Abstract
Constitutive heterochromatin is composed mainly of repetitive elements and represents the typical inert chromatin structure in eukaryotic cells. Approximately half of the mammalian genome is made of repeat sequences, such as satellite DNA, telomeric DNA, and transposable elements. As essential genes are not present in these regions, most of these repeat sequences were considered as junk DNA in the past. However, it is now clear that these regions are essential for chromosome stability and the silencing of neighboring genes. Genetic and biochemical studies have revealed that histone methylation at H3K9 and its recognition by heterochromatin protein 1 represent the fundamental mechanism by which heterochromatin forms. Although this molecular mechanism is highly conserved from yeast to human cells, its detailed epigenetic regulation is more complex and dynamic for each distinct constitutive heterochromatin structure in higher eukaryotes. It can also vary according to the developmental stage. Chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis is a powerful tool to investigate the epigenetic regulation of eukaryote genomes, but non-unique reads are usually discarded during standard ChIP-seq data alignment to reference genome databases. Therefore, specific methods to obtain global epigenetic information concerning repetitive elements are needed. In this review, we focus on such approaches and we summarize the latest molecular models for distinct constitutive heterochromatin types in mammals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Constitutive heterochromatin and repetitive DNA elements
In eukaryotic cells, the genome is packaged by proteins into a structure termed “chromatin.” In 1928, E. Heitz was the first to observe intensely stained and condensed chromatin areas in interphase cells and termed these structures “heterochromatin.” Decondensed regions, or euchromatin, contain the vast majority of activated genes, whereas heterochromatin is gene-poor, and genes in heterochromatic areas are usually silenced. Heterochromatin can be divided in constitutive and facultative heterochromatin. Constitutive heterochromatin is formed at repeated sequence regions and its structural organization is very stable while facultative heterochromatin forms in specific situations, such as developmental processes and during tissue differentiation. For instance, facultative heterochromatin is detected at the inactivated X chromosome and at genes that regulate cell identity. Heterochromatin is characterized by histone hypoacetylation (important for chromatin compaction) and DNA methylation. A distinctive feature of the two heterochromatin types is the methylation of a specific histone H3 residue. Indeed, constitutive heterochromatin is typically defined by trimethylation of lysine 9 of histone H3 (H3K9me3), whereas facultative heterochromatin is enriched in H3 lysine 27 trimethylation (H3K27me3). These histone marks on the histone H3 tail are recognized by specific reader proteins, and upon their binding, chromatin conformation transitions to a more compact form. Heterochromatin protein 1 (HP1) is an evolutionarily conserved non-histone protein that is enriched at constitutive heterochromatin segments. Genetic and biochemical analyses using model organisms showed that HP1 N-terminal domain, called chromodomain, recognizes H3K9me3 and that this binding is essential for the establishment of heterochromatin conformation. On the other hand, the chromodomain of the polycomb protein (Pc) preferentially binds to H3K27me3. Thus, while specific mechanisms and outputs also are involved (reviewed in (Jost et al. 2012; Trojer and Reinberg 2007), the two heterochromatin types rely on similar mechanisms that are highly conserved in metazoans.
Constitutive heterochromatin is composed mainly of repetitive elements. Nearly 50% of the human genome is made of repetitive or repeat-derived sequences (Gregory 2005). The silencing state of these regions has an important role in maintaining chromosome stability by regulating proper chromosome segregation and preventing recombination or transposition activity. In this review, we first summarize the current methods to obtain global epigenetic information on repetitive elements and then discuss the organization of three representative constitutive heterochromatin regions: pericentromeres, telomeres, and retrotransposable elements.
Traditional approaches to analyze heterochromatin organization
Genetic screening using metastable epialleles (i.e., alleles that are expressed differently in genetically identical individuals due to epigenetic modifications) led to key discoveries on the molecular mechanisms underlying constitutive heterochromatin formation. In the 1930s, Muller described an interesting Drosophila melanogaster mutant with a variegated eye phenotype (i.e., red and white patches instead of the normal red eye color) upon X-ray-induced mutagenesis (Muller 1930). This phenotype is the results of the silencing of the white gene in some cells and is caused by a chromosomal inversion that places the white gene locus adjacent to pericentromeric heterochromatin. This effect was termed position effect variegation (PEV) because gene expression depends on its position relative to heterochromatin: the closest it is to heterochromatin, the more likely it will be silenced. Random mutagenesis was then used to identify suppressors or enhancers of this phenotype and led to the identification of heterochromatin-associated factors. Some suppressor of variegation Su(var) factors are conserved from fission yeast to vertebrates and have central functions in heterochromatin formation (Elgin and Reuter 2013). Particularly, Su(var)3–9, the first identified histone methyltransferase, catalyzes histone H3K9 methylation, and Su(var)2–5 encodes HP1 (James et al. 1989; Rea et al. 2000; Lachner et al. 2001). In mammals, a similar genetic screening strategy using a transgenic mouse line that expresses green fluorescent protein in a variegated manner was developed and its modifiers are called Mommes (Modifiers of murine metastable epialleles) (Blewitt et al. 2005; Daxinger et al. 2013). As Momme and Su(Var) are two distinct group of genes.
These genetic screen-based approaches have been very useful for identifying new factors involved in silencing. However, to be identifiable in a diploid system, a mutant must have a dominant effect while remaining viable, and this excludes many genes. Moreover, functionally redundant genes cannot be identified using single-gene loss-of-function approaches. Biochemical analyses using common domain searches or physical protein–protein interaction studies have nicely complemented the genetic efforts to uncover heterochromatin factors and how they work. Major factors have been identified by using these traditional approaches. However, their specific function at each constitutive heterochromatin locus remains unclear. Recently, we and other groups have developed novel strategies to address these questions.
Approaches for sequencing repeat regions
High-throughput sequencing is one of the most popular tools in current epigenetic research. Deep sequencing analysis of immunoprecipitated chromatin (ChIP-seq), methylated DNA (methyl-seq), and DNase-cleaved DNA (DNase-seq) is now routinely employed to uncover global epigenome information. To annotate next generation sequencing (NGS) data, a mapping step is required where the sequenced reads are compared to a reference genome dataset. However, in this step, multi-matched reads are discarded and ignored in further analyses. Moreover, almost all repeat regions are longer than the standard read length, and thus, epigenetic information on constitutive heterochromatin is mostly excluded from standard analyses. Thus, the epigenetic profile of heterochromatin regions in various cell types and environments remains far less documented than that of regions made of uniquely mapped reads. In the future, the read length limitation might be partly overcome by novel sequencing technologies, for instance, single-molecule real-time sequencing with a higher read length (up to 20 kb). Currently, if the samples to study are suspected to contain repeat elements, it is possible to do ChIP-seq analyses and use a random mapping program to account for multi-matched reads. This method allows the random assignment of mappable reads to best match locations and averaging counts over multiple occurrences of exactly the same read match. Computational analyses of repetitive DNA using NGS data are reviewed more in detail in Treangen and Salzberg (2012). Moreover, by using annotated databases of repetitive DNA (e.g., Dfam, Repbase, and RepeatMasker), it is possible to analyze the enrichment and to categorize sequenced data with each repeat annotation (Day et al. 2010). However, tandem repeats are still difficult to analyze by NGS. Although it is possible to compare their tag counts, their quantitativity is uncertain because extreme GC content sequences show a biased PCR amplification efficiency, like AT-rich satellite DNA (Bulut-Karslioglu et al. 2012). This is one of the reasons why filter hybridization using a dot (or slot) blot apparatus remains a method of choice for quantitative comparison of tandem repeat DNA or RNA. Nevertheless, the constant improvement of sequencing sensitivity and library preparation methods might completely solve these issues in the near future.
Proteomic approaches to characterize tandem repeat regions
Proteomics is another valuable tool to analyze the epigenetic profile of a given region. Although ChIP-seq is a powerful method to assess the interactions of known proteins with DNA, a proteomics approach can identify unexpected factors, including proteins with unknown functions, and thus has no bias. Mass spectrometric (MS) analysis of specific chromatin loci is very useful for understanding heterochromatin organization. Classically, in vitro affinity capture approaches are used to identify interactors of specific DNA sequences (Wierer and Mann 2016). This strategy has been adapted to modified histone peptides and reconstituted nucleosomes (Bartke et al. 2010; Vermeulen et al. 2010). These approaches have the advantage to eliminate the contribution of DNA sequences or other modifications. MS analysis of ChIP material (ChIP-MS) has been used to identify specific histone modifications or factors associated with chromatin proteins (Wang et al. 2013; Engelen et al. 2015). However, the quality of the data obtained with this approach depends essentially on the antibody quality, and the information on the co-precipitating genomic loci remains limited. To investigate the protein composition of a specific locus, we developed a chromatin purification method that we called proteomics of isolated chromatin segments or PICh (Dejardin and Kingston 2009). In brief, PICh combines DNA in situ hybridization and pull-down assays. The chromatin of fixed cells is solubilized by sonication like in a standard ChIP method and then hybridized with locked nucleic acid (LNA) that contains a probe against specific DNA sequences. Chromatin-probe hybrids are isolated by exploiting the affinity of the biotin linked to the probe for streptavidin. After cross-linking reversal, proteins are analyzed by SDS-PAGE and are identified and quantified by MS. We already reported results for major satellites and telomeric repeat regions using this standard PICh method (Dejardin and Kingston 2009; Saksouk et al. 2014). These regions can be efficiently isolated because they are highly abundant in the genome (0.01–3% of the genome). Thus, repeatedness, which is a problem for sequencing approaches, is a great advantage in PICh-based analyses. We then developed a quantitative PICh (qPICH) method by combining PICh and the stable isotope labeling of amino acids in cell culture (SILAC) technology. SILAC can compare distinct samples in a quantitative manner and its association with PICh helped to obtain an unprecedented understanding of the molecular mechanisms underlying pericentromeric heterochromatin organization (Saksouk et al. 2014). Furthermore, MS-based analyses can identify and quantify posttranslational modifications of proteins, including histones. Therefore, PICh is also a method of choice to determine the histone code of a given region. However, a major PICh limitation is the target relative abundance (i.e., not less than 0.001% of the input, as PICh enrichment factor reaches ~10.000-fold). This makes the purification of single-copy loci from the mammalian genome virtually impossible using this approach. Recently, we developed end-targeting PICh (ePICh) to characterize low-abundance genomic regions (Ide and Dejardin 2015). We optimized ePICh efficiency by employing a different chromatin solubilization method and by modifying the probe design. Using ePICh, we could identify factors that bind to the promoter region of ribosomal RNA genes. Therefore, we believe that PICh-based methods are a key tool for the investigation of repetitive DNA-containing heterochromatin.
Molecular basis of constitutive heterochromatin formation
The fundamental mechanism of constitutive heterochromatin formation is H3K9me3 deposition and its recognition by HP1. Histone hypoacetylation and DNA methylation also are common features of mammalian constitutive heterochromatin (Trojer and Reinberg 2007). However, the detailed mechanisms of constitutive heterochromatin establishment and maintenance are different for each region and developmental stage. In mammals, SUV39H1/2 (Su(var)3–9 homolog in mammals) and SETDB1 are the histone methyltransferases that catalyze H3K9me3 and, thus, have a central role in constitutive heterochromatin formation (Greer and Shi 2012). Importantly, Suv39h 1/2 double-knockout mouse are viable (albeit born to sub-Mendelian ratios and sterile), whereas Setdb1 deletion leads to early embryonic death (Peters et al. 2001; Dodge et al. 2004; Matsui et al. 2010). Thus, only SETDB1 has non-redundant functions in heterochromatin organization. In the following sections, we summarize the latest models of constitutive heterochromatin formation at pericentromeric, telomeric, and TE regions.
Pericentromeric heterochromatin conformation (Fig. 1)
Vertebrate centromeric and pericentromeric regions consist of tandem repetitive DNA often referred to as satellite DNA (the term was coined on the basis of the visible accessory bands observed when digested genomic DNA is analyzed by density gradient sedimentation). In primates, centromeres are made of higher-order arrays of 171 bp α-satellite DNA (also called alphoid DNA) (Waye and Willard 1986). Pericentromeric regions also contain α-satellite repeats, but their organization is not always in a head-to-tail orientation and each satellite block contains other simple repeat sequences and also transposable elements. Thus, it is very difficult to distinguish centromeric and pericentromeric chromatin only on the basis of their DNA sequence (Schueler and Sullivan 2006). Conversely, mouse centromeres and pericentromeres are made of different satellite repeat sequences: the 123-bp minor satellite and the 234-bp major satellite DNA, respectively (Vissel and Choo 1989; Kipling et al. 1991). Furthermore, mouse pericentromeric heterochromatin can be easily detected as DAPI-densed chromatin regions under a microscope named chromocenters, unlike that of primates. Hence, the mouse has been extensively used as a model organism for investigating pericentromeric heterochromatin organization and functions in mammals.
Schematic representation of the pericentromeric heterochromatin structure. In wild-type mouse embryonic stem (WT mES) cells, pericentromeres are made of 234-bp-long major satellite tandem repeats. Pericentromeric heterochromatin regions are enriched in typical heterochromatic histone marks, such as trimethylation of histone H3 at lysine 9 (H3K9me3) and of histone H4 at lysine 20 (H4K20me3). Their deposition is catalyzed by SUV39H and SUV4-20H, respectively, two proteins of the suppressor of variegation family. The H3K9me3 mark functions as a binding module for heterochromatin protein 1 (HP1) to form a more compact chromatin structure. Cytosine methylation is also a typical epigenetic modification of pericentromeric heterochromatin. DNA methyltransferase 3A (DNMT3A) and DNMT3B have de novo methylation activity and DNMT1 functions as maintenance DNA methyltransferase. DNMT3L does not have the catalytic domain for DNA methylation, but it is required for DNMT3A/3B activity. Transcription factors, such as PAX3 and PAX9, are also important for heterochromatin formation. In addition, satellite RNA is transcribed by RNA polymerase II. These factors might promote de novo heterochromatin formation. In Suv39h knockout (KO) mice, no H3K9me3 is detected at pericentromeric regions, but the polycomb repressive complex 2 (PRC2) and the BEND3-NuRD complex-mediated heterochromatin pathway support heterochromatin formation. Furthermore, in the absence of DNA methylation (Dnmt KO mES cells), PRC2 and BEND3-NuRD components are accumulated at pericentromeric heterochromatin. At the zygotic stage, paternal pericentromeric heterochromatin is characterized by DNA hypomethylation and low H3K9me3 levels. The BEND3- and PRC-mediated silencing systems are active at this stage
Open (less condensed) chromatin regions are more frequent in pluripotent cells than in differentiated cells, and pericentromeric loci that contain major satellite sequences also follow this re-organization during cell differentiation (Meshorer and Misteli 2006; Meshorer et al. 2006; Gaspar-Maia et al. 2011). Furthermore, special heterochromatin structures have been observed in specific cell types, such as in senescent cells (senescence-associated heterochromatic foci or SAHF) and rod photoreceptor cells of nocturnal mammals (inverted pattern of heterochromatin/euchromatin distribution in the nucleus) (Narita et al. 2003; Solovei et al. 2009). Although modifications in the association between nuclear lamina and heterochromatin are the main cause of these special structures (Sadaie et al. 2013; Solovei et al. 2013), the mechanisms underlying this global chromatin spatial reorganization are still unclear. It has been suggested that heterochromatin structural organization can change during development and that specific heterochromatin structures can be formed through different pathways.
Experiments in fission yeast showed that H3K9me3 deposition catalyzed by the only SUV39 protein is essential for pericentromeric heterochromatin silencing (Lachner et al. 2001; Bannister et al. 2001; Nakayama et al. 2001) (Fig. 1). In Suv39h1/2 double-knockout mES cells, H3K9me3 and HP1 at major satellite regions are greatly reduced (Bulut-Karslioglu et al. 2012; Martens et al. 2005), while the chromocenters persist, suggesting that chromocenter formation is not strictly dependent on a functional heterochromatin. PICh analysis suggests that SUV39H is the only histone methyltransferase for H3K9me3 at pericentromeric regions (Saksouk et al. 2014). Intriguingly, the PICh results also demonstrated that in SUV39H-deficient cells, the Pc repressive complex 2 (PRC2 in Fig. 1) and H3K27me3 are recruited (features of facultative heterochromatin). Similarly, in cells without methylated DNA following the triple knockout of the genes encoding the DNA methyltransferases (DNMT) Dnmt1, Dnmt3a, and Dnmt3b, H3K9me3 level is reduced and BEND3 is enriched at major satellite regions. BEND3 then allows the recruitment of PRC2 and of the NuRD histone deacetylation complex, which is involved in heterochromatin formation. Importantly, at the early pronuclear stage after embryo fertilization, loss of the H3K9me3 mark, BEND3 recruitment, and Pc-mediated silencing are observed at paternal pericentromeric heterochromatin regions (Fig. 1) (Saksouk et al. 2014; Santos et al. 2005; Dejardin 2015).
H4K20me3 is another typical histone modification at pericentromeric heterochromatin regions (Schotta et al. 2004). Its deposition is catalyzed by the histone methyltransferases SUV4-20H1 and 2, and its pericentromeric localization is mediated by interaction with HP1, thus positioning this mark downstream of H3K9me3. However, H4K20me3 molecular function during pericentromeric heterochromatin formation is unclear.
How SUV39H finds its specific target remains a challenging question. SUV39H has a chromodomain in its N-terminal region that binds to H3K9me3 (Lachner et al. 2001). However, H3K9me3 is not a specific mark of pericentromeric regions, and the initial recruitment of SUV39H at pericentromeres cannot be explained only by this interaction. An alternative model involves non-coding RNA (ncRNA)-mediated recruitment (Fig. 1). Heterochromatin is a transcriptionally inert chromatin structure, but RNA polymerase-dependent ncRNA transcription occurs also at pericentromeric regions (Saksouk et al. 2015). Several chromodomain proteins have nucleic acid-binding activity; thus, ncRNA-mediated SUV39H or HP1 recruitment is a possible model (Nishibuchi and Nakayama 2014). In addition, transcription factors that directly bind to specific motifs in satellite sequences (PAX3 and PAX9 in Fig. 1) are important for SUV39H-dependent heterochromatin formation; thus, transcription factors or their binding proteins might also contribute to SUV39H recruitment to pericentromeric regions (Bulut-Karslioglu et al. 2012; Dejardin 2015).
We already performed studies on pericentromeric heterochromatin-binding factors and demonstrated that many more factors than those identified by previous genetic screening studies are involved in heterochromatin formation (Saksouk et al. 2014). However, our observations were made in mES cells. We believe that PICh analyses using other cell types might uncover new information about heterochromatin organization.
Telomeric and subtelomeric heterochromatin (Fig. 2)
Telomeres, which are located at the tip of all linear chromosomes, also are made of tandem-repeated sequences. Telomere regions play an important role in shielding the chromosome ends from the DNA repair pathway. In vertebrates, telomeres consist of the simple tandem repeat “TTAGGG” and telomere size ranges from a couple of kilobase to >100 kb, depending on the organism, cell type, and age (Moyzis et al. 1988; Riethman 2008). Constitutive heterochromatin is formed at telomeres and their flanking subtelomeric regions to prevent homologous recombination. Subtelomeric regions also are enriched in telomeric repeat sequences, which are interspersed with some other simple repeat sequences and genes. Telomere dysfunctions and subtelomeric and telomeric region rearrangements can lead to genome instability and cancer and to the development of other human diseases and mental retardation (Mefford and Trask 2002; O’Sullivan and Karlseder 2010).
Schematic representation of the telomeric and subtelomeric heterochromatin structure. The shelterin complex (TRF1, TRF2, TIN2, RAP1, TPP1, and POT1) protects telomeric regions from the DNA repair machinery. Both subtelomeric and telomeric regions are enriched in H3K9me3. ATRX- and DAXX-mediated H3.3 incorporation is important for regulating telomere length and recombination. DNA methylation is observed also at subtelomeric regions. Telomeric non-coding RNA (TERRA) is expressed from the subtelomeric region and contributes to the establishment of the proper telomeric structure
In mammals, the multi-protein shelterin complex has a main function in the regulation of telomere length and protection. The shelterin complex consists of six individual proteins: telomeric repeat-binding factor 1 (TRF1), TRF2, repressor and activator protein 1 (RAP1), TRF1interacting nuclear protein 2 (TIN2), protection of telomeres 1 (POT1), and POT1- and TIN2-interacting protein (TPP1). TRF1 and TRF2 bind to telomeric double-stranded DNA, whereas POT1 attaches to telomeric Goverhangs (Fig. 2). These DNA-binding modules and TPP1–TIN2 interaction promote the formation of T-loops that are crucial for chromosome end protection and telomere length regulation (O’Sullivan and Karlseder 2010). Histones are also an important component of telomeric chromosome regions, and telomeres harbor the H3K9me3-silencing mark. In SUV39H1/2-deficient mice, telomere maintenance is defective (Garcia-Cao et al. 2004). Although recent ChIP-seq analyses suggested that SETDB1 is enriched at telomere regions (Karimi et al. 2011; Elsasser et al. 2015), its function in telomere regulation is enigmatic. SUV4-20H activity is also important for telomere elongation, but our PICh experiments using mES cells did not show H4K20me3 accumulation at telomeric regions (Saksouk et al. 2014). DNA methylation also is a typical mark at subtelomeric regions, and DNA hypomethylation leads to telomere maintenance defects (Gonzalo et al. 2006). These heterochromatic modifications may contribute to the regulation of the expression of telomere repeat-containing RNA (TERRA) (Fig. 2). TERRA consists of subtelomeric-derived sequences and G-rich telomeric repeats, and its length ranges from 0.1 to 10 kb (Azzalin et al. 2007). TERRA is required for the recruitment of the shelterin complex and of HP1 through direct binding to TRF2 and origin recognition complex subunit 1 (ORC1) (Deng et al. 2009), and proper TERRA expression is important for telomere maintenance.
Recent studies on heterochromatin formation in telomeric regions and at retrotransposons highlighted the importance of death domain-associated protein (DAXX)-dependent incorporation of the histone variant H3.3 in heterochromatin (Elsasser et al. 2015; Goldberg et al. 2010; Elsasser et al. 2012). DAXX forms heterocomplexes with the SWI/SNF-like chromatin-remodeling protein α-thalassemia/mental retardation X-linked (ATRX). The ATRX-DNMT3-DNMT3L (ADD) domain of ATRX can bind to trimethylated H3K9 (and to unmodified H3K4) (Iwase et al. 2011), and this might support DAXX-mediated H3.3 incorporation into heterochromatic regions. As H3.3 is also enriched at transcriptionally active regions, it is not clear how it contributes to heterochromatin formation. In H3.3-deficient cells, H3K9me3 levels and ATRX recruitment are reduced at telomeres; conversely, TERRA expression is increased (Udugama et al. 2015). Further analyses are required to understand H3.3 molecular function in heterochromatin formation.
Telomerase activity is detected in about 85% of all human cancers to avoid progressive telomere shortening. Among the remaining 15%, most can maintain telomere length through a recombination and amplification mechanism called alternative lengthening of telomeres (ALT) (Cesare and Reddel 2010). ALT is observed particularly in brain tumors and soft tissue sarcomas, but rarely in common tumor types, such as breast, prostate, lung, and colon cancer. One of the features of ALT-positive cancer cells is the association of promyelocytic leukemia nuclear bodies (PML-NB) with telomeres. Moreover, ALT-positive cancer cells typically show complex karyotypes and highly heterogeneous telomere lengths. Intriguingly, our ChIP-seq analysis suggests that ALT-positive telomeres contain “TCAGGG” repeats and are aberrantly enriched in orphan nuclear receptors of the NR2C/F classes. Specifically, NR2C/F proteins can bind to “TCAGGG” repeats present at ALT-positive telomeres and then bridge their target loci, thus clustering and re-localizing ALT-positive telomeres. Consequently, telomeric sequences can be inserted elsewhere in the genome, and this promotes genomic instability in ALT-positive cancers (Marzec et al. 2015). Although it seems that ATRX/DAXX dysfunction is the trigger to gain the ALT system (Heaphy et al. 2011), the underlying molecular mechanism and association with other ALT-specific features remain unclear.
Silencing of endogenous retrotransposable elements (Fig. 3)
Transposable elements (TEs) are the most abundant repeat elements in mammalian genomes and are interspersed throughout the genome. Active TEs contribute to genetic variation and evolution through insertion into gene coding or regulatory regions. However, this insertional activity may also lead to genome instability, thus the silencing mechanism via heterochromatin formation is important for the host organism survival. TEs are classified in DNA transposons and RNA-mediated transposons (or retrotransposons). Retrotransposons are further categorized in long terminal repeat (LTR) retrotransposons and non-LTR retrotransposons. LTR retrotransposons are also called endogenous retroviruses (ERVs). Some LTR retrotransposon types are still transcriptionally active, such as the intracisternal A particle (IAP) that is also the most abundant ERV in the mouse genome. Long-interspersed nucleotide elements (LINEs) and short-interspersed nucleotide elements (SINEs) are representative non-LTR retrotransposons in the mammalian genome (Padeken et al. 2015).
Schematic representation of the heterochromatin structure of a representative endogenous retrovirus (ERV) region. ERVs are suppressed by H3K9me3, DNA methylation, and/or the PIWI pathway. These three major pathways function redundantly, but the preferred pathway can change according to the cell type and developmental stage (see table). SETDB1 is the main histone methyltransferase for H3K9me3 deposition at ERV regions. KRAB-ZFP and KAP1 are required for SETDB1 activity and stable ERV repression. ATRX- and DAXX-mediated H3.3 incorporation also is important for heterochromatin formation at retrotransposable elements. DNA methylation and/or PIWI-mediated heterochromatin formation also can support repression of ERV activity
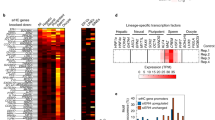
The mechanism of retrotransposon silencing is quite complex because different pathways are involved for each ERV class and developmental stage. Several studies demonstrated that loss of DNA methylation leads to derepression of IAP-type ERVs (class II) in mouse somatic cells (Davis et al. 1989; Jackson-Grusby et al. 2001). However, in mES cells that lack the DNA methyltransferases DNMT1, DNMT3A, and DNMT3B, the effect of DNA hypomethylation on the ERV loci is modest, although DNA methylation also occurs in wild-type ES cells (Karimi et al. 2011; Tsumura et al. 2006). Conversely, SETDB1-mediated H3K9me3 deposition is essential for repression of ERV classes I and II in ES cells, but not in somatic cells (Matsui et al. 2010). LINE elements (non-LTR retrotransposons) are silenced by SUV39H and DNA methylation in mES cells (Bulut-Karslioglu et al. 2014). In addition, SETDB1 could play a crucial role in silencing ERV activity during the developmental stages characterized by DNA hypomethylation, such as early embryonic and germ line development (Wu and Zhang 2010). Indeed, Setdb1 conditional knockout in the mouse causes defective ERV repression in early embryos and during gametogenesis (Eymery et al. 2016; Liu et al. 2014). Interestingly, forward genetic studies using mouse and human cells suggest that exogenous genes integrated adjacent to ERV, and thus silenced, as well as epigenetically repressed endogenous genes are derepressed by dysfunction of SETDB1 and its binding factors (called the HUSH complex) (Daxinger et al. 2013; Tchasovnikarova et al. 2015). Thus, SETDB1 has a major role in the establishment and propagation of the heterochromatin structure in these regions. Krüppel-associated box domain-zinc finger proteins (KRAB-ZFP) and KRAB-ZFP-associated protein 1 (KAP1, also known as tripartite motif-containing protein 28, TRIM28) are required for SETDB1 recruitment to target loci (Rowe et al. 2010). As mentioned before, ATRX/DAXX function is also important for ERV suppression, especially in conditions of global DNA hypomethylation (Elsasser et al. 2015; He et al. 2015). Specifically, in these conditions, ATRX and DAXX recruitment to targets (including ERVs) is promoted through H3.3 incorporation activity and they might facilitate H3K9me3 deposition by recruiting histone methyltransferases. However, while SETDB1 function is indispensable for silencing a subset of ERVs in primordial germ cells (PGCs), some H3K9me3-marked ERVs are not derepressed in PGCs in which Setdb1 has been knocked out (Liu et al. 2014). This means that there are other pathways for silencing retrotransposable elements in germ line cells. One alternative mechanism involves the PIWI pathway. This small RNA-mediated silencing system is highly conserved in eukaryotes, and direct interactions of argonaute family proteins and small RNAs are important for targeting mRNAs for degradation. PIWI, a Drosophila argonaute protein, is essential for germ line development. PIWI-interacting RNAs (piRNA) are small non-coding RNA molecules (mostly 24–32 nucleotides in length) that consist mainly of transposon sequences (Ishizu et al. 2012). PIWI and piRNAs promote transposon repression through transcript degradation and recruitment of heterochromatin factors, such as HP1 and linker histone H1, to target loci (Brower-Toland et al. 2007; Iwasaki et al. 2016). In the mouse, the PIWI-homolog MIWI family of proteins is important for germ line cell development and retrotransposon activity repression (Deng and Lin 2002; Aravin et al. 2006). Therefore, these alternative pathways are important for transposable activity silencing and the preferred pathway may change according to the cell type and developmental stage (see table in Fig. 3). It is not known what factor(s) regulates this preference. However, we believe that the novel technologies for identifying specific chromosome-interacting proteins will provide clues to answer the remaining questions.
Conclusions
Recent studies have shown that the mechanisms to regulate constitutive heterochromatin formation and maintenance dynamically change in various situations. Particularly, multiple pathways are used to maintain the heterochromatin structure at retrotransposable regions. Although ChIP-seq can be adapted for the analysis of repeat sequences, this approach is limited to the investigation of factors that are already known to be involved in heterochromatin formation. Re-analysis of past deep sequencing data and new studies using technologies that allow the sequencing and analysis of repeat sequences might help better understanding heterochromatin organization. Besides the rapid development of novel DNA sequencing methods, MS sensitivity is also increasing dramatically. In our recent study using PICh, we identified more than 100 proteins associated with pericentromeric and more than 600 with telomeric repeats. Although, they were mostly already known suppressors of variegation and modifiers of murine metastable epialleles, the role of some of the discovered interacting proteins in the regulation of heterochromatin formation is unknown. This means that traditional genetic approaches (e.g., loss of function experiments) have a limited use for elucidating the specific role of novel multi-functional proteins or redundant genes. Therefore, new strategies must be developed to uncover the mechanisms underlying constitutive heterochromatin organization.
Abbreviations
- ADD:
-
ATRX-DNMT3-DNMTT3L
- ALT:
-
Alternative lengthening of telomeres
- ATRX:
-
α-Thalassemia/mental retardation X-linked
- BEND3:
-
BEN domain containing 3
- DAXX:
-
Death domain-associated protein
- ERV:
-
Endogenous retrovirus
- HUSH:
-
Human-silencing hub
- HP1:
-
Heterochromatin protein 1
- IAP:
-
Intracisternal A particle
- KAP1:
-
KRAB-ZFP-associated protein 1
- KRAB-ZFP:
-
Krüppel-associated box domain-zinc finger proteins
- LINE:
-
Long-interspersed nucleotide element
- LTR:
-
Long terminal repeat
- Mommes:
-
Modifiers of murine metastable epialleles
- NuRD:
-
Nucleosome remodeling and deacetylase complex
- ORC:
-
Origin recognition complex
- Pc:
-
Polycomb
- PEV:
-
Position effect variegation
- PICh:
-
Proteomics of isolated chromatin segment
- piRNA:
-
PIWI-interacting RNA
- PIWI:
-
P-element-induced wimpy testis
- POT1:
-
Protection of telomeres 1
- RAP1:
-
Repressor and activator protein 1
- SAHF:
-
Senescence-associated heterochromatic foci
- SETDB1:
-
SET domain bifurcated 1
- Su(var):
-
Suppressor of variegation
- TERRA:
-
Telomere repeat-containing RNA
- TIN2:
-
TRF1interacting nuclear protein 2
- TPP1:
-
POT1- and TIN2-interacting protein
- TRF1:
-
Telomeric repeat-binding factor 1
References
Aravin A et al (2006) A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442(7099):203–207
Azzalin CM et al (2007) Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318(5851):798–801
Bannister AJ et al (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410(6824):120–124
Bartke T et al (2010) Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143(3):470–484
Blewitt ME et al (2005) An N-ethyl-N-nitrosourea screen for genes involved in variegation in the mouse. Proc Natl Acad Sci U S A 102(21):7629–7634
Brower-Toland B et al (2007) Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev 21(18):2300–2311
Bulut-Karslioglu A et al (2012) A transcription factor-based mechanism for mouse heterochromatin formation. Nat Struct Mol Biol 19(10):1023–1030
Bulut-Karslioglu A et al (2014) Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol Cell 55(2):277–290
Cesare AJ, Reddel RR (2010) Alternative lengthening of telomeres: models, mechanisms and implications. Nat Rev Genet 11(5):319–330
Davis CM et al (1989) Activation and demethylation of the intracisternal a particle genes by 5-azacytidine. Cell Differ Dev 27(2):83–93
Daxinger L et al (2013) An ENU mutagenesis screen identifies novel and known genes involved in epigenetic processes in the mouse. Genome Biol 14(9):R96
Day DS et al (2010) Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol 11(6):R69
Dejardin J (2015) Switching between epigenetic states at pericentromeric heterochromatin. Trends Genet 31(11):661–672
Dejardin J, Kingston RE (2009) Purification of proteins associated with specific genomic loci. Cell 136(1):175–186
Deng W, Lin H (2002) Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2(6):819–830
Deng Z et al (2009) TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35(4):403–413
Dodge JE et al (2004) Histone H3-K9 methyltransferase ESET is essential for early development. Mol Cell Biol 24(6):2478–2486
Elgin SC, Reuter G (2013) Position-effect variegation, heterochromatin formation, and gene silencing in drosophila. Cold Spring Harb Perspect Biol 5(8):a017780
Elsasser SJ et al (2012) DAXX envelops a histone H3.3-H4 dimer for H3.3-specific recognition. Nature 491(7425):560–565
Elsasser SJ et al (2015) Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature 522(7555):240–244
Engelen E et al (2015) Proteins that bind regulatory regions identified by histone modification chromatin immunoprecipitations and mass spectrometry. Nat Commun 6:7155
Eymery A et al (2016) The methyltransferase Setdb1 is essential for meiosis and mitosis in mouse oocytes and early embryos. Development 143(15):2767–2779
Garcia-Cao M et al (2004) Epigenetic regulation of telomere length in mammalian cells by the Suv39h1 and Suv39h2 histone methyltransferases. Nat Genet 36(1):94–99
Gaspar-Maia A et al (2011) Open chromatin in pluripotency and reprogramming. Nat Rev Mol Cell Biol 12(1):36–47
Goldberg AD et al (2010) Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140(5):678–691
Gonzalo S et al (2006) DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol 8(4):416–424
Greer EL, Shi Y (2012) Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet 13(5):343–357
Gregory TR (2005) Synergy between sequence and size in large-scale genomics. Nat Rev Genet 6(9):699–708
He Q et al (2015) The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell 17(3):273–286
Heaphy CM et al (2011) Altered telomeres in tumors with ATRX and DAXX mutations. Science 333(6041):425
Ide S, Dejardin J (2015) End-targeting proteomics of isolated chromatin segments of a mammalian ribosomal RNA gene promoter. Nat Commun 6:6674
Ishizu H et al (2012) Biology of PIWI-interacting RNAs: new insights into biogenesis and function inside and outside of germlines. Genes Dev 26(21):2361–2373
Iwasaki YW et al (2016) Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol Cell 63(3):408–419
Iwase S et al (2011) ATRX ADD domain links an atypical histone methylation recognition mechanism to human mental-retardation syndrome. Nat Struct Mol Biol 18(7):769–776
Jackson-Grusby L et al (2001) Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27(1):31–39
James TC et al (1989) Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of drosophila. Eur J Cell Biol 50(1):170–180
Jost KL et al (2012) Heterochromatin and gene positioning: inside, outside, any side? Chromosoma 121(6):555–563
Karimi MM et al (2011) DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8(6):676–687
Kipling D et al (1991) Mouse minor satellite DNA genetically maps to the centromere and is physically linked to the proximal telomere. Genomics 11(2):235–241
Lachner M et al (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410(6824):116–120
Liu S et al (2014) Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes Dev 28(18):2041–2055
Martens JH et al (2005) The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J 24(4):800–812
Marzec P et al (2015) Nuclear-receptor-mediated telomere insertion leads to genome instability in ALT cancers. Cell 160(5):913–927
Matsui T et al (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464(7290):927–931
Mefford HC, Trask BJ (2002) The complex structure and dynamic evolution of human subtelomeres. Nat Rev Genet 3(2):91–102
Meshorer E, Misteli T (2006) Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol 7(7):540–546
Meshorer E et al (2006) Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 10(1):105–116
Moyzis RK et al (1988) A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci U S A 85(18):6622–6626
Muller HJ (1930) Types of visible variations induced by X-rays in Drosophila. J Genet 22:299–334
Nakayama J et al (2001) Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292(5514):110–113
Narita M et al (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113(6):703–716
Nishibuchi G, Nakayama J (2014) Biochemical and structural properties of heterochromatin protein 1: understanding its role in chromatin assembly. J Biochem 156(1):11–20
O’Sullivan RJ, Karlseder J (2010) Telomeres: protecting chromosomes against genome instability. Nat Rev Mol Cell Biol 11(3):171–181
Padeken J et al (2015) Repeat DNA in genome organization and stability. Curr Opin Genet Dev 31:12–19
Peters AH et al (2001) Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107(3):323–337
Rea S et al (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406(6796):593–599
Riethman H (2008) Human telomere structure and biology. Annu Rev Genomics Hum Genet 9:1–19
Rowe HM et al (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463(7278):237–240
Sadaie M et al (2013) Redistribution of the Lamin B1 genomic binding profile affects rearrangement of heterochromatic domains and SAHF formation during senescence. Genes Dev 27(16):1800–1808
Saksouk N et al (2014) Redundant mechanisms to form silent chromatin at pericentromeric regions rely on BEND3 and DNA methylation. Mol Cell 56(4):580–594
Saksouk N et al (2015) Constitutive heterochromatin formation and transcription in mammals. Epigenetics Chromatin 8:3
Santos F et al (2005) Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol 280(1):225–236
Schotta G et al (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18(11):1251–1262
Schueler MG, Sullivan BA (2006) Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7:301–313
Solovei I et al (2009) Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137(2):356–368
Solovei I et al (2013) LBR and Lamin a/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152(3):584–598
Tchasovnikarova IA et al (2015) GENE SILENCING. Epigenetic silencing by the HUSH complex mediates position-effect variegation in human cells. Science 348(6242):1481–1485
Treangen TJ, Salzberg SL (2012) Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 13(1):36–46
Trojer P, Reinberg D (2007) Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell 28(1):1–13
Tsumura A et al (2006) Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells 11(7):805–814
Udugama M et al (2015) Histone variant H3.3 provides the heterochromatic H3 lysine 9 tri-methylation mark at telomeres. Nucleic Acids Res 43(21):10227–10237
Vermeulen M et al (2010) Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142(6):967–980
Vissel B, Choo KH (1989) Mouse major (gamma) satellite DNA is highly conserved and organized into extremely long tandem arrays: implications for recombination between nonhomologous chromosomes. Genomics 5(3):407–414
Wang CI et al (2013) Chromatin proteins captured by ChIP-mass spectrometry are linked to dosage compensation in Drosophila. Nat Struct Mol Biol 20(2):202–209
Waye JS, Willard HF (1986) Structure, organization, and sequence of alpha satellite DNA from human chromosome 17: evidence for evolution by unequal crossing-over and an ancestral pentamer repeat shared with the human X chromosome. Mol Cell Biol 6(9):3156–3165
Wierer M, Mann M (2016) Proteomics to study DNA-bound and chromatin-associated gene regulatory complexes. Hum Mol Genet 25(R2):R106–R114
Wu SC, Zhang Y (2010) Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol 11(9):607–620
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible editors: Nick Gilbert and Davide Marenduzzo
Rights and permissions
About this article
Cite this article
Nishibuchi, G., Déjardin, J. The molecular basis of the organization of repetitive DNA-containing constitutive heterochromatin in mammals. Chromosome Res 25, 77–87 (2017). https://doi.org/10.1007/s10577-016-9547-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-016-9547-3