Abstract
Recently, matrix norm \(l_{2,1}\) has been widely applied to feature selection in many areas such as computer vision, pattern recognition, biological study and etc. As an extension of \(l_1\) norm, \(l_{2,1}\) matrix norm is often used to find jointly sparse solution. Actually, computational studies have showed that the solution of \(l_p\)-minimization (\(0<p<1\)) is sparser than that of \(l_1\)-minimization. The generalized \(l_{2,p}\)-minimization (\(p\in (0,1]\)) is naturally expected to have better sparsity than \(l_{2,1}\)-minimization. This paper presents a type of models based on \(l_{2,p}\ (p\in (0, 1])\) matrix norm which is non-convex and non-Lipschitz continuous optimization problem when \(p\) is fractional (\(0<p<1\)). For all \(p\) in \((0, 1]\), a unified algorithm is proposed to solve the \(l_{2,p}\)-minimization and the convergence is also uniformly demonstrated. In the practical implementation of algorithm, a gradient projection technique is utilized to reduce the computational cost. Typically different \(l_{2,p}\ (p\in (0,1])\) are applied to select features in computational biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
In many applications, such as computer vision, handwriting character recognition, medical diagnosis and etc., \(l_{2,1}\) matrix norm has received increasing attention to select features for its joint sparsity. The underlying assumption is that features in high dimensional data are related to each other but only a few of informational features contribute to discrimination. Existing schemes employ various strategies to capture the feature relatedness and select the most discriminative features, then take into account in the learning formulation. The involved models and methods are frequently constructed in \(l_1\)-norm framework. Actually, extensive computational studies [2–5, 12] have showed that using \(l_p\)-norm \((0<p<1)\) can find sparser solution than using \(l_1\)-norm. Naturally, one can expect mixed \(l_{2,p}\)-norm \((0<p\le 1)\) based minimization to have better sparsity pattern than \(l_{2,1}\)-norm. A similar \(l_p-l_q\) (\(0<p\le 1,\ 1\le q\le 2\)) penalty for sparse linear and multiple kernel multi-task learning has been considered in [13]. But the induced optimization problems have to be separately solved by different algorithms according to the convex (\(p=1\)) and non-convex (\(0<p<1\)) cases. This brings computational difficulty to freely vary \(p\) and \(q\). This paper presents a generalized model based on \(l_{2,p}\) (\(p\in (0, 1]\)) matrix normFootnote 1. When \(p\) is a positive fraction (\(0<p<1\)), the involved optimization problem is neither convex nor Lipschitz continuous. When \(p=1\), it is just the well defined \(l_{2,1}\)-minimization. Inspired by the work in [1, 2], we will develop a unified algorithm to solve the mixed \(l_{2,p}\)-norm based minimization for all \(p\) in \((0, 1]\). To the best of our knowledge, this presentation has the innovations as follows. (1) The general model based on mixed \(l_{2,p}\ (p\in (0,1])\) norm is more adaptive than \(l_{2,1}-\)norm to offer better sparsity for different data structures. (2) The unified algorithm and its unform convergence for \(p\in (0, 1]\) provides algorithmic support in pursuing more sparse patterns. (3) In the implementation of unified algorithm, a gradient projection scheme is utilized to reduce the computational cost. The running CPU time comparisons confirm the numerical economy of this technique. (4) Typical \(p\in (0, 1]\) are tested in \(l_{2,p}\)-minimization, the experiments in bioinformatics study provide empirical evidence that some \(0<p<1\) are alternatives in constructing better sparse patterns than \(p=1\) .
The rest of the paper is organized as follows. Section 2 states some necessary notations and induces the generalized model based on \(l_{2,p}\)-norm (\(p\in (0,1]\)). Section 3 develops a unified approach to solve the specially mixed optimization problem, and the convergence analysis is also established. Section 4 considers a gradient projection technique for solving the subproblems inexactly. Some experiment results are reported in the Sect. 5. Conclusions and further extensions are discussed in the last section.
2 \(l_{2,p}\)-Norm based minimization
We employ the notations as usual. Matrices are written as uppercase letters while vectors are written as lowercase letters. For example, \(A=(a_{ij})_{d\times n}\) denotes a real \(d\times n\) matrix, \(a^i\in R^n (i=1,\cdots , d)\) and \(a_j\in R^d (j=1,\cdots , n)\) are the \(i-\)th row and \(j-\)th column of \(A\) respectively.
For any \(x\in R^d\), several useful vector norms are given as follows,
where \(p\in (0,1)\). Actually, neither \(l_0\) nor \(l_p\) (\(0<p<1\)) is a well defined norm because the both definitions do not satisfy the norm axioms.
\(l_{2,1}\)-norm of matrix was firstly introduced in [6] which can be considered as a generalization of \(l_1\) vector norm to matrix,
Now we generalize the definition of \(l_{2,1}\)-norm to mixed \(l_{2,p}\)-norm as follows
Note that \(l_{2,p}\)-norm (\(0<p<1\)) is not a valid norm, and neither convex nor Lipschitz continuous. This properties challenge researchers to solve the related optimization problems.
Given observation data \(\{a_1,a_2,\cdots ,a_n\}\subseteq R^d\) and corresponding outputs \(\{b_1,b_2, \cdots ,b_n\}\subseteq R^c\). Traditional least square regression for discrimination solves the following optimization for unknown \(X\in R^{d\times c}\)
where \(X\) contains the projection matrix and bias vector for simplicity. \(R(X)\) denotes regularization and \(\alpha >0\) is the regularization parameter. It is well known that the square-norm residual is sensitive to outliers, hence Nie et. al. [1] proposed to use a robust \(l_{2,1}\)-norm loss function
In this paper, we like to use the generalized version
For any \(p\in (0, 1]\), the noise magnitude of distant outlier in (6) is no more than that in (5). Thus the model (6) is expected to be more robust than (5).
Joint sparse regularization \(R(X)\) is usually chosen
Theoretically, \(R_\triangle (X)\) are mostly preferred for its desirable sparsity. But \(R_\triangledown (X)\) is practically chosen more often for the computational sake. Under certain conditions, \(R_\triangledown (X)\)-regularization is equivalent to \(R_\triangle (X)\)-regularization. Here we chose the generalized norm in the sense
Hence the feature selection from high-dimensional data can be concluded as a mixed optimization problem based on \(l_{2,p}\)-norm (\(p\in (0,1]\)),
where \(\alpha =\gamma ^p\) is the regularization parameter. When \(p=1\), problem (9) is reduced to the popular \(l_{2,1}\)-norm based minimization proposed in [1]. But when \(0<p<1\), it is a non-convex and non-Lipschitz continuous minimization, the algorithm in [1] can not be directly applied. As far as we know, very few scheme is presented to uniformly solve this specially mixed problem. Therefore, it is necessary to develop an unified approach to efficiently solve problem (9) for all \(p\in (0, 1]\).
3 Main results
Denote \(A=[a_1,a_2,\cdots ,a_n]\in R^{d\times n}\) and \(B=[b_1,b_2,\cdots ,b_n]^T\in R^{n\times c}\), the problem (9) can be rewritten as
Let \(E=\frac{1}{\gamma }(A^TX-B)\), the unconstrained optimization problem (10) becomes
It can be easily proved that \(\Vert \left[ \begin{array}{l}E \\ X\end{array}\right] \Vert _{2,p}^p=\Vert E\Vert _{2,p}^p+\Vert X\Vert _{2,p}^p\). If we denote
where \(m=n+d\), problem (11) can be reformulated as
The Lagrangian function of optimization problem (13) is
where \(\Lambda \in R^{n\times c}\) is Lagrangian multiplier matrix, and \(Tr(\cdot )\) stands for trace operator. \(Y^\star \) is the KKT point of problem (13) if and only if there exists a \(\Lambda ^\star \in R^{n\times c}\) such that
where
is induced from \(Y^\star \). After simple reformulation, (15) is equivalent to
Although \(D_{\star }\) is necessary in equation (15), only \(D_\star ^{-1}\) is involved to compute \(Y^{\star }\) in formula (17). If we fix \(Y\) and \(D\) in (16) and (17) alternatively, an iterative algorithm to solve problem (13) can be designed as follows.
Algorithm 3.1
(Solving problem (13))
-
1.
Start: Given \(M\in R^{n\times m}\), \(B\in R^{n\times c}\) and set \(D_1=I_m\).
-
2.
For \(k=1,2,\cdots \) until convergence do : \( \begin{array}{l} Y_{k+1}=D_{k}^{-1}M^T(MD^{-1}_{k}M^T)^{-1}B,\\ Update\ D_{k+1}^{-1}\ with\ diagonal\ entries: \ \Vert y_{k+1}^i\Vert _{2}^{2-p}, i=1, 2,\cdots , m. \end{array} \)
Remark 3.1
If \(D, Y\) are decided as in (16) and (17), it can be easily derived that \(Tr(Y^TDY)=\Vert Y\Vert _{2,p}^p\) for \(0<p\le 1\).
Denote \(\{Y_{k}\}\) the matrix sequence generated by Algorithm 3.1, now let us show its convergence. The first lemma is apparent so the proof is omitted.
Lemma 3.1
If \(\phi (t)=\frac{2}{2-p}t-\frac{p}{2-p}t^{\frac{2}{p}}-1\) for \(p\in (0, 1]\), then \(\phi (t)\le 0\) in \((0, +\infty )\) and \(t=1\) is the unique maximum point.
Lemma 3.2
Suppose that \(y^i_k\), \(y^i_{k+1}\) is the \(i-\)th row of \(Y_k\), \(Y_{k+1}\) respectively, then for any \(p\) in \((0, 1]\)
Equalities in (18) hold if and only if \(\Vert y_{k+1}^i\Vert _{2}^p=\Vert y_{k}^i\Vert _{2}^p\) for \(i=1,2,\cdots , m\).
Proof
Let \(t=\frac{\Vert y_{k+1}^i\Vert _{2}^p}{\Vert y_{k}^i\Vert _{2}^p}\) in \(\phi (t)\), then \(\phi (\frac{\Vert y_{k+1}^i\Vert _{2}^p}{\Vert y_{k}^i\Vert _{2}^p})\le 0\), that is
Note that \(\Vert y_{k+1}^i\Vert _{2}^p=\Vert y_{k}^i\Vert _{2}^p\) for \(i=1,2,\cdots , m\) is sufficient and necessary to let the equality in (19) happen. Multiplying the two sides of formula (19) with \((1-\frac{p}{2})\Vert y_k^i\Vert _2^p\), we have
which is also an equivalent formula of (18).\(\square \)
Theorem 3.1
\(\Vert Y_{k}\Vert _{2,p}^p\) monotonically decreases with respect to iteration \(k\) until the matrix sequence \(\{Y_{k}\}\) converges to the KKT point of problem (13).
Proof
From remark 3.1 and the construction of Algorithm 3.1, we can easily verify that \(Y_{k+1}\) is the optimal solution to
So we have
which is to say
On the other hand, formula (18) in Lemma 3.2 shows
Combining (23) and (24), we have
which is \(\Vert Y_{k+1}\Vert _{2,p}^p\le \Vert Y_{k}\Vert _{2,p}^p\).
If \(\Vert Y_{k+1}\Vert _{2,p}^p=\Vert Y_{k}\Vert _{2,p}^p\) happens for some \(k\), since \(Y_{k+1}\) is constructed such that (22) and (23) hold, hence from (24) we easily derive
Notice that (18) is valid for each \(1\le i\le m\), the equalities in Lemma 3.2 have to exist which means \(\Vert y_{k+1}^i\Vert _{2}^p=\Vert y_{k}^i\Vert _{2}^p\) for \(i=1,2,\cdots , m\). Hence \(D_{k+1}^{-1}\) equals \(D_{k}^{-1}\) in Algorithm 3.1, \(Y_{k+1}\) and \(D_{k+1}\) satisfy the stationary condition (17). Algorithm 3.1 generates a matrix sequence such that the objective function value monotonically decreases until it converges to the KKT matrix of problem (13). When \(p=1\), the convergence matrix is also the global minimizer of problem (13). \(\square \)
Remark 3.2
To some extent, Algorithm 3.1 offers an alternative to solve \(l_p\) (\(0<p<1\)) regularized problems when the number of columns in \(Y\) is \(1\).
Remark 3.3
Algorithm 3.1 is a unified approach to solve problem (13) for any \(p\in (0, 1]\). This scheme provides algorithmic support to freely adapt \(p\) for better sparsity pattern in different data structures.
4 Computational details and practical algorithm
In Algorithm 3.1, each step has to compute \((MD^{-1}_{k-1}M^T)^{-1}\) which is expensive especially for high dimensional data. We notice that in the \(k-\)th iteration of Algorithm 3.1, \(Y_{k+1}\) solves (21) exactly. Actually, subproblem (21) is a quadratic programming with linear equality constraints, and there are varieties of efficient methods to solve it iteratively.
Now let us solve the subproblem (21) inexactly. Here we employ the gradient projection method in [14]. Suppose that \(Y_k\) has been generated as an approximate solution to the \((k-1)-\)th subproblem. The next approximate matrix \(Y_{k+1}\) to the \(k-\)th subproblem (21) will be constructed from \(Y_k\)
where \(\alpha _k\) is the step length and \(S_k\) is the line search matrix. If only the objective function value \(f_k(Y_{k+1})\) has sufficient reduction compared with \(f_k(Y_{k})\), the convergence will be guaranteed. Because the last approximation \(Y_k\) is feasible, that is \(MY_k=B\), \(Y_{k+1}\) is feasible if and only if \(MS_k=0\). In the gradient projection method [14], \(S_k\) is chosen to be the projection of \(-\nabla f_k(Y_k)\) on the null subspace of \(M\), where
is the steepest direction matrix from \(Y_k\). Let \(P\) denote the projection operator from \(R^{m\times c}\) to \(Null(M)\), then \(S_k:=-PD_kY_k\). Different \(P\) results in different numerical algorithm. Here we choose
where \((M^T)^+=(MM^T)^{-1}M\). Since \(M^T\) has full column rank, \((MM^T)^{-1}\) is well defined. If we have the QR decomposition of \(M^T\) in the form
where \(Q=[Q_1\ \ Q_2]\) is an orthogonal matrix and \(R\in R^{n\times n}\) is an invertible upper triangular matrix, then
It is easily verified that \(P\) is an orthogonal projection operator from \(R^{m\times c}\) to its subspace \(Null(M)\).
After the line search matrix \(S_k\) is fixed, the step length \(\alpha _k\) can be computed by solving the following minimization
The \(\varphi (\alpha )\) can be detailedly rewritten as
Matrices \(D_k\) and \(P\) are obviously symmetric and positive semi-definite, hence
Then the optimal step length to (31) is
The objective function value reduction of subproblem (21) can be evaluated
which is sufficient to guarantee the convergence of matrix sequence \(\{Y_k\}\).
Based on the computational details (Eqs. (27)–(34)), a one-step gradient projection method for solving problem (13) can be concluded as follows.
Algorithm 4.1
(One-step gradient projection method for problem (13))
-
1.
Start: Given \(M\in R^{n\times m}\) and \(B\in R^{n\times c}\).
-
2.
QR decompose \(M^T=Q_1R\), where \(Q_1\in R^{m\times n}\) and \(R\in R^{n\times n}\).
-
3.
Compute \(P=I_m-Q_1Q_1^T\) and \(Y_1=Q_1R^{-T}B\).
-
4.
For \(k=1,2,\cdots \) until convergence do : \( \begin{array}{l} D_k=\hbox {diag}\{\Vert y_{k}^1\Vert _{2}^{p-2}, \Vert y_{k}^2\Vert _{2}^{p-2}, \cdots , \Vert y_{k}^m\Vert _{2}^{p-2}\},\\ S_k=-PD_kY_k,\\ \alpha _k=-\frac{Tr(S_k^TD_kY_k)}{Tr(S_k^TD_kS_k)},\\ Y_{k+1}=Y_k+\alpha _kS_k. \end{array} \)
Remark 4.1
The \(k-\)th iteration in Algorithm 4.1 is essentially the steepest descent method over the subspace \(Null(M)\). Since \(M^T\) has full column rank, any accumulation point of \(\{Y_k\}\) will be the KKT point of problem (13) [14] .
Remark 4.2
If \(y_{k}^i=0\) happens in some iteration, then \(D_k\) can not be well updated and Algorithm 4.1 breaks down. Here we treat \(D_k\) in two natural ways. One is setting the \(i-\)th diagonal element \(\{D_k\}_{ii}=0\) which can be considered as the generalized inverse of \(D_k^{-1}\). The other one is to give a perturbation \(\epsilon >0\) such that \(\{D_k\}_{ii}=({\sqrt{y^i_k(y^i_k)^T+\epsilon }})^{p-2}\ne 0\).
The computational cost comparison between two algorithms can be easily analyzed as follows. With the same outer loop, Algorithm 4.1 substitutes one-step gradient projection to the matrix inverse \((MD^{-1}_{k-1}M^T)^{-1}\) in Algorithm 3.1. In each iteration, the time complexities are \(\mathcal {O}(n^2m+cmn)+\mathcal {O}(n^3)\) flops in Algorithm 3.1 and \(\mathcal {O}(cm^2)\) flops in Algorithm 4.1. Here \(m,\ n,\ c\) denote the numbers of dimension, samples and selected features respectively. Except for extremely small sample data, Algorithm 4.1 will be faster than Algorithm 3.1 which is also confirmed by the CPU time comparison in the next section.
5 Experimental results
In our experiments, four public data sets in biological study are used. Brief description about all data sets is given as follows.
- ALLAML:
-
is Leukemia gene microarray data, originally obtained by Golub et. al. [8]. There are 7129 genes, 72 samples containing two classes, acute lymphocytic leukemia (ALL) and acute mylogenous leukemia (AML).
- GLIOMA:
-
contains four classes, cancer glioblastomas, non-cancer glioblastomas, cancer oligodendrogliomas and non-cancer oligodendrogliomas [9]. There are total \(50\) samples and each class has \(14, 14, 7, 15\) samples respectively. Each sample has \(12625\) genes.
- LUNG:
-
cancer data is available at [10]. There are \(12533\) genes and total \(203\) samples in two classes, malignant pleural mesothelioma and adenocarcinoma of the lung.
- Prostate-GE:
-
data set has \(12600\) genes. It contains two classes, tumor and normal. \(52\) samples are tumor and \(50\) samples are normal. The dataset is available in [11].
Those data have high dimensional features around ten thousand but most of features are redundant even noise for classifications. Features with strong discrimination power always lie in a lower dimension subspace. It is important and necessary to select the most informative features for knowledge discovery and practical diagnosis in medicinal field. Before applied Algorithms 3.1 or 4.1 to feature selection, all data sets are performed the same preprocessing as in [7] to remove the redundant genes. Then the data sets are standardized to be zero-mean and normalized by standard deviation.
To demonstrate the effect of different \(l_{2,p}\) matrix pseudo norms in feature selection, we implement typical \(l_{2,p}\)-norm based optimization problems for \(p=0.25, 0.5, 0.75\) and \(1\). Using the selected top \(20,\ 40,\ 60,\ 80\) features respectively, SVM classifiers are individually performed on four data sets with fivefold crosses. Based on Theorem 3.1, the reduction of objective function value estimates the convergence precision. To eliminate the magnitude influence of different data, we employ \(\rho _k:=\frac{\Vert Y_{k}\Vert _{2,p}^p-\Vert Y_{k+1}\Vert _{2,p}^p}{\Vert Y_k\Vert _{2,p}^p}\le 10^{-5}\) as the stopping criterion of two algorithms. Under the same running environment, Algorithm 3.1 and 4.1 have the same classification accuracy which are reported in Tables 1, 2. The bold values indicate the best ones.
The experimental procedure indicates that four \(l_{2,p}\)-norm (\(p=0.25,\ 0.5,\ 0.75\) and \(1\)) based minimizations do select different features, hence result in distinct classification performances. The parameter \(p\) in \((0,1]\) balances the sparsity and convexity of optimization problem (13). The closer to \(0\) the \(p\) is, the sparser the representation is. While \(p\) is very near to \(1\), the model is almost convex. The classification error comparisons show that non-convex \(l_{2,p}\) \((0<p<1)\) matrix norms provide alternatives to \(l_{2,1}\)-norm. Especially, \(p=0.5\) empirically outperforms \(p=1\) for choosing better sparse patterns in various situations.
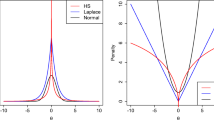
To validate the consistent efficiency of the unified algorithms solving nonconvex \(l_{2,p}\) (\(0<p<1\)) pseudo norm optimization problems as well as the convex \(l_{2,1}\)-norm based minimization, we present the convergence behavior curve of Algorithm 3.1 with exact solution to subproblem (21). Actually, the convergence behaviors for each \(l_{2,p}\)-norm case are similar with different numbers of top features. We display the change of \(\rho _k\) with respect to iterations in the case of \(80\) features (see Fig. 1 ). It can be seen that all experiments on four data sets uniformly get the expected accuracy within around \(20\) steps.
In practical implementation, especially for relatively large sample data, Algorithm 4.1 is preferred to Algorithm (3.1) for its economical computation. We still choose the top \(80\) features as a typical example to compare the efficiency of two algorithms. Under the same precisions (\(10^{-3}\) and \(10^{-5}\)) of \(\rho _k\), the running CPU time of two algorithms on four data sets is listed in Table 3. Algorithm 3.1 and Algorithm 4.1 are abbreviated by Alg 3.1 and Alg 4.1 respectively. All experiments are performed in the same running environment. In most of situations, Algorithm 4.1 is more time-saving than Algorithm 3.1 which especially supports Algorithm 4.1 in large scale applications.
6 Conclusions
In this paper, a type of minimizations based on \(l_{2,p}\ (p\in (0, 1])\) matrix pseudo norm is presented. A unified algorithm is designed to solve the mixed optimization problems and the convergence is also uniformly ensured. To refine the algorithm implementation, a gradient projection is applied to inexactly solve the subproblems. Experiment results on gene express data sets validate the unified performance of the proposed method. This scheme provides more choices of \(p\in (0, 1]\) to fit variety of jointly sparse requirements.
Notes
\(\Vert \cdot \Vert _{2,p}\) (\(0<p<1\)) is not a valid matrix norm because it does not admit the triangular inequality. Here we call it matrix norm for convenience.
References
Nie, F.P., Huang, H., Cai, X., and Ding, C.: Efficient and robust feature selection via joint \(l_{2,1}\)-norms minimization. Twenty-Fourth Annual Conference on Neural Information Processing Systems, pp. 1–9. (2010)
Candès, Emmanuel J., Wakin, Michael B., Boyd, Stephen P.: Enhancing sparsity by reweighed \(l_1\) minimization. J. Fourier Anal. Appl. 14(5), 877–905 (2008)
Chartrand, R.: Exact reconstructions of sparse signals via nonconvex minimization. IEEE Signal Process. Lett. 14(10), 707–710 (2007)
Chartrand, R., and Yin, W.: Iteratively reweighed algorithms for compressive sensing. 33rd International Conference on Acoustics, Speech, and Signal Processing, pp. 3869–3872 ( 2008)
Chen, X.J., Xu, F.M., Ye, Y.Y.: Lower bound theory of nonzero entries in solutions of \(l_2-l_p\) minimization. SIAM J. Sci. Comput. 32(5), 2832–2852 (2010)
Ding, C., Zhou, D., He, X.F., and Zha, H.Y.: \(R1-\)PCA: Rotational invariant \(L_1-\)norm principal component analysis for robust subspace factorization. Proceedings of the 23th International Conference on Machine Learning, pp. 281–288 (2006)
Dudoit, S., Fridly, J., Speed, T.P.: Comparison of discrimination methods for the classification of tumors using gene expression data. J. Am. Stat. Assoc. 97(457), 77–87 (2002)
Golub, T.R., Slonim, D.K., Tamayo, P., Huard, C., Gaasenbeek, M., Mesirov, J.P., Coller, H., Loh, M.L., Downing, J.R., Caligiuri, M.A., Bloomfield, C.D., Lander, E.S.: Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286(5439), 531–537 (1999)
Nutt, C.L., Mani, D.R., Betensky, R.A., Tamayo, P., Caincross, J.G., Ladd, C., Pohl, U., Hartmann, C., Mclaughlin, M.E.: Gene expression-based classification of malignant gliomas correlates better with servival than histological classification. Cancer Res. 63, 1602–1607 (2003)
Gordon, G.J., Jensen, R.V., Hsiao, L.L., Gullans, S.R., Blumenstock, J.E., Ramaswamy, S., Richards, W.G., Sugarbaker, D.J., Bueno, R.: Translation of microarray data into clinically relevant cancer diagnoistic tests using gene expression ratios in lung cancer and mesothelioma. Cancer Res. 62(17), 4963–4967 (2002)
Singh, D., Febbo, P.G., Ross, K., Jackson, D.G., Manola, J., Ladd, C., Tamayo, P., Renshaw, A.A., D’Amico, A.V., Richie, J.P., Lander, E.S., Loda, M., Kantoff, P.W., Golub, T.R., Sellers, W.R.: Gene expression correlates of clinical prostate cancer behavior. Cancer Cell 1(2), 203–209 (2002)
Xu, Z.B., Zhang, H., Wang, Y., Chang, X.Y., Yong, L.: \(L_{\frac{1}{2}}\) regularizer. Sci. China 52(6), 1159–1169 (2010)
Rakotomamonjy, A., Flamary, R., Gasso, G., Canu, S.: \(l_p-l_q\) Penalty for sparse linear and sparse multiple kernel multitask learning. IEEE Transac. Neural Netw. 22(8), 1307–1320 (2011)
Rosen, J.B.: The gradient projection method for nonlinear programming. Part 1 Linear constraints. J. SIAM 8, 181–217 (1960)
Acknowledgments
The first author thanks Dr. Zhang Hongchao for his helpful suggestions on this paper.
Author information
Authors and Affiliations
Corresponding author
Additional information
The work is supported by the NSFC11001128, NSFC61035003, NSFC61170151, NSFC11071117 and the Fundamental Research Funds for the Central Universities (No. NZ2013306 and NZ2013211).
Rights and permissions
About this article
Cite this article
Wang, L., Chen, S. & Wang, Y. A unified algorithm for mixed \(l_{2,p}\)-minimizations and its application in feature selection. Comput Optim Appl 58, 409–421 (2014). https://doi.org/10.1007/s10589-014-9648-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10589-014-9648-x