Abstract
RbAp46/48, histone chaperone, is a family of evolutionarily conserved WD40 repeat-containing proteins, which are involved in various chromatin-metabolizing processes, but their in vivo functional relevance is yet unclear. In order to examine the biological role of pRbAp48 in chicken DT40 cells, we generated a tetracycline-inducible system for conditional RbAp48-knockout cells. Depletion of RbAp48 led to delayed S phase progression associated with slow DNA synthesis and nascent nucleosome formation, followed by accumulation in G2/M phase, finally leading to cell death. Prior to cell death, these cells exhibited aberrant mitosis such as highly condensed and abnormal chromosome alignment on the metaphase plate, leading to chromosome missegregation. Depletion of RbAp48 also caused dissociation of heterochromatin protein 1 (HP1) from pericentromeric heterochromatin. Furthermore, depletion of RbAp48 from cells led to elevated levels of acetylation and slightly decreased levels of methylation, specifically at Lys-9 residue of histone H3. These results suggest that RbAp48 plays an important role in chromosome stability for proper organization of heterochromatin structure through the regulation of epigenetic mark.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In eukaryotic nuclei, the genomic DNA is wrapped around histone octamers to form nucleosome core particles, basic repeating units of chromatin, with the aid of histone chaperone proteins through replication-coupled or replication-independent pathways (Adam et al. 2014). Since nucleosomal histones are thought to serve as a barrier to prevent transacting factors involved in DNA metabolism from accessing DNA, histone chaperone proteins play important roles in chromatin assembly, DNA replication, transcriptional regulation, and DNA damage repair (Adam et al. 2014; De Koning et al. 2007). Chromatin assembly factor-1 (CAF-1), a well-conserved protein complex consisting of three polypeptides of p150, p60, and p48, was originally purified from human cells as a factor that promotes de novo nucleosome assembly on replicating SV40 DNA (Smith and Stillman 1989). Of these, p48 subunit, initially identified as a retinoblastoma (Rb)-binding protein (RbAp48), is a member of the histones H3/H4 chaperons containing WD40 repeats β-propeller structure (Verreault et al. 1996). The highly related protein RbAp46 is a subunit of the histone-acetyltransferase-1 (HAT1) complex, which acetylates newly synthesized histone H4 on lysines 5 and 12 prior to nucleosome assembly (Verreault et al. 1998). In addition to roles in nucleosome assembly process, both RbAp48 and RbAp46 are also found in several other complexes involved in the regulation of chromatin structure, including NuRD (nucleosome remodeling histone deacetylase complex), histone deacetylase (HDAC)-containing complexes (Sin3), and histone methyltransferase (HMT)-containing complex (polycomb repressive complexes 2, PRC2) (Zhang et al. 1999, 1997; Muller et al. 2002). Since RbAp48/p46 proteins have the ability to bind directly to histones H4 and H3 (Nowak et al. 2011). they are proposed to act as escorting factors that connect these complexes to their substrate nucleosomes, and are implicated in multiple aspects of cellular events involved in histone metabolizing process depending on the complex’s composition.
Physiological functions of RbAp46/48 family have been well studied in model organisms such as yeast, plants, and Drosophila. In Saccharomyces cerevisiae, the RbAp46/p48 homologs, Msi1 and Hat2, participate in silencing of the telomere and/or in the maintenance of transcriptionally silent heterochromatin structure (Kelly et al. 2000; Kaufman et al. 1997). Arabidopsis RbAp46/p48 homolog, Msi1, functions to regulate development and flowering timing through epigenetic regulation (Hennig et al. 2003). In Drosophila, Nurf55, the RbAp46/p48 homolog, is also critical for development, cell viability, and proliferation in several tissues (Anderson et al. 2011). In mammals, despite the accumulation of knowledge about the role of RbAp48/p46-containing complexes such as CAF-1, NuRD, and PRC2, in influencing various cellular functions, including cellular development and tumorigenesis, there is little information concerning the role of RbAp48/p46 in these complexes and their in vivo functional relevance.
In this paper, we describe the development of a genetic system for assessing the role of RbAp48 in the chicken DT40 B cell line. Due to the necessity of RbAp48 for the viability of DT40 cells, we generated a conditional RbAp48-knockout cells. Depletion of RbAp48 caused decreased DNA synthesis, chromatin instability on newly replicated DNA, and was subsequently accompanied by the accumulation of various mitotic defects, resulting in aberrant chromosomal segregation. These results demonstrate for the first time that RbAp48 plays essential roles for nascent chromatin assembly in vivo and maintenance of chromosome stability in vertebrate cells.
Materials and methods
Plasmid constructs
Using chicken RbAp48 cDNA as a probe, the genomic DNA clone specific for RbAp48 was isolated on the screening of a DT40 genomic DNA library in a Lambda FIXII as described (Takami et al. 1999). To generate targeting vectors, p∆p48/neo and p∆p48/hisD, for disruption of the RbAp48 gene, we cloned the 8.0-kb SacI/BamHI fragment into pBluescript II (Stratagene) and replaced the 2.0-kb StuI/NsiI fragment, containing the region of intron 1 to exon 4, with the cassette of the neomycin-resistant gene or that of the hisD gene driven by the β-actin promoter (Takami et al. 2007). Probe 5′ was the 1.0-kb SacI fragment derived from 5′-upstream of the RbAp48 gene. Probes neo and hisD were derived from neo and hisD as described (Takami et al. 2007). To construct the ptetHAp48 vector expressing HA-tagged pRbAp48 under the control of the tet operator (tetO) and CMV minimal promoter, the PCR-amplified cDNA product, encoding HA epitope-tagged pRbAp48, was inserted into the pUHD10-3 plasmid (Gossen and Bujard 1992). ptTA-bleo construct, which contains the tet-responsive transactivator gene and the bleomycin-resistant gene driven by the β-actin promoter, was as described (Sanematsu et al. 2006). Targeting constructs for CENP-H-GFP-Hyg and HP1γ-DsRed-puro were described previously (Fukagawa et al. 2001; Takami et al. 2007).
Cell cultures, transfection, and isolation of transfectants
DT40 cells and all subclones were cultured as described (Takami et al. 2007). At the indicated times, cells were counted to determine the growth rate. Transfection by electroporation was carried out as described (Takami et al. 2007). Drug-resistant colonies were selected on 96-well microtiter plates in medium containing appropriate concentrations of several drugs, i.e., 2 mg of G-418 (GIBCO-BRL) per ml, 0.3 mg of phleomycin (Sigma) per ml, 0.8 mg of histidinol (Sigma) per ml, or 0.4 μg of puro (Sigma) per ml. Doxycycline (tet) (Sigma) was used at a concentration of 100 ng per ml to suppress the expression of the tet-responsive HAp48 transgene.
Western blotting
Cells (5 × 106) were lysed in 200 μl of SDS buffer, and aliquots (20 μl) were separated by 7.5 % SDS-PAGE, followed by electro-blotting onto a PVDF membrane filter as described (Takami et al. 2007). Total histones were extracted by H2SO4 acid, precipitated by TCA (Barman et al. 2006). and separated by 12.5 % SDS-PAGE, followed by electro-blotting as above. The filter was probed with a 1/1000 dilution of anti-RbAp48 antiserum (Transduction Laboratories), anti-HA antibodies (12CA5, Boehringer), and anti-histone antibodies (Millipore) followed by incubation with a 1/1000 dilution of secondary antibodies (horseradish peroxidase conjugated anti-mouse IgG or anti-rabbit IgG antibodies (Dako)). The signal was developed using a Super Signal™ CL-HRP system (Pierce, Rockford, IL) and visualized with a LAS-1000 (Fuji Film).
Flow cytometric analysis of cell cycles
Flow cytometric analyses were carried out using a FACS Calibur (Becton-Dickinson, Mountain View, CA) as described (Takami et al. 2007). For two-dimensional cell cycle analyses, cells were cultured in the presence of bromodeoxyuridine (BrdU) for 10 min, fixed in 70 % ethanol, and stained with a fluorescein isothiocyanate (FITC)-labeled anti-BrdU antibody (Pharmingen) and propidium iodide (PI). For synchronization of the mitotic phase, cells were cultured for the indicated times in the presence or absence of tet, treated with nocodazole (500 ng/ml) for 6.5 h, and then released from the block by washing with medium three times and cultured further. At 2 h intervals, cells were collected, fixed in 70 % ethanol, stained with PI, and analyzed.
Analysis of chromatin structure in vivo
Cells were cultured with or without tet for the indicated times. Collected cells were adjusted to 1 × 107/ml in pre-warmed medium and pulse-labeled by the addition of 50 μCi/ml of 3H-thymidine (PerkinElmer Life Sciences) for 10 min. Nuclei were prepared as follows. Cells were washed with cold PBS twice, suspended in NB (10 mM Tris–HCl, pH 7.5, 0.1 mM EDTA, 2 mM magnesium acetate, 2 mM CaCl2, 1 mM DTT, protease inhibitor cocktail (Sigma)), and incubated in the presence of 0.2 % NP-40 for 10 min on ice. The resultant nuclei were washed with NB twice, suspended at 20 A260/ml (A260 was measured in 2 M NaCl and 5 M urea) in NB, and then digested at 37 °C for 7 min with micrococcal nuclease (MNase) (Sigma) to the final concentration of 0.2, 0.068, or 0.022 units/ml. The reactions were stopped by adding of 10 mM EDTA and 0.5 % SDS, and DNA was purified by incubation with 100 μg/ml of proteinase K for 2 h at 37 °C, followed by phenol-chloroform extraction and ethanol precipitation. DNA was electrophoresed in a 1.4 % agarose gel and transferred to a Hybond N+ membrane. For the detection of 3H-labeled DNA, blots were directly exposed on a BAS screen exclusive for tritium and visualized using a Mac BAS-1000 (Fuji Film). The same filter was hybridized with 32P-labeled probes chHAT1-5′ and HDAC-5′, respectively, which correspond to the 5′ promoter regions of the chicken HAT1 and HDAC1 genes. Signals were detected using a Mac BAS-1000.
Immunofluorescence microscopy
Immunofluorescence staining of whole cells was performed as described (Fukagawa et al. 2001). Briefly, DT40 cells were cytospun onto slides at 1000 rpm for 5 min, fixed in 3 % paraformaldehyde in Hepes buffer for 15 min, permeabilized with 0.1 % NP-40 in PBS for 15 min, and blocked in 0.5 % BSA in PBS for 5 min three times. The primary antibodies were incubated for 1 h at 37 °C in the blocking buffer at appropriate dilutions (1/50 dilution for FITC-conjugated anti-alpha-tubulin antibody (Sigma), 1/500 dilution of anti-chicken CAF-1p150 and p60 antibodies, and 1/500 dilution of anti-PCNA (PC10; Sigma-Aldrich)) (Takami et al. 2007). To detect the primary antibodies, slides were incubated in a 1/1000 dilution of AlexaFluor488- or 594-conjugated anti-mouse or anti-rabbit IgG antibodies (Molecular probe) for 1 h and then counterstained with DAPI at 0.2 μg/ml. For Fig. 4c, DNA was counterstained with PI at 0.1 μg/ml and images of alpha-tubulin were obtained with a Leica TCS-4 confocal scanning microscope equipped with a 100 × /NA 1.4 plan oil immersion objective. Other images in Figs. 3, 4 and 5 were obtained using a CCD camera (Orca-ER, Hamamatsu) mounted on a Zeiss Axiovert equipped with a Plan Apo 60 × /NA 1.3 oil immersion objective and processed computationally in IPlab systems.
Fluorescence in situ hybridization (FISH) was performed using the painting probe specific for chicken chromosome 3 as described previously (Fukagawa et al. 2001). Metaphase chromosome spreads were prepared by the addition of a hypotonic solution and fixed in methanol/acetic acid (3:1). The biotin-labeled probe was detected with FITC-conjugated avidin. Chromosomes were counterstained with DAPI at 0.2 μg/ml.
Live cell images were acquired automatically at multiple locations by using Nikon BioStation IM. The conditional RbAp48-knockout cells expressing both CENP-H-GFP and HP1-DsRed were plated onto 35 mm coverglass bottom dishes with or without doxycycline (tet) for 36 h in CO2 incubator and then transferred to fluorescence, and phase contrast images were acquired with ×40 objective lens and obtained every 20 min for a period of 36 to 60 h after initial incubation with or without doxycycline (tet).
Results
Depletion of RbAp 48 alters cell growth and the cell-cycle distribution
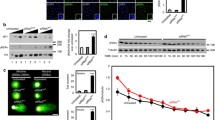
Despite the nearly ubiquitous expression of both RbAp46 and RbAp48 in all cell types, chicken DT40 cells showed marginal expression of RbAp46 mRNA at substantially lower levels than RbAp48 mRNA (data not shown), suggesting the prevalent role of RbAp48 in this cell line. In order to investigate the in vivo role of RbAp48, two disruption constructs were designed based on the genomic DNA as shown in Fig. 1a. To avoid the predicted lethality of RbAp48-deficient cells, we generated RbAp48-conditional knockout cells. We first obtained RbAp48 –/+clones, in which one allele was disrupted by the neo disruption construct (Fig. 1b). Next, we produced RbAp48 –/+ , tetHAp48 cell clones, in which N-terminal HA-tagged RbAp48 was expressed under the control of a tet-responsive promoter. After confirmation of the tet-repressive expression of HA-RbAp48 (Fig. 1c), one clone (cl3-tet2) was transfected for the second round of disruptions of the remaining allele (Fig. 1b). We obtained RbAp48 –/– , tetHAp48 clone (cl3-tet2-36), referred to conditional RbAp48-knockout cells, in which levels of the HA-RbAp48 protein were essentially same with that of endogenous RbAp48 protein in DT40 cells, but upon the addition of tet, it declined to below detection levels by 24 h (Fig. 2b).
Generation of conditional RbAp48-deficient cells. a Genomic organization and schematic diagram of the homologous recombination resulting in disruption of the first and second alleles of RbAp48. Two targeting vectors, p∆p48/neo and p∆p48/hisD constructs, are shown. The locations of probes 5′, neo and hisD, are indicated by bars. b Southern blotting of homologous recombination events in RbAp48 loci. The HindIII fragments were analyzed with probe 5′, neo or hisD. c Depletion of the HA-tagged pRbAp48 protein, denoted as HAp48. HAp48 proteins in mutants (cl3-tet2 and cl3-tet4) grown in the presence (+) or absence (−) of tet for 24 h were detected by Western blotting with anti-HA antibody
Effects of RbAp48-deficiency on cell growth and cell cycle. a Growth curve. Two RbAp48-deficient subclones, cl 3-tet2-35 (RbAp48 -/+ , HAp48) denoted as #35 and cl 3-tet2-36 (RbAp48 -/- , HAp48) denoted as #36, were cultured at 1 × 105 cells per ml in the absence or presence of tet. After 24 h, the cells were transferred to fresh medium at 1 × 105 cells per ml and then continuously grown. b Depletion of the HA-tagged pRbAp48 protein. Cell extracts were separated by SDS-10 % PAGE and analyzed by Western blotting with anti-RbAp48 at the indicated times after the addition of tet. The level of endogenous RbAp48 was also determined with serial dilutions of the cell extract of DT40 cells. c Effects of tet on the cell cycle and DNA synthesis in cl3-tet2-36. At the indicated times after the addition of tet, cells were pulse-labeled with BrdU for 10 min and stained with FITC-anti-BrdU (y-axis, log scale) for the detection of BrdU incorporation, and with PI for the detection of the DNA content (x-axis, linear scale). d Effects of depletion of RbAp48 on cell-cycle progression. Cells were cultured in the absence (−tet) or presence (+tet) of tet, then synchronized at the metaphase by the addition of nocodazole for 6.5 h before the indicated times. After release from the M-phase block, cells were collected at 2 h intervals, fixed, and stained with PI, and DNA content was analyzed by flow cytometry
In the absence of tet, the growth rate of the conditional RbAp48-knockout cells was similar to that of RbAp48 –/+ , tetHAp48 clone and DT40 cells, with an approximate doubling time of 11–12 h (Fig. 2a). Upon the addition of tet, RbAp48-knockout cells began to grow slowly at 36 h, and finally, almost all of the cells were dead by 72–84 h (Fig. 2a), indicating that RbAp48 is essential for viability of DT40 chicken B cells.
To assess the effects of RbAp48-depletion on cell cycle, DNA synthesis, and cell death, we performed the two-dimensional FACS analyses after pulse labeling with BrdU (Fig. 2c). Upon RbAp48-depletion, the amount of BrdU incorporated into individual cells was slightly reduced at 24 h and was greatly reduced by 48–72 h after the addition of tet (see the level of the y-axis and compare the shapes of the BrdU arc), indicating an impaired DNA synthesis in S phase cells. In addition, a cell population with sub-G1 DNA content, indicative of apoptosis, became apparent at 48 h and increased thereafter (Fig. 2c).
For closer examination of cell cycle progression, cells were synchronized at mitotic (M) phase by nocodazole, and the DNA content was monitored by FACS at 2 h intervals after removal of the drug. In the absence of tet, cells proceeded to G1/S and G2/M phases by 2 and 8 h, respectively (Fig. 2d left). On the other hand, after 30 h of incubation with tet, the progression from M to G1 phase was not affected, but delayed progression through S phase was observed (Fig. 2d center). Moreover, after 36 h of incubation with tet, in addition to S phase delay, the accumulation of M phase cells was also observed (Fig. 2d right). These findings together indicate that depletion of RbAp48 causes the delay in S phase progression accompanied by impaired DNA replication, and prolonged depletion of RbAp48 prevents mitotic exit (M to G1 transition).
RbAp48 contributes to replication-coupled nucleosome assembly in vivo
Previous study revealed close association of DNA replication with nucleosome formation onto replicated DNA by CAF-1 (Takami et al. 2007). Although RbAp48 is a subunit of the CAF-1 complex and has been shown to be necessary for the nucleosome assembly onto newly replicated DNA in vitro system (Verreault et al. 1996; Kadyrova et al. 2013), in vivo contribution of RbAp48 to this process is still unclear. We therefore analyzed stability of nascent chromatin structure during DNA replication by pulse labeling DNA with 3H-thymidine, followed by MNase digestion assay. After gel electrophoresis of MNase-digested samples, DNA was stained with ethidium bromide (EtBr) to evaluate bulk chromatin structure, and pulse-labeled DNA on the blot was detected using 3H-radioimaging. To assess chromatin structure of the specific genome region, the same blot was subject to Southern hybridization with 32P-labeled probes HAT1-5′ and HDAC1-5′, which correspond to the promoter regions of the HAT1 and HDAC1 genes, respectively. MNase sensitivity of bulk and specific regions were almost unchanged in RbAp48-depleted cells (24–48 h) compared to that in non-depleted cells (0 h) (Fig. 3a, c, d), indicating that the global chromatin structure is not significantly impaired until at least 48 h after treatment with tet. In contrast, substantial MNase sensitivity was observed in the chromatin onto newly replicated DNA within 24 h treatment with tet (Fig. 3b). These findings not only indicate that the RbAp48-depletion specifically compromises chromatin onto newly replicated DNA but barely influences the general stability of overall chromatin, but also that RbAp48 is absolutely required for CAF-1 activity in vivo as a part of CAF-1 complex.
Effects of depletion of RbAp48 on chromatin structure. At the indicated times after the addition of tet, conditional RbAp48-knockout cells were collected and pulse-labeled with 3H-thymidine. Isolated nuclei were treated with MNase at 0.2, 0.068, or 0.022 units/ml (from left to right at each time in each panel). Purified DNAs were electrophoresed in a 1.4 % agarose gel, stained with EtBr (a), and transferred to a Hybond N+ membrane. The membrane was directly autoradiographed (b) and hybridized with 32P-labeled probe HAT1-5′, corresponding to the promoter region of HAT1 (c). After dehybridization, the membrane was rehybridized with 32P-labeled probe HDAC1-5′, corresponding to the promoter region of HDAC1 (d). e Conditional RbAp48-knockout cells were cultured in the presence of tet for the indicated times and total cell extracts were separated by SDS-8 % PAGE and analyzed by Western blotting with the indicated antibodies. f Conditional RbAp48-knockout cells were cultured in the absence or presence of tet for 36 h and attached onto slides followed by pre-extraction with CSK containing Triton X-100 (0.4 %) before fixation. After staining with anti p150 or p60 antibodies, DNA was counterstained with DAPI (blue). p150 and p60 signals were detected with Alexa 594-conjugated second antibody (red)
To assess the in vivo roles of RbAp48 in CAF-1 function, we examined the stability of other CAF-1 subunits, p150 and p60, and their cellular localization in RbAp48-depleted cells. As shown Fig. 3e, upon depletion of RbAp48, both protein levels of p150 and p60 were reduced to approximately half of those in non-depleted cells within 36 h after addition of tet, indicating obvious degradation of p150 and p60 proteins in RbAp48-depleted cells. Furthermore, immunofluorescence studies showed that discrete nuclear foci for CAF-1 p150 and p60, which are known to colocalize with proliferating cell nuclear antigen (PCNA) foci through the interaction of the p150 subunit with PCNA, indicative of replication factories, were diffused throughout the cells within 30 h after addition of tet, although some small residual foci for p150 remained (Fig. 3f). These results together suggest that RbAp48 is not only required for stability of other CAF-1 subunits but also for their stable association to newly replicated region.
Prolonged depletion of RbAp48 causes mitotic abnormalities
Since the prolonged depletion of RbAp48 resulted in the delayed M to G1 progression (Fig. 2c), we cytologically examined the time-course kinetics of the mitotic index and cell viability by staining with DAPI. While viable cells started to decrease from 48 h, the mitotic index increased from about 5 to 12 % at 36 to 48–60 h (Fig. 4a). On the other hand, population of the cells at the anaphase or telophase decreased slightly from 36 h, indicating that the prolonged depletion of RbAp48 impedes metaphase progression to anaphase where mitotic cells exhibit a high incidence of highly condensed, misaligned chromosomes (Fig. 4b(c), c(c)–(e)) and lagging chromosomes (Fig. 4b(d), 4c(f)). Staining with anti-alpha-tubulin also revealed that numerous mitotic cells had multipolar (>2) spindles (Fig. 4c(d)), probably resulting from extra centrosome amplification, leading to abnormal cell division. In addition, FISH analysis showed that cells with aberrant number of chromosome 3 were increased gradually from 48 h after addition of tet (Fig. 4d). These results indicate that the prolonged RbAp48-depletion induces chromosome segregation errors as a result of various mitotic defects.
Prolonged RbAp48-deficiency induces aberrant chromosome alignment and chromosome missegregation. a Conditional RbAp48-knockout cells were cultured in the presence of tet for the indicated times, fixed, and stained with DAPI. Populations of the indicated cell types were counted under a fluorescence microscope. At least 1000 cells were counted each time. The numbers of ana/telophase cells are indicated as percentages of those per total mitotic cells. b Cells were cultured in the absence or presence of tet for 48 h, fixed, and stained with DAPI. Left panels a and b represent well-ordered chromosomes at the metaphase and anaphase, respectively, in the absence of tet. Panels c and d represent misaligned and condensed chromosomes at the metaphase and anaphase bridge at the anaphase, respectively, in the presence of tet. Right panel indicates quantitation of aberrant mitotic cells by addition of tet at time 0. We scored the number of aberrant metaphase cells in total of over 100 mitotic cells. c Cells were fixed and stained with FITC-conjugated anti-alpha-tubulin antibodies (green), and DNA was counterstained with PI (red). Fluorescence images were visualized using a confocal scanning microscope. Panels a and b show cells at the metaphase and anaphase, respectively, in the absence of tet. Panels c to e or f show cells at the metaphase or anaphase, respectively, in the presence of tet. Panel c also shows multiple spindle pole bodies. d Cells were cultured in the presence of tet for the indicated times. FISH analysis was performed on metaphase chromosome spreads, using the painting probe specific for chromosome 3 (green). DNA was counterstained with DAPI (blue). Panel a indicates normal diploid cells (2N) with two chromosome 3; panel b indicates cells with one allelic loss of chromosome 3; and panel c indicates cells showing polyploidy (4N). Cells with aberrant number of chromosomes 3 (2 < n > 2) were counted on at least 300 metaphase plates. The results are summarized in right panel
Depletion of RbAp48 causes defects in pericentric heterochromatin
It is well known that proper organization of kinetochore structure and pericentric heterochromatin is necessary for stable metaphase chromosome positioning and the faithful chromosome segregation (Kwon et al. 2007; Fukagawa and Earnshaw 2014). Therefore, we examined localizations of centromere proteins and heterochromatin proteins 1 (HP1) by tagging the endogenous CENP-H and HP1γ with fluorescent protein (CENP-H-GFP and HP1γ-DsRed) in RbAp48-knockout cells(Takami et al. 2007). As shown in Fig. 5a, centromeric localization of CENP-H (indicated by dotted signals) appeared not to be significantly altered within 60 h after the addition of tet in both interphase nuclei (Fig. 5a) and the mitotic chromosome (date not shown), suggesting that kinetochore structure was not severely compromised in RbAp48-depleted cells. However, it is not clear whether such kinetochore is precisely functional or not. On the other hand, HP1γ signals, which mainly localized to discrete foci overlapping with centromeric CENP-H foci, indicative for pericentric heterochromatin region, became dissociated from pericentric region throughout the cell nucleus at 60 h after the addition of tet (Fig. 5a and Supplementary files 1 and 2). These results suggest that the prolonged RbAp48-deficiency causes impaired structures of pericentric heterochromatin domains, probably affecting the kinetochore functions.
Depletion of RbAp48 causes dissociation of heterochromatin protein 1 (HP1) from pericentromeric heterochromatin and increased acetylation of histone H3. a Localization of HP1γ and CENP-H in interphase cells. Conditional RbAp48-knockout cells, endogenously tagged with DsRed and EGFP into respective last exons of HP1γ and CENP-H alleles, were cultured in the absence (−tet) or presence (+tet) of tet for 60 h. Cells were fixed and counterstained with DAPI (blue). Signals for HP1γ-Ds-Red (red) and CENP-H-GFP (green) were directly observed. b Conditional RbAp48-knockout cells were cultured in the absence or presence of tet. At the indicated times after the addition of tet, total histone proteins were acid-extracted and separated by SDS-12.5 % PAGE and analyzed by Western blotting with the indicated antibodies (right). Serial dilutions of the histone extracts were used to confirm data integrity
It is well documented that histone modifications function as major determinant for formation of heterochromatin, and that histone chaperone protein, RbAp48, is a component of several complexes involved in histone modifications such as HDAC-containing complexes and HMT-containing complex (PRC2) (Zhang et al. 1997; Muller et al. 2002). In addition, histone H3 lysine 9 (H3K9) methylation has been shown to serve as a specific binding site for HP1 (Kwon and Workman 2008). Therefore, we examined the effect of RbAp48-depletion on histone modifications using acid extracted total histone proteins by Western blotting. As shown in Fig. 5b, while global acetylation levels of histone H4(K8, K12, and K16) remained constant but acetylation levels of histone H3K9 and H3K27 were significantly increased with time of RbAp48-depletion. Conversely, repressive histone methylation marks, di-and tri-methylation levels of H3K9, and tri-methylation levels of H3K27 were only slightly decreased or almost unchanged in RbAp48-depleted cells. These results suggest that RbAp48 may control histone modification in heterochromatin via several chromatin complexes containing histone-modifying enzyme, and hyperacetylation of H3K9 and H3K27 in RbAp48-depleted cells appears to provoke dissociation of HP1 from pericentromeric heterochromatin.
Discussion
Although some studies with RNAi knockdown approach suggested biological roles for RbAp48 in chromatin organization and transcriptional regulation in mammalian cell lines (Hayashi et al. 2004; Scuto et al. 2007). To better understand the physiological functions of RbAp48 in other cell types, we genetically created conditional RbAp48-knockout DT40 cells. Using this system, we clearly demonstrated that RbAp48 is essential for the viability of chicken B cells and showed that depletion of RbAp48 causes defects in DNA replication and S phase progression accompanied by impaired chromatin assembly specifically onto newly replicated DNA. Similar phenotypes have been observed in DT40 cells deficient in other CAF-1 subunits, p150 or p60 (Takami et al. 2007). thus suggesting that these defects in S phase caused by RbAp48-depletion are mainly due to dysfunction of CAF-1. Indeed, during the depletion of RbAp48, CAF-1p150 and p60 proteins, which have central role for assembly of new histones into replicating DNA, became unstable and lost their ability of recruitment to replication foci. Although the fine functional role of RbAp48 in CAF-1 complex are still unclear, our results indicated for the first time that trimeric subunits formation is critical for in vivo CAF-1 function that promote nucleosome formation onto newly replicated DNA in vertebrate cells. Incomplete histone assembly onto replicated DNA probably leads to regional and stochastic disturbances of chromatin formation triggering various cellular defects, such as impaired DNA replication resulting from slowdown of replication fork progression, and leading to delayed S phase progression. Consistent with this prediction, a recent study indicates that efficient histone assembly onto daughter strand regulates fork progression by coupling histone supply and unloading PCNA (Mejlvang et al. 2014).
In addition to defects in S phase, we also found that prolonged deficiency of RbAp48 causes aberrant mitosis, accompanied with multiple centrosomes, hypercondensed, misaligned chromosomes, and chromosome bridges (Fig. 4b, c) that occurred only after passage through one or two cell cycles following the loss of RbAp48, suggesting a possible involvement of impaired epigenetic regulation. Centromere component proteins have been well known to play important roles in the proper chromosome segregation (Fukagawa and Earnshaw 2014). Previous genetic studies also showed that the loss of functional heterochromatin protein, such as Drosophila HP1 and Swi6p, a HP1 homolog in Saccharomyces pombe, results in structural defects of centromere accompanied by chromosome missegregations (Nestorov et al. 2013). In addition, the interactions of CAF-1p150 with HP1 are thought to relate with replication of heterochromatin domains in late S phase (Quivy et al. 2004; De Koning et al. 2007). In this study, we showed that although extensive changes in localization of centromeric protein, CENP-H, could not be observed, HP1γ was clearly dissociated from pericentric heterochromatin region in prolonged RbAp48-deficient cells (Fig. 5a), suggesting that the kinetochore function was at least partially compromised along with impaired pericentric heterochromatin. Interestingly, these phenotypes were not seen in CAF-1p150 or p60-knockout DT40 cells (Takami et al. 2007). suggesting additional roles of RbAp48 in organizing pericentric heterochromatin domain, which are independent of CAF-1 function. In this context, RbAp48 is implicated in a role for CENP-A loading to centromere to organize functional kinetochore (Hayashi et al. 2004). and in histone modification through different chromatin regulatory complexes including histone deacetylase (Mi2/NuRD, Sin3 complex) or histone methyltransferase (PRC2 complex) (Zhang et al. 1999; Muller et al. 2002). In general, constitutive heterochromatin is signified by repressive histone code modifications, deacetylation, and methylation of histone H3 lysine 9 (H3K9), which is crucial for its organization by creating a binding site for HP1 proteins (Kwon and Workman 2008). Intriguingly, we showed that depletion of RbAp48 induced a time-dependent increase in the global acetylation states of H3K9 along with dissociation of HP1 from pericentric chromatin, while by contrast, the methylation states of H3K9 were only slightly reduced, suggesting that H3K9 methylation is not sufficient to recruit HP1 to heterochromatin. This observation is consistent with previous data suggesting that other than primary methyl-H3K9 binding, stable HP1 recruitment to heterochromatin requires secondary protein-protein and/or -noncoding RNA interaction (Kwon and Workman 2008). In addition, it has been shown that treatment of human cells with HDAC inhibitors, such as trichostatin A (TSA) and FR901228 (DP), which cause hyperacetylation of H3K9 without its demethylation, resulted in dissociation of HP1 from pericentric heterochromatin, decreased pericentromeric targeting of Aurora B kinase, followed by assembly of deficient kinetochores in M-phase (Robbins et al. 2005). Taken together, these results support the idea that RbAp48 plays an important role for proper organization of pericentric heterochromatin structures by regulating histone acetylation at these regions through tethering histone-modifying enzyme complexes, and that impaired pericentric hetrochromatin in RbAp48-deficient cells may trigger aberrant mitosis, resulting in mis-segregation of chromosomes. However, we cannot exclude the possibility that effects of RbAp48-depletion on mitotic progression may be a result of possible changes in expression of several genes involved in chromosome stability and/or mitotic process through the altered chromatin modification.
In conclusion, we speculate that the crucial roles of RbAp48 in cell viability and proliferation in S phase is most likely due to its indispensable role in CAF-1 activity, and RbAp48-guided chromatin regulatory complexes other than CAF-1 may also confer pericentromeric domains with a functional configuration to recruit heterochromatin binding proteins such as HP1 and other partner molecules via histone modification, which is critical for chromosome integrity. Further studies will be required to reveal which of these complexes participate in CAF-1-independent functions of RbAp48 in DT40 cells. Our findings may help to understand the potential link between replication-coupled nucleosome assembly, DNA replication, chromosome segregation, and histone modification in vertebrate cells. Further analysis of conditional RbAp48-knockout cells will improve our understanding of its roles in multiple cellular aspects involved in histone metabolizing processes, including DNA replication, checkpoint response, DNA repair, and maintenance of chromosome integrity in vertebrate cells.
Abbreviations
- CAF-1:
-
Chromatin assembly factor
- HP1:
-
Heterochromatin protein 1
- Rb:
-
Retinoblastoma
- HAT:
-
Histone-acetyltransferase
- HDAC:
-
Histone deacetylase
- HMT:
-
Histone methyltransferase
References
Adam S, Polo SE, Almouzni G (2014) How to restore chromatin structure and function in response to DNA damage—let the chaperones play. FEBS J 281:2315–2323
Anderson AE, Karandikar UC, Pepple KL, Chen Z, Bergmann A, Mardon G (2011) The enhancer of trithorax and polycomb gene Caf1/p55 is essential for cell survival and patterning in Drosophila development. Development 138:1957–1966
Barman HK, Takami Y, Ono T, Nishijima H, Sanematsu F, Shibahara K, Nakayama T (2006) Histone acetyltransferase 1 is dispensable for replication-coupled chromatin assembly but contributes to recover DNA damages created following replication blockage in vertebrate cells. Biochem Biophys Res Commun 345:1547–1557
De Koning L, Corpet A, Haber JE, Almouzni G (2007) Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol 14:997–1007
Fukagawa T, Earnshaw WC (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30:496–508
Fukagawa T, Mikami Y, Nishihashi A, Regnier V, Haraguchi T, Hiraoka Y, Sugata N, Todokoro K, Brown W, Ikemura T (2001) CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J 20:4603–4617
Gossen M, Bujard H (1992) Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A 89:5547–5551
Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M (2004) Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell 118:715–729
Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W (2003) Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130:2555–2565
Kadyrova LY, Rodriges Blanko E, Kadyrov FA (2013) Human CAF-1-dependent nucleosome assembly in a defined system. Cell Cycle 12:3286–3297
Kaufman PD, Kobayashi R, Stillman B (1997) Ultraviolet radiation sensitivity and reduction of telomeric silencing in Saccharomyces cerevisiae cells lacking chromatin assembly factor-I. Genes Dev 11:345–357
Kelly TJ, Qin S, Gottschling DE, Parthun MR (2000) Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol Cell Biol 20:7051–7058
Kwon SH, Workman JL (2008) The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol Cell 26:217–227
Kwon MS, Hori T, Okada M, Fukagawa T (2007) CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell 18:2155–2168
Mejlvang J, Feng Y, Alabert C, Neelsen KJ, Jasencakova Z, Zhao X, Lees M, Sandelin A, Pasero P, Lopes M, Groth A (2014) New histone supply regulates replication fork speed and PCNA unloading. J Cell Biol 204:29–43
Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O’Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila polycomb group repressor complex. Cell 111:197–208
Nestorov P, Tardat M, Peters AH (2013) H3K9/HP1 and polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr Top Dev Biol 104:243–291
Nowak AJ, Alfieri C, Stirnimann CU, Rybin V, Baudin F, Ly-Hartig N, Lindner D, Muller CW (2011) Chromatin-modifying complex component Nurf55/p55 associates with histones H3 and H4 and polycomb repressive complex 2 subunit Su(z)12 through partially overlapping binding sites. J Biol Chem 286:23388–23396
Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G (2004) A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J 23:3516–3526
Robbins AR, Jablonski SA, Yen TJ, Yoda K, Robey R, Bates SE, Sackett DL (2005) Inhibitors of histone deacetylases alter kinetochore assembly by disrupting pericentromeric heterochromatin. Cell Cycle 4:717–726
Sanematsu F, Takami Y, Barman HK, Fukagawa T, Ono T, Shibahara K, Nakayama T (2006) Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J Biol Chem 281:13817–13827
Scuto A, Zhang H, Zhao H, Rivera M, Yeatman TJ, Jove R, Torres-Roca JF (2007) RbAp48 regulates cytoskeletal organization and morphology by increasing K-Ras activity and signaling through mitogen-activated protein kinase. Cancer Res 67:10317–10324
Smith S, Stillman B (1989) Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell 58:15–25
Takami Y, Kikuchi H, Nakayama T (1999) Chicken histone deacetylase-2 controls the amount of the IgM H-chain at the steps of both transcription of its gene and alternative processing of its pre-mRNA in the DT40 cell line. J Biol Chem 274:23977–23990
Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T (2007) Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell 18:129–141
Verreault A, Kaufman PD, Kobayashi R, Stillman B (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87:95–104
Verreault A, Kaufman PD, Kobayashi R, Stillman B (1998) Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol 8:96–108
Zhang Y, Iratni R, Erdjument-Bromage H, Tempst P, Reinberg D (1997) Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89:357–364
Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D (1999) Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev 13:1924–1935
Acknowledgments
This work was supported in part by Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Ryoko Masuya and Nahoko Nagamatsu-Yamamoto for their technical assistance and Radha Madhyastha and HarishKumar Madhyastha for useful comments and critical reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interest of any sort with anyone.
Additional information
Responsible Editor: Job Dekker.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary file 1
The live-cell observation was started at 36 h after plating the conditional RbAp48-knockout cells, endogenously tagged with DsRed and EGFP in the normal condition (without tet), and time-lapse images were taken every 20 min for the next 23 h. Time is shown as hours:minutes after addition of tet. Left panel, phase contrast; center panel, CENPH-GFP; right panel, HP1γ–DsRed. (AVI 2162 kb)
Supplementary file 2
The live-cell observation was started at 36 h after plating the conditional RbAp48-knockout cells, endogenously tagged with DsRed and EGFP in the presence of tet, and time-lapse images were taken every 20 min for the next 23 h. Time is shown as hours:minutes after addition of tet. Left panel, phase contrast; center panel, CENPH-GFP; right panel, HP1γ–DsRed. (AVI 2345 kb)
Rights and permissions
About this article
Cite this article
Satrimafitrah, P., Barman, H.K., Ahmad, A. et al. RbAp48 is essential for viability of vertebrate cells and plays a role in chromosome stability. Chromosome Res 24, 161–173 (2016). https://doi.org/10.1007/s10577-015-9510-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-015-9510-8