Abstract
We searched for candidate target genes in metastatic gastric cancer, using comparative genomic hybridization (CGH) and mRNA expression array analysis of endoscopic biopsy samples collected from 32 patients. Recurrent amplicons included 17q21.2 (36,569,293–37,307,055), 8q24.13-q24.21 (126,357,475–130,159,285), and 20q13.33 (60,211,249–61,382,787). In this paper, we focused on the 1.1-Mb genomic region containing 24 genes in chromosome 20q13.33 (from 60,211,249 to 61,382,787), the third most frequent amplicon that was amplified in three of 32 patients (9.4 %), with log2 tumor/reference ratios ranging from 0.6 to 1.5. Of three genes in the 20q13.33 amplicon, ADRM1 was chosen for functional analyses. ADRM1 knockdown suppressed the proliferation of two human gastric cancer cells, SNU-601 and SNU-216. Overexpression of Adrm1 promoted cell proliferation of conditionally-immortalized, mouse ImSt gastric epithelial cells, with increased S1 phase fraction and decreased expression of p21(Cip1). These results collectively indicate that ADRM1 promoted gastric epithelial cell proliferation by cell cycle progression. Therefore, ADRM1 is a candidate target gene in the chromosome 20q13.33 amplicon that may possibly be linked to development of gastric cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastric cancer is the second most common cause of cancer deaths worldwide [1]. Cytotoxic chemotherapy is the mainstay treatment for metastatic gastric cancers, but the prognosis is generally poor. In addition to cytotoxic chemotherapy, HER2- and VEGFR2-targeted agents are currently available therapies shown to improve the prognosis of metastatic gastric cancers [2]. Given limited treatment options, there is an unmet need for novel targeted therapies to improve the prognosis of metastatic gastric cancer patients. Since gene amplifications are more relevant therapeutic targets for gastric cancers than single nucleotide variations or fusion genes [2], it will be a highly valuable study to identify novel target genes in recurrent amplicons in human gastric cancer. We previously reported the results of our expression array-based studies to identify biomarkers for chemotherapy resistance [3, 4]. In this parallel study, we sought to identify candidate target genes in metastatic gastric cancer patients, using array comparative genomic hybridization (CGH) analysis of the same endoscopic biopsy samples, and identified several recurrent copy number aberrations. Agilent array CGH technology, which was used in our previous studies [3], was used to evaluate copy number status of gastric cancer biopsy samples.
In this integrative CGH and mRNA microarray study, we evaluated copy number aberrations identified in 32 metastatic gastric cancer patients whose mRNA expression microarray data are also available [3]. We found that chromosome 17q21.2, 8q24.13-q24.21, and 20q13.33 were most recurrent amplification loci in our gastric cancer samples. Chromosome 20q13 is frequently amplified in gastric cancer [5, 6] and linked to high proliferative potential of tumors [7]. In contrast to 17q or 8q amplicon, 20q13 amlicon has not been fully defined for target genes [7]. Therefore, we focused on the 1.1-Mb amplicon in chromosome 20q13.33, which was found to be the third most frequent amplicon in our study patients. Among the 24 genes in the amplicon, ADRM1/RPN13 demonstrated strong mRNA overexpression in gastric cancers and became the focus of this study. ADRM1 is a 19S proteasomal cap-associated protein that interacts with a deubiquitinating enzyme UCH37 [8]. ADRM1 has been proposed as a candidate driver gene in the 20q13.33 amplicon in ovarian and colorectal cancers [9, 10]. As a result of functional experiments, here we demonstrate that ADRM1 amplification may contribute to gastric carcinogenesis by cell cycle progression, providing evidences that ADRM1 is a candidate target gene in gastric cancer.
Materials and methods
Tissue samples were collected from metastatic gastric cancer patients by endoscopy before chemotherapy (cisplatin in combination with either fluorouracil or capecitabine) and frozen in liquid nitrogen from 2001 to 2006, under a protocol approved by the Institutional Review Board (IRB) of the National Cancer Center Hospital in Goyang, Korea. All patients signed IRB-approved informed consent forms. Tissue samples were collected and processed for total RNA and genomic DNA extraction as previously described [3]. Macrodissection was performed to ensure ≥50 % tumor content in all of the RNA and DNA samples. In parallel, grossly-normal gastric mucosa tissue samples were collected from the antrum of 21 healthy volunteers. Expression and CGH array analyses were performed using Affymetrix HG-U133A (Santa Clara, CA) and Agilent 4x44k HD-CGH (Agilent Technologies, Santa Clara, CA) arrays, respectively. For array CGH, human genomic DNA from multiple anonymous female donors (Promega, Madison, WI) was used as the reference DNA [3]. Mean log2 tumor/reference ratio >0.5 for 5 or more consecutive probes was identified as the copy number gain.
For proliferation assays, 3 × 105 shEGFP-transduced and shADRM1-transduced SNU-601 cells were grown for 4 days in RPMI media with 10 % fetal bovine serum (FBS) on 150 mm dishes, and cells were counted daily using Countess (Invitrogen, Carlsbad, CA). Also, 1 × 104 empty vector-transduced and Adrm1-overexpressing, immortalized mouse gastric epithelial ImSt cells were grown for 4 days in RPMI media with 10 % FBS on 12 well plates. MTT [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assays were performed each day. Cell numbers and optical densities were compared using repeated measure ANOVA tests. Cell cycle analyses were performed using FACS. Fluorescence in situ hybridization (FISH) analyses were performed on formalin-fixed, paraffin-embedded (FFPE) slides for ADRM1-amplified gastric cancers using Cy3-labeled probes for ADRM1 and FITC-labeled probes for 20p11.21. ADRM1 immunohistochemisty (IHC) analyses were performed on ADRM1-amplified gastric cancers and also on a commercially available tissue microarray (TMA) slide containing 54 gastric cancers. Detailed experimental methods were described in Supplementary Information.
Results
ADRM1 gene amplification in gastric cancer tissue samples
Thirty two patients with metastatic gastric cancer were studied (Table 1). Median age was 62 years (range 27–74). There were 26 (81 %) males, and 13 Lauren intestinal histologic types (41 %). Median percentage of tumor nuclei was 60 % (range 50–90).
Recurrent amplicons in our study patients included 17q21.2 (36,569,293–37,307,055 (n = 5)), 8q24.13-q24.21 (126,357,475–130,159,285 (n = 4)), and 20q13.33 (60,211,249–61,382,787 (n = 3)) (Tables 2 and S1). In contrast to 17q or 8q amplicon, 20q amlicon has not been fully defined for target genes, although it is frequently amplified in gastric cancer. Therefore, this study focused on the 1.1-Mb amplicon in chromosome 20q13.33 containing 24 genes (60,211,249–61,382,787), the third most frequent amplicon (Table 2). Log2 tumor/reference ratio of the 20q13.33 amplicon ranged from 0.6 to 1.5. Two of the three patients with 20q13.33 amplification co-amplified 8q24.13-q24.21 (MYC locus).
It was postulated that target genes in the 20q13.33 amplicon may be overexpressed in mRNA and protein in corresponding tissue samples. To search for candidate target genes in the 20q13.33 amplicon, we compared the biopsy tissue mRNA expression microarray data between the three patients harboring 20q13.33 amplification and 21 healthy volunteers [4]. Three of 24 genes in the common amplicon in chromosome 20q13.33 (ADRM1, TCFL5, and C20orf20) were overexpressed by >twofold (p < 0.001) in three patients harboring 20q13.33 amplification, compared with healthy volunteers (Tables 3 and S2). We applied an additional filter to this gene list, using 2,446 genes differentially expressed between pre-chemotherapy samples and post-chemotherapy samples collected at the time of disease progression in chemotherapy responders (acquired resistance signature) [4]. Of three genes overexpressed in cancer patients, only ADRM1 was included in the acquired resistance signature (1.3-fold overexpression in chemoresistant samples compared with pretreatment samples) [4]. Therefore, ADRM1 amplification was subject to subsequent functional analyses.
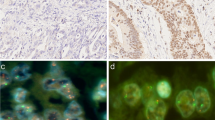
ADRM1 gene amplification was confirmed by FISH in all of the ADRM1-amplified patient tissue samples (Fig S1). When ADRM1 immunohistochemistry was performed on FFPE slides for these tumors to confirm correlation between ADRM1 amplification and protein overexpression, all of three ADRM1-amplificated samples overexpressed nuclear ADRM1 protein in cancer cells compared with surrounding histologically-normal gastric mucosal epithelial cells (Fig. 1a) and 54 gastric cancers of a TMA slide with unknown ADRM1 copy number status (p = 0.076) (Fig. 1b). Interestingly, among 54 gastric cancers represented in TMA, there was trend of the higher ADRM1 nuclear grade with positive lymph node involvement (Fig. 1c; p = 0.022).
Immunohistochemical analyses for ADRM1 in tissue samples of three ADRM1-amplified cases. a ADRM1 immunohistochemistry images of three ADRM1-amplified gastric cancers. Histologically-normal gastric mucosa surrounding two tumors is also shown. b Comparison of the percentage of nuclear ADRM1 positivity between three ADRM1-amplified gastric cancers and gastric cancers represented in tissue microarray with unknown ADRM1 copy number status. c ADRM1 nuclear grade of gastric cancers represented in tissue microarray according to lymph node involvement. Chi square test was used to calculate the p value
According to mRNA expression array results from the whole study patients, ADRM1 mRNA expression level (201281_at) was significantly higher in 32 gastric cancers than in 21 normal mucosal biopsy samples (p < 10−7; 2.3-fold). When the expression analysis was extended to a separate group of 92 metastatic gastric cancer patients whose samples were not subject to CGH array analysis [3], ADRM1 was also overexpressed in 92 cancer patients than in 21 healthy volunteers (p < 10−7; 2.2-fold).
ADRM1 promoted cell proliferation by cell cycle progression
Then, we moved on to evaluate functional impact of ADRM1 in proliferation of gastric epithelial cells. ADRM1 knockdown suppressed the monolayer growth of SNU-601 human gastric cancer cells (p for repeated measure ANOVA = 0.005) (Fig. 2a, b). ADRM1 knockdown decreased the S phase fraction from 55 to 49 % in SNU-601 cells (p = 0.005) (Fig. 2c). Western blot analyses demonstrated the modest increase in protein level of p21(Cip1) (Fig. 2a). ADRM1 knockdown suppressed the proliferation of another gastric cancer cell line, SNU-216 (p = 0.034; Fig S2).
Proliferation and FACS assays after ADRM1 modulation. a Western blot analyses. SNU-601 clones stably expressing lentiviral shADRM1 vector demonstrated undetectable expression level of ADRM1 and modest overexpression of p21(Cip1), as compared with control vector-transduced SNU-601 clones. b Growth curves of control-transduced and shADRM1-transduced SNU-601 cells. Error bars indicate SD from triplicate cell counting experiments. Student t test was used to compare cell number at each day (*p < 0.05; **p < 0.01). c FACS cell cycle analyses on control-transduced and shADRM1-transduced SNU-601 cells on Day 4. Percentage (mean ± SD) of each phase is also shown (lower panel). Differences in S phase fraction were compared using Student t test (**p < 0.01 and ***p < 0.001). d Western blot analyses. ImSt clones stably overexpressing Adrm1 demonstrate ecreased expression level of p21(Cip1), as compared with empty vector-transduced ImSt cells. e Growth curves of empty vector-transduced and Adrm1-overexpressing ImSt cells. Error bars indicate SD from quadruplicate MTT experiments. Student t test was used to compare cell number at each day (*p < 0.05; **p < 0.001). f FACS cell cycle analyses on empty vector-transduced and Adrm1-overexpressing ImSt cells on Day 1. Percentage (mean ± SD) of each phase is also shown (lower panel). Differences in S phase fraction were compared using Student t test (***p < 0.001)
ImSt is a conditionally immortalized stomach epithelial cell line isolated from the gastric epithelium of transgenic mice with a temperature-sensitive mutation of the simian virus 40 (SV40) large tumor antigen [11]. This cell line was chosen for Adrm1 overexpression experiments because of the premalignant phenotype (Fig. 2d). Overexpression of Adrm1 enhanced the monolayer growth potential of ImSt cells (p for repeated measure ANOVA = 0.0002; Fig. 2e). S phase fraction of ImSt cells increased from 26 to 28 % after Adrm1 overexpression (p = 0.0002; Fig. 2f). Protein expression level of p21(Cip1) was decreased after Adrm1 overexpression (Fig. 2d), and increased after Adrm1 knockdown (Fig S3). These results collectively indicate that ADRM1 promotes gastric epithelial cell proliferation by cell cycle progression.
We recently established a primary cultured mouse gastric cancer cell line, NCC-S1, and the lung metastatic derivative cells, NCC-S1M, from a spontaneous gastric adenocarcinoma of a Villin-Cre;Smad4 F/F ;Trp53 F/F ;Cdh1 F/+ mouse (Park JW et al., manuscript under review). ADRM1 mRNA and protein expressions were higher in metastatic derivative NCC-S1M than in parental gastric cancer cell line NCC-S1 (Fig S4), with no alteration in Adrm1 gene copy number status (A_53_P148343) according to array CGH analyses (data not shown).
Since ADRM1 is a substoichiometric subunit of the 26S proteasome, we planned to explore if some other proteasome subunits are overexpressed in ADRM1-amplified cells. According to real-time PCR analyses on four selected gastric cancer cells, the copy number gain of ADRM1 was documented in SNU-216 cells only (Fig. 3a). According to Western blot analyses, protein expression of ADRM1 in SNU-216 cells was higher than the other gastric cancer cell lines without ADRM1 gene amplification (Fig. 3b). Compared with cell lines without ADRM1 amplification, ADRM1-amplified SNU-216 cells also express higher protein levels of a proteasome 26S subunit RPN2/S1 [12], E2 ubiquitin-conjugating enzyme UBE2C [13], and ADRM1-interacting partner UCH37 [14–16] (Fig. 3b).
ADRM1 copy number status and expression of related proteins in human gastric cancer cell lines. a LINE1–normalized genomic DNA copy number status of ADRM1 in four selected human gastric cancer cell lines, according to real-time PCR. Error bars indicate SD from triplicate experiments. b Western blot analyses on ADRM1 and selected proteins functionally related to ADRM1 in the same sets of cell lines
Discussion
Copy number aberrations have been exclusively studied using resected gastric cancer specimens so far, given the lack of available biopsy tissue samples from metastatic patients. In this regard, this is a unique study to evaluate the association of copy number aberration with gastric carcinogenesis. Despite relatively high frequency of chromosome 20q13.33 amplification in gastric cancer [5–7], target genes in this genomic region have not been clearly defined [7]. Chromosome 20q is frequently amplified in common solid tumors, and deletion of this arm is very rare [17–20]. According to a study performed by Tabach et al., 20q13 amplification is linked to high proliferative potential, and ADRM1, UBE2C, CSE1L, RPN2, C20orf45, MYBL2, TOMM34, AURKA, RAE1, PFDN4, PSMA7, RPS21 and VAPB are suggested as key cancer initiating genes, given overexpression in several cancers with advanced stage and poor prognosis [7].
Our study is the first to propose a possible role of ADRM1 amplification in gastric cancer. ADRM1 has been proposed as a candidate driver gene in the 20q13.33 amplicon in ovarian and colorectal cancers [9, 10]. ADRM1 overexpression increases proliferation, migration, and growth in soft agar in ADRM1-amplified OAW42 ovarian cancer cell lines, and knockdown of ADRM1 results in apoptosis with dysregulation of CDK-activating kinase assembly factor MAT1 [9]. Knockdown of Adrm1 by shRNA in human colon carcinoma RKO cells inhibits their anchorage-independent growth, cell migration as well as cell proliferation through inducing apoptosis and cell cycle arrest at the G1 phase [10].
ADRM1 is a 19S proteasomal cap-associated protein that interacts with UCH37, a deubiquitinating enzyme [8]. Our data suggests that ADRM1 gene amplification may contribute to gastric carcinogenesis by cell cycle progression through enhanced degradation of cell cycle checkpoint proteins such as p21(Cip1). These results are consistent with a recent study performed by Anchoori et al., in which the pharmacologic inhibition of ADRM1 by RA190 reduces growth of multiple myeloma and ovarian cancer xenografts by stabilizing p53 and p53-regulated genes, such as p21, through proteasome inhibition [21].
In our 32 study patients, ADRM1 was the third most frequent target for gene amplification, and the amplification was accompanied by ADRM1 overexpression. ADRM1 overexpression promoted monolayer growth of ImSt cells, while ADRM1 knockdown led to modest, but significant suppression of proliferation of gastric cancer cells. Therefore, ADRM1 may be a candidate target gene for chromosome 20q13.33 gene amplification possibly linked to the development of gastric cancer.
Abbreviations
- CGH:
-
Comparative genomic hybridization
- EGFP:
-
Enhanced green fluorescent protein
- ANOVA:
-
Analysis of variance
- FISH:
-
Fluorescence in situ hybridization
- FFPE:
-
Formalin-fixed: paraffin-embedded
- FITC:
-
Fluorescein isothiocyanate
- FACS:
-
Fluorescence-activated cell sorting
- TMA:
-
Tissue microarray
References
Alberts S, Cervantes A, van de Velde C (2003) Gastric cancer: epidemiology, pathology, and treatment. Ann Oncol 14(Suppl 2):ii31–ii36
Kim HK, Green JE (2014) Predictive biomarker candidates for the response of gastric cancer to targeted and cytotoxic agents. Pharmacogenomics 15(3):375–384
Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Yamada Y, Arao T, Nishio K, Michalowski A, Green JE (2012) Three-gene predictor of clinical outcome for gastric cancer patients treated with chemotherapy. Pharmacogenomics J 12(2):119–127
Kim HK, Choi IJ, Kim CG, Kim HS, Oshima A, Michalowski A, Green JE (2011) A gene expression signature of acquired chemoresistance to cisplatin and fluorouracil combination chemotherapy in gastric cancer patients. PLoS ONE 6(2):e16694
Cheng L, Wang P, Yang S, Yang Y, Zhang Q, Zhang W, Xiao H, Gao H, Zhang Q (2012) Identification of genes with a correlation between copy number and expression in gastric cancer. BMC Med Genomics 5:14
Fan B, Dachrut S, Coral H, Yuen ST, Chu KM, Law S, Zhang L, Ji J, Leung SY, Chen X (2012) Integration of DNA copy number alterations and transcriptional expression analysis in human gastric cancer. PLoS ONE 7(4):e29824
Tabach Y, Kogan-Sakin I, Buganim Y, Solomon H, Goldfinger N, Hovland R, Ke XS, Oyan AM, Kalland KH, Rotter V, Domany E (2011) Amplification of the 20q chromosomal arm occurs early in tumorigenic transformation and may initiate cancer. PLoS ONE 6(1):e14632
Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453(7194):481–488
Fejzo MS, Dering J, Ginther C, Anderson L, Ramos L, Walsh C, Karlan B, Slamon DJ (2008) Comprehensive analysis of 20q13 genes in ovarian cancer identifies ADRM1 as amplification target. Genes Chromosomes Cancer 47(10):873–883
Chen W, Hu XT, Shi QL, Zhang FB, He C (2009) Knockdown of the novel proteasome subunit Adrm1 located on the 20q13 amplicon inhibits colorectal cancer cell migration, survival and tumorigenicity. Oncol Rep 21(2):531–537
Yan F, Cao H, Chaturvedi R, Krishna U, Hobbs SS, Dempsey PJ, Peek RM Jr, Cover TL, Washington MK, Wilson KT, Polk DB (2009) Epidermal growth factor receptor activation protects gastric epithelial cells from Helicobacter pylori-induced apoptosis. Gastroenterology 136(4):1297–1307
Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M (2008) Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453(7194):548–552
Okamoto Y, Ozaki T, Miyazaki K, Aoyama M, Miyazaki M, Nakagawara A (2003) UbcH10 is the cancer-related E2 ubiquitin-conjugating enzyme. Cancer Res 63(14):4167–4173
Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, Washburn MP, Conaway RC, Conaway JW, Cohen RE (2006) Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol 8(9):994–1002
Hamazaki J, Iemura S, Natsume T, Yashiroda H, Tanaka K, Murata S (2006) A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J 25(19):4524–4536
Qiu XB, Ouyang SY, Li CJ, Miao S, Wang L, Goldberg AL (2006) hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J 25(24):5742–5753
Tsafrir D, Bacolod M, Selvanayagam Z, Tsafrir I, Shia J, Zeng Z, Liu H, Krier C, Stengel RF, Barany F, Gerald WL, Paty PB, Domany E, Notterman DA (2006) Relationship of gene expression and chromosomal abnormalities in colorectal cancer. Cancer Res 66:2129–2137
Hodgson JG, Chin K, Collins C, Gray JW (2003) Genome amplification of chromosome 20 in breast cancer. Breast Cancer Res Treat 78:337–345
Alers JC, Krijtenburg PJ, Vis AN, Hoedemaeker RF, Wildhagen MF, Hop WC, van Der Kwast TT, Schröder FH, Tanke HJ, van Dekken H (2001) Molecular cytogenetic analysis of prostatic adenocarcinomas from screening studies: early cancers may contain aggressive genetic features. Am J Pathol 158:399–406
Mahlamaki EH, Barlund M, Tanner M, Gorunova L, Hoglund M, Karhu R, Kallioniemi A (2002) Frequent amplification of 8q24, 11q, 17q, and 20q-specific genes in pancreatic cancer. Genes Chromosomes Cancer 35:353–358
Anchoori RK, Karanam B, Peng S, Wang JW, Jiang R, Tanno T, Orlowski RZ, Matsui W, Zhao M, Rudek MA, Hung CF, Chen X, Walters KJ, Roden RB (2013) A bis-benzylidine piperidone targeting proteasome ubiquitin receptor RPN13/ADRM1 as a therapy for cancer. Cancer Cell 24(6):791–805
Acknowledgments
This work was supported by the Proteogenomic Research Program and 2013K000429 funded by the Korean Ministry of Science, ICT, and Future Planning and by National Cancer Center Grant 1210051 and 1410850.
Conflict of interest
Authors have nothing to disclose.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jang, S.H., Park, J.W., Kim, H.R. et al. ADRM1 gene amplification is a candidate driver for metastatic gastric cancers. Clin Exp Metastasis 31, 727–733 (2014). https://doi.org/10.1007/s10585-014-9663-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-014-9663-4