest (Posada and Crandall 1998).
Genealogical relationships among all sampled haplotypes throughout the Indo-Pacific were reconstructed as a haplotype network using the statistical parsimony method of Templeton et al. (1992) as implemented in TCS, version 1.06 (Clement et al. 2000). This method estimates the maximum number of substitutions to connect parsimoniously two haplotypes with 95% confidence and is useful for inferring relationships among genes with low levels of divergence.
Genotypic variation and population differentiation
Allelic diversity, observed (HO) and expected heterozygosity (HE) were calculated at each of six loci for the population samplings on Palau, Australia, Papua New Guinea, and Indonesia. HE was computed using the estimate of Nei and Roychoudhury (1974) as implemented in Microsatellite Analyzer (Dieringer and Schlotterer 2002). Deviation from Hardy–Weinberg (H-W) equilibrium was assessed using exact tests based on the Markov chain method of Guo and Thompson (1992) as implemented in GENEPOP 3.3 (1000 dememorization, 1000 batches and 10,000 iterations; Raymond and Rousset 1995). The alternative null hypothesis of heterozygote deficiency was also tested using the same methodology. Linkage disequilibrium was investigated for all pairs of loci using GENEPOP 3.3 (Raymond and Rousset 1995). Type I error rates for tests of linkage disequilibrium and departure from H-W expectations were corrected for multiple comparisons using the sequential Bonferroni procedure (Rice 1989).
Levels of nuclear DNA differentiation among populations were estimated by pairwise population comparisons of θ, an analogue of Fst (Weir and Cockerham 1984) calculated in GENETIX (Belkhir et al. 2001) and Rho, an unbiased Rst estimator implemented in RSTCALC (Goodman 1997). Correspondence of geographically separated populations as discrete genetic units was further tested using the Bayesian method of Pritchard et al. (2000) as implemented in Structure. Specifically, the number of populations (K) reconstructed from the total sample was estimated using the average of three iterations of a Markov chain Monte Carlo simulation (MCMC) including 1.0 × 106 repetitions following an initial burning of 5.0 × 104 repetitions. The Structure analysis assumes that all potentially contributing populations have been representatively sampled. As this is clearly not the case in the current study, the purpose of the analysis was not to infer the number of sub-populations within the dataset but rather to use this method as a qualitative estimate of the genetic distinctiveness of the sampled populations.
The genetic distinctiveness of the sampled populations was further assessed as the proportion of individuals correctly assigned to the population from which they were originally sampled using the exclusion-simulation test of the partial Bayesian assignment method of Rannala and Mountain (1997) as implemented in GENECLASS (Cornuet et al. 1999). The self-assignment tests were conducted following simulation of 10,000 randomly generated genotypes. Individuals with a likelihood <5% of belonging to their sampled population were not assigned to that locality.
Demographic history
Evidence for a population bottleneck in the Palauan population was assessed using the microsatellite dataset and the Cornuet and Luikart (1996) test for excess heterozygosity as enacted in Bottleneck (Piry et al. 1999). As rare alleles, which contribute minimally to heterozygosity, are the first to be lost during a population contraction, this approach relies on the supposition that a recently reduced population will exhibit an HE excess relative to an equilibrium population with an equivalent number of alleles (Piry et al. 1999). Analyses were run assuming loci conformed to the two-phase mutation model (TPM) with 95% single-step mutations and a variance of 12 for multi-step mutations, consistent with recommendations by Piry et al. (1999). The Wilcoxon sign test was used to determine if the number of loci exhibiting heterozygosity excess was significant. Despite the low power afforded by the sampling of six loci, the test of Cornuet and Luikart (1996) was performed in order to detect general patterns associated with the genotypic variation recovered in this population.
Results
Haplotypic variation
A single mtDNA control region (CR) haplotype was recovered among the 39 C. porosus individuals sampled from Palau. Nine additional mtDNA CR haplotypes were identified from exemplar individuals from Papua New Guinea (PNG), Borneo, Sulawesi, Philippines, Cambodia, Thailand, Australia, and the Bismarck and Solomon Islands, respectively. Sequence divergence among C. porosus haplotypes from different sampling sites ranged from 0.00% to 1.28% based on HKY + G distances. The Palau haplotype exhibited intra-specific sequence divergences ranging from 0.17% (Borneo) to 0.90% (Cambodia, Thailand). This relatively low level of intra-specific sequence divergences across a wide geographical area is in stark contrast to those exhibited between the Palau haplotype and haplotypes recovered from exemplar individuals from C. siamensis (8.31%), C. novaeguineae (8.85%), and C. mindorensis (11.1%).
Genealogical relationships
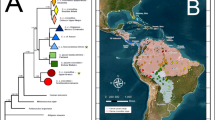
A single haplotype network was reconstructed within which all haplotypes had a 95% probability of being parsimoniously connected (Fig. 2). Overall, there was little geographic structure to the recovered relationships, punctuated by the lack of clustering of haplotypes sampled from multiple localities on Australia (E, J), PNG (H, I) and the Philippines (A, C) (Fig. 2). Haplotype A, sampled from two localities on Mindanao Island in the Philippines, was central to the network, connected by a single step to haplotypes recovered from such diverse areas as Borneo (B), Sulawesi (D), and the Bismarck (C) and Solomon Islands (D). The Palau haplotype (G) was most closely related to haplotype B sampled on Borneo, differing by a single nucleotide substitution (Fig. 2). This limited sampling provides only an initial reconstruction of the phylogeographic context of Palauan C. porosus. In fact, a forthcoming study of the phylogeographic structure of C. porosus throughout the Indo-Malay archipelago and western Pacific found the Palau haplotype (G) to be identical to a widespread haplotype recovered from distinct localities in Australasia. Specifically, of 170 samples sequenced from throughout the region, the Palauan haplotype was present in Sulawesi (4/12), east Kalimantan in Borneo (1/20) and the Northern Territory of Australia (5/26) (data not shown; Gratten, in preparation).
Haplotype network constructed under statistical parsimony for all C. porosus haplotypes recovered across the Indo-Malay Archipelago and western Pacific Ocean. Geographic origin of each haplotype is indicated on the map to the left, overlaying the C. porosus distribution in the Indo-Pacific (shaded in dark gray). The above reconstruction is based on the maximum number of substitutions to connect parsimoniously two haplotypes with 95% confidence, with open circles representing hypothesized intermediate haplotypes not sampled. Sampling areas include: Australia (AUS), Bismarck Islands (BIS), Borneo (BOR), Cambodia (CAM), Palau (PAL), Papua New Guinea (PNG), Philippines (PHI), Solomon Islands (SOL), Sulawesi (SUL), and Thailand (THA). Number of individuals exhibiting sampled haplotype included in parentheses
Genotypic variation
Genotypes were generated at six loci for all individuals sampled on Palau (n = 39) and in Queensland, Australia (n = 5). These data were augmented by genotypes for the same loci from 153 individuals across seven populations distributed throughout PNG and across Sulawesi and the Sunda Shelf regions of Indonesia (Gratten 2003). No significant deviation from H-W equilibrium was detected for any locus in any sampled population following sequential Bonferroni correction. Furthermore, there was no evidence of non-random association of genotypes (P > 0.05) in any of the pairwise tests for linkage disequilibrium performed for all possible pairwise comparisons of the sampled loci.
Allelic diversity and expected hetorozygosities did not deviate significantly across populations, ranging from 3.17 (NSP) to 5.17 (sPNG) alleles per locus and from 0.456 (nwPNG) to 0.622 (AUS), respectively (Table 2). The percentage of private alleles per population, used as a measure of relative uniqueness, averaged 6.79% and ranged from 0.00% (nePNG, NB, SUL), to comparatively high proportions for the SUN (19.23%) and AUS (21.74%) populations (Table 2). Consequently, levels of allelic diversity (4.33), heterozygosity (0.575), and uniqueness (3.85% private alleles) in the Palauan C. porosus were not markedly different from any other sampled population (Table 2).
There was no evidence for a genetic bottleneck in the Palauan C. porosus population according to the test of Cornuet and Luikart (1996) for heterozygosity excess. This test assumes that in a stable population at mutation-drift equilibrium, an equal number of loci are expected to show heterozygosity excess and deficit, whereas in a recently bottlenecked population this ratio should be skewed significantly towards heterozygosity excess. Regarding the Palau population, none of the sampled loci exhibited heterozygosity excess and in fact, all were found to exhibit a deficit of heterozygosity ranging from slight (Cj18; −0.004) to highly significant (Cj104; −5.108). The probability of observing a heterozygosity deficit at all six loci by chance alone is very small (P < 0.015). This is the opposite of what is expected after a genetic bottleneck, but is consistent with either recent population expansion or admixture.
Population differentiation
The Palauan population of C. porosus was significantly genetically differentiated from all other sampled populations, on the basis of both θ and Rho (Table 3). Levels of differentiation for the Palauan population were comparable to those observed among other pairwise population comparisons, all of which were significant (Table 3).
Population self-assignment
Overall, 82.2% of individuals were correctly assigned to the population from which they were sampled according to the exclusion-simulation test (Cornuet et al. 1999) of the partial Bayesian assignment method of Rannala and Mountain (1997). Under this test, all but eight individuals sampled on Palau were correctly assigned to their population of origin (79.5%). Of particular note, two of eight individuals collected on but not assigned to Palau were assigned to the population on Sulawesi while the other six were not assigned to any sampled population. This level of self-assignment on Palau is consistent with values found in populations sampled on Papua New Guinea and Indonesia, which ranged from 63.2% (SUN) to 84.4% (nwPNG).
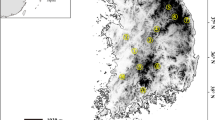
Six partitions (K = 6) were reconstructed from the total sample by the Bayesian method of Pritchard et al. (2000). The purpose of this analysis, however, was not to delineate the absolute number of groupings within the data insomuch as to assess the genetic distinctiveness of the sampled populations. In that regard, the Palauan C. porosus population constituted one of the distinct partitions revealed in this analysis, qualitatively exhibiting levels of heterogeneity and genetic admixture similar to that shown by the other recovered groupings (Fig. 3).
STRUCTURE plot indicating the genetic composition of sampled C. porosus populations according to the Bayesian method of Pritchard et al. (2000). Colors represent the relative contribution of each of six genetic partitions recovered from the data for each individual (column) in each sampled population. Population acronyms are as in Table 2
Discussion
In the absence of direct knowledge of the species identity and source population(s) of Crocodylus introduced to Palau in the 1930’s, population genetic approaches enable the indirect assessment of potential hybridization and introgression between Palauan C. porosus and non-native crocodilians. Several results from the current study argue against the historical introduction of non-native Crocodylus species to Palau. First, all 39 C. porosus individuals sampled on Palau shared a single mtDNA control region haplotype that was identical to previously sampled haplotypes observed in other saltwater crocodiles (Gratten, unpublished), and distantly related to those identified in several other Indo-Pacific species of Crocodylus (C. mindorensis, C. novaeguineae, C. siamensis). This represents good evidence that no maternal introgression has occurred between native and non-native species. Due to its maternal inheritance, however, mitochondrial DNA does not provide any information regarding potential one-way hybridization events between introduced male Crocodylus sp. and female C. porosus. The nuclear microsatellite markers utilized in the current study revealed a very low percentage of alleles shared between Palauan C. porosus and exemplars of potentially introduced C. mindorensis (4.6%), C. novaeguineae (0.0%), and C. siamensis (3.4%). This low frequency of allelic overlap between non-native species and Palauan C. porosus is consistent with no history of introgression. That said, the limited sampling of non-native species in the current study confers little power to detect male-mediated introgression if it had occurred. Future efforts should be made to generate larger comparative microsatellite data sets for non-native Crocodylus species that were potentially introduced to Palau to more definitely address this question.
Palauan C. porosus were fixed for a single mtDNA haplotype that was shared with other localities in Australasia. Despite this low level of mtDNA diversity, the Palauan C. porosus population was significantly genetically differentiated from other populations sampled across the Indo-Malay Archipelago and western Pacific Ocean on the basis of nuclear DNA markers.
There was no evidence for a genetic bottleneck in the Palauan C. porosus population, despite reports of a significant population contraction over the course of the past century (Ross 1998). Although the power of Cornuet and Luikart’s (1996) test to detect heterozygosity excess in the Palauan population is quite limited due to the small number of loci sampled, the results are consistent with those of Gratten (2003), who found no evidence for genetic bottlenecks in other Indo-Pacific populations of C. porosus that are also known to have experienced marked declines in recent times (Ross 1998). Interestingly, even though only six loci were sampled, all exhibited a deficit of heterozygosity, indicating an excess of rare alleles. This pattern is actually the opposite of what is expected following a bottleneck, and is indicative of either recent population expansion, or the recent influx of genetically distinct alleles due to immigration or admixture (Luikart and Cornuet 1998). Historical expansions dating to the Pleistocene have been detected for C. porosus maternal lineages in the majority of sampled populations throughout the Indo-Pacific (nwPNG, nePNG, NB, NSP, SUN), in spite of their shared history of recent decline (Gratten 2003). It is presently unknown whether a similar historical expansion occurred in the Palauan population, but it is unlikely that a similar event could explain the observed heterozygosity deficit because this effect is transient, lasting only 0.2–4 Ne generations (Cornuet and Luikart 1996; Luikart and Cornuet 1998).
Given that the Palauan population is understood to have experienced a strong demographic decline in the past century, and that rare alleles are likely to have been lost during this period, an excess of rare alleles may be a legacy of introductions of non-native C. porosus in the recent past, perhaps during the commercial farming initiatives in the 1930s. For the heterozygosity deficit to be a consequence of the introduced non-native C. porosus and subsequent admixture with the endemic population, the patterns observed from the mitochondrial and nuclear DNA data would require one or more of the following: (1) deliberate or natural introduction of C. porosus from localities that share the single haplotype recovered on Palau; (2) exclusive importation of males from non-Palauan populations; and/or (3) selective breeding of introduced males with native females. Although no direct evidence exists to support any of these scenarios, the revealed pattern of rare allele excess as well as the genotypic assignment of two Palauan individuals to the Sulawesi population indirectly suggest a recent history of admixture, likely linked to the introduction of non-native C. porosus to Palau.
Gratten (2003) recognized all populations throughout the sampled range of C. porosus in the Indo-Malay Archipelago and western Pacific Ocean as a single evolutionarily significant unit (ESU; Moritz 1994) based on the lack of deep phylogeographic structure across the region. At a finer level, Gratten (2003) proposed that the nwPNG, nePNG, nPNG islands, sPNG, Sulawesi and Sunda Shelf populations constitute distinct management units (Moritz 1994) on the basis of concordant mtDNA and microsatellite allelic frequency differences. Like all other populations throughout the Indo-Pacific, C. porosus on Palau should be included in the single ESU for saltwater crocodiles, given the recovery of a fixed, but geographically widespread haplotype.
Although Palauan C. porosus exhibited significant population differentiation relative to all other sampled populations, the identification of a potential genetic signature of admixture with non-native saltwater crocodiles precludes its delineation as a distinct management unit at this time. Given the difficulty of sampling saltwater crocodiles and the extent of their range, more comprehensive population surveys of C. porosus throughout the Indo-Pacific would provide greater context for investigating the extent of variation identified on Palau, as well as the formative processes, both natural and human-mediated, underlying the recovered patterns. Moreover, enhanced population samplings on Palau are critical for better assessing the genetic consequences of early 20th century introductions, including the potential for improved detection of individuals not impacted by the history of farming operations. Future studies of Palauan C. porosus will be further aided by the availability of a large number of additional microsatellite loci that are currently being optimized for this group (Glenn, pers. com). As a representative population at the distal end of the saltwater crocodile distribution in the Indo-Pacific, the Palauan C. porosus population remains vulnerable to disturbance as a consequence of its small size, thereby warranting continued protection of mangrove and other critical habitats, minimization of commercial hunting, and increased vigilance preventing the introduction of non-native crocodilians.
References
Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F (2001) Genetix, logiciel sous Windows TM pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UPR 9060. Université de Montpellier II, Montpellier (France)
Brazaitis P, Eberdong J, Brazaitis PJ (2003) The Saltwater Crocodile, Crocodylus porosus in the Republic of Palau: results of field investigations: 2 to 20 June 2003. A Special Report to the United States Fish and Wildlife Service and The Nature Conservancy, 103 pp (unpublished)
Clement M, Posada D, Crandall KA (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
Cornuet JM, Piry S, Luikart G, Estoup A, Solignac M (1999) New methods employing multilocus genotypes to select or exclude populations as origins of individuals. Genetics 153:1989–2000
Crombie RI, Pregill GK (1999) A checklist of the herpetofauna of the Palau Islands (Republic of Belau), Oceania. Herpetological Monographs 13:29–80
Dieringer D, Schlotterer C (2003) Microsatellite Analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes 3:167–169
FitzSimmons NN, Tanksley S, Forstner MRJ, Louis EE, Daglish R, Gratten J, Davis S (2000) Microsatellite markers for Crocodylus: new genetic tools for population genetics, mating system studies and forensics. In: Gordon C. Grigg, Frank Seebacher, Craig Franklin (eds) Crocodile biology and evolution. Surrey Beatty and Sons, Brisbane, Australia, pp 51–57
Gratten J (2003) The molecular systematics, phylogeography and population genetics of Indo-Pacific Crocodylus. Dissertation, University of Queensland
Goodman SJ (1997) Rst Calc: a collection of computer programs for calculating estimates of genetic differentiation and gene flow from microsatellite data and determining their significance. Mol Ecol 6:881–885
Guo SW, Thompson EA (1992) Performing the exact test of Hardy–Weinberg proportions for multiple alleles. Biometrics 43:805–811
Luikart G, Cornuet JM (1998) Empirical evaluation of a test for identifying recently bottlenecked populations from allele frequency data. Conserv Biol 12:228–237
Messel H, King FW (1991) Survey of the crocodile populations of the Republic of Palau, Caroline Islands, Pacific Ocean, 8–24 June 1991: A report to the government of the Republic of Palau, Koror, Palau, 49 pp (unpublished)
Moritz C (1994) Defining “Evolutionary Significant Units” for conservation. Trends Ecol Evol 9:373–375
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Nei M, Roychoudhury AK (1974) Sampling variances of heterozygosity and genetic distance. Genetics 76:379–390
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Heredity 90:502–503
Posada D, Crandall KA (1998) MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9221
Ray DA, Densmore L (2002) The crocodilian mitochondrial control region: general structure, conserved sequences, and evolutionary implications. J Exp Zool 294:334–345
Raymond M, Rousset F (1995) GENEPOP (v. 1.2): a population genetics software for exact tests and ecumenicism. J Heredity 86:248–249
Rice WR (1989) Analysing tables of statistical tests. Evolution 43:223–225
Ross JP (ed) (1998) Crocodiles. Status survey and conservation action plan, 2nd edn. IUCN/SSC Crocodile Specialist Group. IUCN, Gland, Switzerland and Cambridge, UK, viii + 167 pp
Russello M, Calcagnotto D, DeSalle R, Amato G (2001) Characterization of microsatellite loci in the endangered St. Vincent Parrot, Amazona guildingii. Mol Ecol Notes 1:162–164
Schneider S, Roessli D, Excoffier L (2000) Arlequin v2.000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Swofford DL (2002) PAUP*: Phylogenetic analysis using Parsimony, v.4.0b10. Illinois Natural History Survey, Champaign
Templeton AR, Crandall KA, Sing CF (1992) A Cladistic analysis of phenotypic associations with haplotypes inferred from restriction endonuclease mapping and DNA sequence data .3. Cladogram estimation. Genetics 132:619–633
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgments
A special thanks to the people of Palau for their hospitality and graciousness over the course of this study. We would like to particularly thank Mr. Joshua Eberdong, and the Bureau of Marine Resources, Republic of Palau, for their extensive field and logistical support and our Palauan field colleagues, Mr. Harvey Kloulechad and Mr. Theodore Ngiramelekei, who took great personal risks in securing the field-sampled materials. Nancy FitzSimmons kindly provided replicate samples for calibrating the genotypic datasets and offered insightful comments that greatly improved this work. Peter John Brazaitis and Julian Dendy assisted us in the field, and Chaz Hyseni and Greg Mulvey aided in the collection of molecular data for this study. We wish to acknowledge the Peabody Museum of Natural History, National Museum of Natural History, and Field Museum of Natural History for providing access to their collections. Additionally, the St. Augustine Alligator Farm, and the Gladys Porter Zoo supplied blood samples from their extensive animal collections. George Amato and Kent Vliet provided assistance in multiple capacities, and Sarah Klain, Bureau of Marine Resources, Koror, Palau graciously prepared the locality map in Figure 1. This work was supported by the U.S. Fish and Wildlife Service’s Pacific Islands Coastal Program Grant # 122005M020.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Russello, M.A., Brazaitis, P., Gratten, J. et al. Molecular assessment of the genetic integrity, distinctiveness and phylogeographic context of the Saltwater crocodile (Crocodylus porosus) on Palau . Conserv Genet 8, 777–787 (2007). https://doi.org/10.1007/s10592-006-9225-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-006-9225-7