Abstract
The water monitor lizard (Varanus salvator macromaculatus (VSA), Platynota) has a chromosome number of 2n = 40: its karyotype consists of 16 macrochromosomes and 24 microchromosomes. To delineate the process of karyotype evolution in V. salvator macromaculatus, we constructed a cytogenetic map with 86 functional genes and compared it with those of the butterfly lizard (Leiolepis reevesii rubritaeniata (LRE); 2n = 36) and Japanese four-striped rat snake (Elaphe quadrivirgata (EQU); 2n = 36), members of the Toxicofera clade. The syntenies and gene orders of macrochromosomes were highly conserved between these species except for several chromosomal rearrangements: eight pairs of VSA macrochromosomes and/or chromosome arms exhibited homology with six pairs of LRE macrochromosomes and eight pairs of EQU macrochromosomes. Furthermore, the genes mapped to microchromosomes of three species were all located on chicken microchromosomes or chromosome 4p. No reciprocal translocations were found in the species, and their karyotypic differences were caused by: low frequencies of interchromosomal rearrangements, such as tandem fusions, or centric fissions/fusions between macrochromosomes and between macro- and microchromosomes; and intrachromosomal rearrangements, such as paracentric inversions or centromere repositioning. The chromosomal rearrangements that occurred in macrochromosomes of the Varanus lineage were also identified through comparative cytogenetic mapping of V. salvator macromaculatus and V. exanthematicus. Morphologic differences in chromosomes 6–8 between the two species could have resulted from pericentric inversion or centromere repositioning.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genome sequences of the domestic chicken (Gallus gallus; International Chicken Genome Sequencing Consortium ICGSC 2004) precipitated advances in comparative genomics between Reptilia and Aves that facilitated comparison of genomic structures between two taxa at the molecular level. Comparison of the chromosome maps of the Chinese soft-shelled turtle (Pelodiscus sinensis), Siamese crocodile (Crocodylus siamensis), Japanese four-striped rat snake (Elaphe quadrivirgata), butterfly lizard (Leiolepis reevesii rubritaeniata), and Australian central bearded dragon (Pogona vitticeps) with G. gallus revealed that syntenies and gene orders have been highly conserved between Aves, Testudines, Crocodylia, and Squamata (Matsuda et al. 2005; Matsubara et al. 2006, 2012; Srikulnath et al. 2009b; Uno et al. 2012; Young et al. 2013) since the Sauropsida (all existing reptiles and birds) diverged from the Synapsida around 320 MYA (Shedlock and Edwards 2009). The draft genome assembly of the green anole (Anolis carolinensis) confirmed that the sytenies and gene orders of the green anole are also highly conserved in the chicken genome (Alföldi et al. 2011).
Squamate reptiles, the most diverse reptilian order, are traditionally classified into three suborders: Serpentes (snakes), Amphisbaenia (worm lizards), and Lacertilia (lizards). The most common chromosome number in snakes is 2n = 36: their karyotypes consist of 16 macro- and 20 microchromosomes. There is relatively little karyotypic variation (2n = 30–42) in snakes, whereas worm lizards show a large variation in chromosome number (2n = 30–50; Olmo and Signorino 2005). The extant lizards can be categorized into six infraorders—Iguania, Gekkota, Scincomorpha, Diploglossa, Dibamia, and Platynota (Uetz 2013)—and they also exhibit a large variation in both chromosome number (2n = 24–46) and chromosome morphology (Olmo and Signorino 2005). Karyotypes with few or no microchromosomes are found in the Lacertidae family of Scincomorpha and in the Gekkota, whereas karyotypes containing many microchromosomes are found in the Iguania, Platynota, Dibamia, Diploglossa, and Scincomorpha excluding the Lacertidae.
Molecular phylogenetic studies suggest that iguanians (Iguania) and anguimorphs (Platynota and Diploglossa) are closely related to snakes, and all are members of the so-called Toxicofera clade, which bear oral toxin-secreting glands (Vidal and Hedges 2005). However, the close genetic relationship of iguanians with snakes and anguimorphs remains controversial (Townsend et al. 2004; Vidal and Hedges 2005; Srikulnath et al. 2010; Wiens et al. 2012). The common chromosome number of Iguania is 2n = 36: these karyotypes consist of 12 macro- and 24 microchromosomes. This differs from the most common karyotype of Serpentes (2n = 36), which comprises 16 macro- and 20 microchromosomes (Olmo and Signorino 2005). Our previous chromosome mapping study (Srikulnath et al. 2009b), which compared chromosomal locations of 54 functional genes between the butterfly lizard (Iguania) and the Japanese four-striped rat snake, revealed that most macrochromosomes were homologous to each other, whereas several chromosomal rearrangements—such as centric fission/fusion of macrochromosomes, telomere-to-telomere tandem fusion between macrochromosomes and between macro- and microchromosomes, and centromere repositioning—had occurred between the species. By contrast, anguimorphs show a large variation in the numbers of both macro- and microchromosomes (2n = 30–44) within the same taxon (Olmo and Signorino 2005). However, the degree of conservation of synteny and gene order of anguimorph chromosomes relative to snake and iguanian chromosomes, and the pattern of karyotype evolution in anguimorphs, remain unknown. Comparative gene mapping in anguimorphs, snakes, and iguanians is necessary to clarify the process of karyotype evolution in the Toxicofera clade.
The monitor lizards (varanid lizards, Varanus sp., Varanidae), an ancient group of anguimorph lizards, inhabit Afro-Arabia, Western to Southeast Asia, the Indonesian Archipelago, Papua New Guinea, and Australia (Ast 2001; Townsend et al. 2004; Amer and Kumazawa 2008). Currently, the Varanidae family comprises 73 extant species (Uetz 2013). According to King and King (1975), the chromosome number of all monitor lizards is 2n = 40: these karyotypes comprise 16 macro- and 24 microchromosomes. The only variation in their karyotypes is in the morphology of the macrochromosomes. The karyotype of the water monitor lizard (Varanus salvator macromaculatus) comprises two pairs of large metacentrics (1st and 2nd), two pairs of medium-sized metacentrics (3rd and 4th), one pair of small acrocentrics (5th), three pairs of small submetacentrics (6th–8th), and 12 pairs of microchromosomes. This is considered the primitive form of varanid lizard karyotypes (King and King 1975; Chaiprasertsri et al. 2013). However, the karyotype of the Savannah monitor lizard (Varanus exanthematicus) differs in several pairs of small macrochromosomes from that of V. salvator macromaculatus (King and King 1975). This suggests that the two monitor lizards might be good models to elucidate the process of chromosomal changes in the lineage of Varanus. Here, we construct a comparative cytogenetic map of functional genes for V. salvator macromaculatus using fluorescence in situ hybridization (FISH), and examine its conserved synteny with L. reevesii rubritaeniata and E. quadrivirgata chromosomes. Using comparative mapping data from these species, we delineate the process of karyotype evolution in the Toxicofera clade. Additionally, we also construct a cytogenetic map of V. exanthematicus chromosomes 6–8, to identify the chromosomal rearrangements that occurred between the two monitor lizard species.
Materials and methods
Specimens, cell culture, and chromosome preparation
Fibroblasts from a male water monitor lizard (V. salvator macromaculatus) used in the work of Chaiprasertsri et al. (2013) were recovered from liquid nitrogen and cultured. A male Savannah monitor lizard (V. exanthematicus) was purchased from a pet shop in Nagoya, Japan. Morphologic identification of the species was performed as previously described (Bennett and Thakoordyal 2003; and data not shown). After intraperitoneal injection of pentobarbital, the heart, lungs, and mesenteries were removed and used for cell culture. The animal’s gender was identified morphologically, and confirmed by its internal anatomy. All experimental procedures using animals conformed to guidelines established by the Animal Care and Use Committee, Nagoya University, Japan. The tissues were minced, and cultured in Gibco® Dulbecco’s modified Eagle’s medium (Life Technologies Corporation, Carlsbad, CA, USA) supplemented with 15 % Gibco® Fetal Bovine Serum (Life Technologies Corporation), 100 μg/ml kanamycin, and 1 % Gibco® Antibiotic-Antimycotic (Life Technologies Corporation). The cultures were incubated at 26 °C in a humidified atmosphere of 5 % CO2 in air. Primary cultured fibroblasts were harvested using trypsin and subcultured. For replication R-banding, fibroblast cultures from the two species were incubated with 5-bromo-2′-deoxyuridine (12 μg/ml; Sigma-Aldrich Corporation, St. Louis, MO, USA) for 12 h, including 45 min of colcemid treatment (120 ng/ml) before harvesting, and chromosome preparations were made following a standard air-drying method. After staining the chromosome slides with Hoechst 33258 (1 μg/ml) for 8 min, the slides were heated at 65 °C for 3 min, and exposed to ultraviolet light at 65 °C for an additional 6 min (Matsuda and Chapman 1995). The slides were kept at −80 °C until use.
Molecular identification of V. exanthematicus
For the molecular identification of V. exanthematicus, partial DNA fragments of the mitochondrial ND2 gene were cloned, and their nucleotide sequences determined. Whole genomic DNA was extracted from the liver of a male V. exanthematicus following a standard salting-out protocol, and used as a template for polymerase chain reaction (PCR). Primers and PCR conditions were previously described (Kumazawa and Endo 2004). The PCR products were cloned using the pGEM®-T-Easy Vector System I (Promega Corporation, Madison, WI, USA), and the nucleotide sequences of the DNA fragments were determined using an ABI 3130 Automated Capillary DNA Sequencer (Applied Biosystems, Life Technologies Corporation). The nucleotide sequences were used to search the National Center for Biotechnology Information (NCBI) database using the BLASTx and BLASTn programs (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and were deposited in the DNA Data Bank of Japan (AB795272; http://www.ddbj.nig.ac.jp/index-e.html). The nucleotide sequences of the V. exanthematicus mitochondrial ND2 gene were aligned with those of other platynotan lizards and outgroups, comprising the banded gecko (Coleonyx variegatus), American alligator (Alligator mississippiensis), Siamese crocodile (C. siamensis), and domestic chicken (G. gallus) taken from the NCBI database (Supplementary Table S1). All unalignable and gap-containing sites were carefully removed, and sequence divergence between the species was estimated using the default parameters of Molecular Evolutionary Genetics Analysis 4 software (Center for Evolutionary Functional Genomics, The Biodesign Institute, Tempe, AZ, USA; Kumar et al. 2004), using uncorrected pairwise distances.
Cross-species chromosome mapping
The chromosomal locations of the 18S–28S and 5S ribosomal RNA (rRNA) genes, telomeric (TTAGGG)n sequences, and 86 functional genes were determined by FISH, as previously described (Matsuda and Chapman 1995; Srikulnath et al. 2009a). cDNA fragments cloned from L. reevesii rubritaeniata (54 genes; Srikulnath et al. 2009a, b), Gekko hokouensis (one gene), Lacerta agilis (30 genes; Srikulnath et al., unpublished data), and E. quadrivirgata (one gene; Matsubara et al., unpublished data; Table 1; Supplementary Table S2), and a 99-base pair (bp) fragment (pCSI5S; EU723235) of the C. siamensis 5S rRNA genes were used for chromosome mapping. Nucleotide sequence homologies of all reptilian cDNA fragments with those of the chicken were examined and the cDNA fragments were confirmed to be homologs of chicken genes. We labeled 250 ng of cDNA or 5S rDNA probe with biotin-16-dUTP (Roche Diagnostics, Basel, Switzerland) by nick translation following the manufacturer’s protocol. After hybridization, the probes were detected with goat anti-biotin antibody (Vector Laboratories, Burlingame, CA, USA), and stained with Alexa Fluor® 488 rabbit anti-goat IgG (H + L) conjugate (Molecular Probes®, Life Technologies Corporation). Slides were subsequently counterstained with 0.75 μg/ml propidium iodide. Hybridization signals were captured using a cooled charge-coupled device (CCD) camera mounted on a Nikon fluorescence microscope (Nikon Corporation, Tokyo, Japan), and processed using software by Nikon Microsystems Imaging Solutions Ltd. (Tokyo, Japan).
Dual-color FISH was performed to compare the chromosomal locations of the 18S–28S rRNA genes and telomeric (TTAGGG)n sequences in V. salvator macromaculatus and V. exanthematicus. A partial 1.8-kb fragment (pCSI1) of the 8.2-kb fragment (EU727190) of the C. siamensis 18S–28S rRNA genes, and a biotin-labeled 42-bp oligonucleotide complementary to (TTAGGG)n sequences (Sigma-Aldrich Corporation), were used as probes. We labeled 250 ng of the 18S–28S rDNA probe with digoxigenin-11-dUTP (Roche Diagnostics) and hybridized to V. salvator macromaculatus and V. exanthematicus chromosomes with the biotin-labeled TTAGGG repeats. After hybridization, the digoxigenin- and biotin-labeled probes were stained with anti-digoxigenin-rhodamine Fab fragments (Roche Diagnostics) and avidin labeled with fluorescein isothiocyanate (avidin-FITC; Vector Laboratories), respectively. Fluorescence hybridization signals were captured using a cooled CCD camera mounted on a Leica DMRA microscope (Leica Microsystems, Wetzlar, Germany), and processed using 550CW-QFISH software by Leica Microsystems Imaging Solutions Ltd. (Cambridge, UK).
Results
Molecular identification of V. exanthematicus
The data set of aligned ND2 fragments showed 761 nucleotides, comprising 63 variable sites and 472 parsimony informative sites. The degree of sequence divergence between the V. exanthematicus ND2 fragment obtained in this study (AB795272) and that previously deposited in the database (AF407496) was 1 % (data not shown). This result confirmed that the animal used in this study was V. exanthematicus, considering that intraspecific variation of ND2 sequences was shown to be about 10 % in lizards (Glor and Laport 2012).
Karyotypes of V. salvator macromaculatus and V. exanthematicus
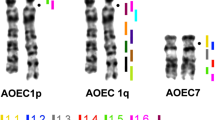
We examined over 20 Hoechst-stained metaphase spreads for one male V. salvator macromaculatus and one male V. exanthematicus. Diploid chromosome numbers were 40 in both species, and morphologic differences were evident in three pairs of chromosomes (6th–8th) between the two species (Fig. 1). The eight pairs of macrochromosomes of V. salvator macromaculatus comprised two pairs of large metacentrics (1st and 2nd), two pairs of metacentrics (3rd and 4th), one pair of acrocentrics (5th), and three pairs of submetacentrics (6th–8th; King and King 1975; Chaiprasertsri et al. 2013), whereas the 6th and 8th chromosomes were acrocentric and the 7th submetacentric in V. exanthematicus (Fig. 1). Our data on the V. exanthematicus karyotype were inconsistent with a previous report (King and King 1975), which described chromosomes 6–8 as acrocentric.
Cross-species chromosome mapping of cDNA clones in V. salvator macromaculatus, and a cytogenetic map of V. salvator macromaculatus
We constructed a cytogenetic map of V. salvator macromaculatus with 86 genes (Figs. 2 and 3). This is the first reported cytogenetic map in anguimorph lizards. Over 30 metaphase spreads were observed for each gene, and hybridization efficiency ranged from approximately 20–80 %. Chromosome homology between V. salvator macromaculatus and the chicken was analyzed using the chicken genome database (http://www.ncbi.nlm.nih.gov/genome/guide/chicken/). Twelve genes mapped on V. salvator macromaculatus (VSA) chromosome 1 localized to chicken (GGA) chromosomes Z, 12, 13, 16, and 18. Eight genes mapped on VSA2 localized to GGA3, 5, 7, and 13, and ten genes on VSA3 were located on GGA1p and 4q. Seven genes on VSA4 localized to GGA2p and 27, and five genes on VSA5 to GGA1q. Nine genes mapped on VSA6 localized to GGA6 and 9, three genes on VSA7 to GGA2q and 13, and six genes on VSA8 to GGA8, 20 and 26. Twenty-six genes mapped on VSA microchromosomes were located on GGA microchromosomes or GGA4p that was derived from a microchromosome of the ancestral avian karyotype (Nishida-Umehara et al. 2007).
Chromosomal locations of cDNA fragments of functional genes in V. salvator macromaculatus. The SOX9 gene was located on chromosome 1q (a), SOX5 on chromosome 3p (c), WAC on chromosome 4q (e), ELMOD1 on chromosome 5 (g), EPHA4 on chromosome 6q (i), HDAC3 on chromosome 7p (k), and USP49 on chromosome 8q (m). b, d, f, h, j, l, n Hoechst-stained patterns of the same metaphase spreads shown in (a), (c), (e), (g), (i), (k), and (m), respectively. Arrows indicate hybridization signals. Scale bars represent 5 μm
Comparative cytogenetic map of V. salvator macromaculatus constructed with 60 functional genes and the 18S–28S and 5S ribosomal RNA genes. The ideograms of V. salvator macromaculatus macrochromosomes were constructed according to Hoechst-stained band patterns. Locations of the genes on V. salvator macromaculatus chromosomes are shown to the right of chromosomes. The chromosome numbers of the chicken (G. gallus, GGA), butterfly lizard (L. reevesii rubritaeniata, LRE), and Japanese four-striped rat snake (E. quadrivirgata, EQU), which show homologies with V. salvator macromaculatus chromosomes, are shown to the left. “no” indicates no data of chromosome homology, and “un” indicates a gene for which the chromosomal location has not been determined. The chromosomal locations of L. reevesii rubritaeniata genes were taken from Srikulnath et al. (2009b). The chromosomal locations of E. quadrivirgata genes were taken from Matsubara et al. (2006, 2012)
Chromosomal locations of the 18S–28S ribosomal RNA genes and telomeric (TTAGGG)n sequences in V. salvator macromaculatus and V. exanthematicus
The 18S–28S rRNA genes localized to the secondary constriction in the proximal region of V. salvator macromaculatus and V. exanthematicus chromosome 1p. Hybridization signals for (TTAGGG)n sequences were observed at the telomeric ends of all chromosomes, and no interstitial signals were found (Fig. 4). To the best of our knowledge, this is the first study to report the chromosomal location of the 18S–28S rRNA genes and (TTAGGG)n sequences in varanid lizards.
Chromosomal locations of the 18S–28S ribosomal RNA genes and (TTAGGG)n sequences in a V. salvator macromaculatus male (a) and V. exanthematicus male (b). Hybridization patterns of rhodamine-labeled 18S–28S ribosomal RNA (rRNA) genes (red) and fluorescein isothiocyanate-labeled (TTAGGG)n sequences (green) on Hoechst-stained metaphase spreads. Arrows indicate the hybridization signals of the 18S–28S rRNA genes. Scale bar represents 10 μm
Comparative cytogenetic map between V. salvator macromaculatus and V. exanthematicus
Hoechst-stained karyotypes of V. salvator macromaculatus and V. exanthematicus revealed morphologic differences in chromosomes 6–8 (Fig. 1), indicating that rearrangements occurred in these chromosomes between the two species. To compare the chromosome structures and delineate the chromosomal rearrangements that occurred in chromosomes 6–8 between V. salvator macromaculatus and V. exanthematicus (VEX), we mapped nine genes to VEX6, three genes to VEX7, and five genes to VEX8, and constructed a comparative cytogenetic map of VSA and VEX (Figs. 5 and 6).
Chromosomal locations of cDNA fragments of functional genes in a V. exanthematicus male. The ADAM12 gene was located on chromosome 6 (a), ENPP2 on chromosome 7q (c), ZNF326 on chromosome 8q (e), and RPN2 on chromosome 8q (g). b, d, f, h Hoechst-stained patterns of propidium iodide-stained metaphase spreads are shown in (a), (c), (e), and (g), respectively. Arrows indicate hybridization signals. Scale bars represent 5 μm
Discussion
Comparison of conserved synteny of V. salvator macromaculatus with other Toxicofera species and the chicken
Comparison of syntenies and gene orders of V. salvator macromaculatus with those of the chicken and two squamate reptiles (L. reevesii rubritaeniata and E. quadrivirgata) revealed that 11 syntenic groups of chicken macrochromosomes (GGA1p, 1q, 2p, 2q, 3, 4q, 5, 6, 7, 8, Z) were highly conserved in eight macrochromosomes and/or chromosome arms of V. salvator macromaculatus (VSA1p, 2, 3, 4q, 5, 6q, 7q, and 8p), as well as L. reevesii rubritaeniata and E. quadrivirgata (Matsubara et al. 2006, 2012; Srikulnath et al. 2009b; Fig. 7). Genes that mapped to microchromosomes in V. salvator macromaculatus localized to chicken microchromosomes or GGA4p, whereas homologous segments of 19 chicken microchromosome-linked genes localized to VSA1q, 2p, 4p, 6q, 7p, and 8q (Table 1). Similarly, all genes that localized to L. reevesii rubritaeniata and E. quadrivirgata microchromosomes were also located on chicken microchromosomes (Matsubara et al. 2006, 2012, unpublished data; Srikulnath et al. 2009b). These results indicate that approximately half of chicken microchromosomes showed homology with macrochromosomes in the three Toxicofera species.
Comparative cytogenetic maps of macrochromosomes among V. salvator macromaculatus, L. reevesii rubritaeniata, and E. quadrivirgata, constructed with 63 functional genes. The chromosome map of L. reevesii rubritaeniata (LRE) was taken from Srikulnath et al. (2009b). The ideograms of E. quadrivirgata (EQU) macrochromosomes were taken from Matsuda et al. (2005), and chromosomal locations of the genes from Matsubara et al. (2006, 2012). V. salvator macromaculatus (VSA) 3, VSA6, VSA7, EQU5, EQU7, and EQUZ are inverted to facilitate comparison
Comparing the chromosome maps of L. reevesii rubritaeniata (LRE) and E. quadrivirgata (EQU) with that of V. salvator macromaculatus, we deduced the process of karyotype reorganization among these species by the most parsimonious explanation of chromosomal rearrangements. VSA5 was homologous to LRE3q and EQU4, and VSA6 to LRE3p and EQU5. The occurrence of VSA5 and VSA6 or EQU4 and EQU5 could be explained by centric fission of an ancestral bi-armed macrochromosome homologous to LRE3 (Figs. 7 and 8a). Alternatively, LRE3 resulted from centric fusion between the acrocentric proto-VSA6 and VSA5, or between the acrocentric proto-EQU4 and EQU5. VSA6 might have resulted from pericentric inversion or centromere repositioning in the acrocentric proto-VSA6, and EQU4 from centromere repositioning in the acrocentric proto-EQU4. Another plausible explanation is that LRE3 derived from the tandem fusion of two ancestral chromosomes, which remain as VSA5 and VSA6 and EQU4 and EQU5, followed by inactivation of one of the two centromeres on the derived dicentric chromosome (Fig. 8b).
Schematic representation of the process of chromosomal rearrangements that occurred in V. salvator macromaculatus chromosomes 3, 5, and 6 (VSA3, VSA5, and VSA6), L. reevesii rubritaeniata chromosomes 3 and 5 (LRE3 and LRE5), and E. quadrivirgata chromosomes 4–7 (EQU4–EQU7). VSA3, VSA6, EQU5, and EQU7 are inverted to facilitate comparison. Chromosomal locations of the genes are shown to the right of chromosomes by arrowheads. The syntenies and gene orders of LRE3p, LRE3q, LRE5p, and LRE5q, their homologous chromosomes, and chromosome arms and/or segments are shown in red, green, yellow, and blue, respectively. a The diagram schematically summarizes the occurrence of LRE3 and VSA5, VSA6, EQU4, and EQU5 by two different processes of chromosomal rearrangements. One is a centric fission event of the bi-armed, followed by subsequent centromere repositioning in acrocentric proto-EQU4, and pericentric inversion or centromere repositioning in acrocentric proto-VSA6. The alternative process is that LRE3 was derived from centric fusion between VSA5 and acrocentric proto-VSA6, or between acrocentric proto-EQU4 and EQU5. The bi-armed VSA6 and EQU4 probably resulted from the same process. Arrows indicate the directions of chromosomal rearrangement. The bidirectional arrow indicates the region where centromere repositioning or pericentric inversion might have occurred. b The diagram schematically summarizes the tandem fusion event between VSA5 and VSA6, or EQU4 and EQU5, followed by inactivation of one of two centromeres on the derived dicentric chromosomes, leading to LRE3. Large arrows indicate the direction of chromosomal rearrangements. Small arrows indicate centromeres that might have been inactivated in LRE3 after the tandem fusions. c Two different processes for the occurrence of LRE5, VSA3, EQU6, and EQU7. In the first, centric fission occurred in VSA3 after a small pericentric inversion, followed by pericentric inversion or centromere repositioning in the fission-derived acrocentric proto-EQU6 and proto-EQU7, leading to the bi-armed EQU6 and EQU7 (Srikulnath et al. 2009b). Alternatively, centric fusion occurred between acrocentric proto-EQU6 and proto-EQU7, followed by pericentric inversion in VSA3, leading to LRE5
EXOC1 and UCHL1 near the centromere of LRE5p localized separately to VSA3q and VSA3p, respectively, and EXOC1 to EQU7p, and UCHL1 to EQU6p (Fig. 8c; Matsubara et al. 2006, 2012; Srikulnath et al. 2009b; this study). This implies that a small pericentric inversion occurred in VSA3, followed by centric fission in the lineage of E. quadrivirgata. The present forms of EQU6 and EQU7 might have resulted from pericentric inversion or centromere repositioning in the fission-derived acrocentric proto-EQU6 and proto-EQU7. Also plausible is that VSA3 resulted from centric fusion of the acrocentric proto-EQU6 and proto-EQU7, followed by pericentric inversion, leading to LRE5.
VSA7 corresponded to LRE4p and EQU3p, and VSA8 to LRE4q and EQU3q. VSA7 and VSA8 might have derived from centric fission of an ancestral bi-armed macrochromosome homologous to LRE4 and EQU3, followed by pericentric inversion or centromere repositioning in the acrocentric proto-VSA7 and proto-VSA8 (Fig. 9). Another plausible mechanism is centric fusion of the acrocentric proto-VSA7 and proto-VSA8, leading to LRE4 and EQU3.
Schematic representation of the process of chromosomal rearrangements that occurred in V. salvator macromaculatus chromosomes 7 and 8 (VSA7 and VSA8), L. reevesii rubritaeniata chromosome 4 (LRE4), and E. quadrivirgata chromosome 3 (EQU3). VSA7 is inverted to facilitate comparison. The syntenies and gene orders of LRE4p and LRE4q and their homologous chromosomes are shown in orange and violet, respectively. The diagram schematically summarizes the centric fission event of EQU3 or LRE4, followed by pericentric inversion or centromere repositioning in acrocentric proto-VSA7 and proto-VSA8. Alternatively, centric fusion of acrocentric proto-VSA7 and proto-VSA8 occurred, leading to the bi-armed EQU3 or LRE4. The bi-armed VSA7 and VSA8 probably resulted from the same process. Arrows indicate the direction of chromosomal rearrangements
VSA1 was homologous to LRE2 and EQU2, VSA2 to LRE1 and EQU1, and VSA4 to LRE6 and EQUZ (Fig. 7). However, microchromosome-linked genes in V. salvator macromaculatus and L. reevesii rubritaeniata, TRIM37 and AMH (GGA19-linked gene homologs) and EEF2 (GGA28-linked gene homolog), localized to the distal ends of the short and long arms of EQU1, respectively (Fig. 7; Table 1). This result suggests that tandem fusion of microchromosomes to macrochromosomes occurred in both the p and q arms of EQU1 after this lineage diverged from the common ancestor of iguanian and anguimorph lizards. The chromosomal location of the GGA12-linked TKT gene in VSA1q differed from that in LRE2q and EQU2q: TKT localized to the proximal regions of LRE2q and EQU2q, although homology with GGA12 was found in the distal regions of chromosome 2q of E. quadrivirgata (Fig. 3) and A. carolinensis (Matsubara et al. 2006, 2012; Alföldi et al. 2011). The order of SOX9 and RUFY1 on LRE2q, EQU2q, and VSA1q did not differ between the three species, suggesting that at least two paracentric inversions occurred sequentially in the ancestral chromosome of LRE2 and EQU2: a large paracentric inversion that first occurred at the breakpoints near the centromere and within the region homologous to GGA12 in the q arm of chromosome 2, followed by a paracentric inversion in the region containing SOX9 and RUFY1. High-resolution gene mapping in this chromosomal region is necessary to confirm this possibility.
Comparison of chromosome structures in V. salvator macromaculatus and V. exanthematicus
The chromosome number of V. salvator macromaculatus and V. exanthematicus was shown to be 2n = 40, which comprises 8 pairs of macrochromosomes and 12 pairs of microchromosomes. This karyotypic feature is conserved throughout the genus Varanus (King and King 1975; Chaiprasertsri et al. 2013). The 18S–28S rRNA genes localized to the secondary constriction in the proximal region of V. salvator macromaculatus and V. exanthematicus chromosome 1p. These results were the same as that of V. acanthurus, in which the same chromosomal region was stained by Ag-NOR staining (King et al. 1982). According to King and King (1975), the karyotypes of varanid lizards can be classified into six groups, based on the morphology of macrochromosomes. The salvator group, containing V. salvator macromaculatus, was located at the basal position of the karyotypic phylogeny of varanid lizards, and V. exanthematicus (gouldii group) was positioned at the terminus of the tree. However, this karyological relationship was not in accordance with previously reported molecular phylogeny (Ast 2001), which indicated that V. exanthematicus was located at the basal position of the tree. King and King (1975) suggested that chromosomes tended to change from bi-armed to acrocentric by pericentric inversions in varanid lizards during their evolution. In this study, the hybridization signal of interstitial telomeric sites that appear to be remnants of chromosomal rearrangements such as fusion or inversion was not found in any chromosomes of the two varanid lizards, but Hoechst-stained karyotypes of V. salvator macromaculatus and V. exanthematicus revealed morphologic differences in chromosomes 6–8. The chromosomal rearrangements that occurred in chromosomes 6–8 between VSA and V. exanthematicus were then identified by comparative mapping with 17 functional genes. Although we could not determine the direction of chromosome reorganization, VEX6 and VEX8 seem to retain the acrocentric proto-VSA6 and proto-VSA8 (Figs. 8 and 9). The syntenies and gene orders of three chromosomes were highly conserved between the two species, whereas HDAC3 on VSA7p localized to VEX7q and ZNF326 on VSA8p to VEX8q, and all genes on the long arm of submetacentric VSA6 localized to acrocentric VEX6 (Fig. 6). The submetacentric VSA6 probably resulted from at least two pericentric inversions: a large pericentric inversion in the region containing ADAM12 and PSAP in VEX6, followed by an additional pericentric inversion. A pericentric inversion probably occurred between the submetacentric VSA7 and VEX7, and the morphologic difference between VSA8 and VEX8 could derive from a pericentric inversion or centromere repositioning. More comparative chromosome mapping data are required to resolve these issues.
Comparison of the cytogenetic maps of V. salvator macromaculatus, L. reevesii rubritaeniata, and E. quadrivirgata, and of V. salvator macromaculatus and V. exanthematicus, in this study revealed patterns of karyotype reorganization in the Toxicofera clade, and also in the lineage of varanid lizards. However, comparative gene mapping for more lacertilian species—especially the Gekkota and Lacertidae, which have karyotypes with few or no microchromosomes—is necessary for precise delineation of the process of karyotype evolution in Squamata. A greater understanding of the chromosome segments conserved among reptiles, and between reptiles and birds, will provide insight into the phylogenetic hierarchy of genome evolution in amniotes.
Abbreviations
- BLAST:
-
Basic Local Alignment Search Tool
- cDNA:
-
Complementary DNA
- EQU:
-
Elaphe quadrivirgata
- FISH:
-
Fluorescence in situ hybridization
- GGA:
-
Gallus gallus
- LRE:
-
Leiolepis reevesii rubritaeniata
- MYA:
-
Million years ago
- rRNA:
-
Ribosomal RNA
- VSA:
-
Varanus salvator macromaculatus
References
Alföldi J, Di Palma F, Grabherr M et al (2011) The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 477:587–591
Amer SAM, Kumazawa Y (2008) Timing of a mtDNA gene rearrangement and intercontinental dispersal of varanid lizards. Gene Genet Syst 83:275–280
Ast JC (2001) Mitochondrial DNA evidence and evolution in Varanoidea (Squamata). Cladistics 17:211–226
Bennett D, Thakoordyal R (2003) The Savannah Monitor Lizard: the truth about Varanus exanthematicus. Viper Press, UK, pp 1–83
Chaiprasertsri N, Uno Y, Peyachoknagul S et al. (2013) Highly species-specific centromeric repetitive DNA sequences in lizards: molecular cytogenetic characterization of a novel family of satellite DNA sequences isolated from the water monitor lizard (Varanus salvator macromaculatus, Platynota). J Hered 104:798–806
Glor RE, Laport RG (2012) Are subspecies of Anolis lizards that differ in dewlap color and pattern also genetically distinct? A mitochondrial analysis. Mol Phylogenet Evol 64:255–260
International Chicken Genome Sequencing Consortium (ICGSC) (2004) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695–716
King M, King D (1975) Chromosomal evolution in the lizard genus Varanus (Reptilia). Aust J Biol Sci 28:89–108
King M, Mengden GA, King D (1982) A pericentric-inversion polymorphism and a ZZ/ZW sex-chromosome system in Varanus acanthurus Boulenger analyzed by G- and C-banding and Ag staining. Genetica 58:39–45
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Kumazawa Y, Endo H (2004) Mitochondrial genome of the Komodo dragon: efficient sequencing method with reptile-oriented primers and novel gene rearrangements. DNA Res 11:115–125
Matsubara K, Tarui H, Toriba M et al (2006) Evidence for different origin of sex chromosomes in snakes, birds, and mammals and step-wise differentiation of snake sex chromosomes. Proc Natl Acad Sci U S A 103:18190–18195
Matsubara K, Kuraku S, Tarui H et al (2012) Intra-genomic GC heterogeneity in sauropsids: evolutionary insights from cDNA mapping and GC3 profiling in snake. BMC Genomics 13:604
Matsuda Y, Chapman VM (1995) Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis 16:261–272
Matsuda Y, Nishida-Umehara C, Tarui H et al (2005) Highly conserved linkage homology between birds and turtles: bird and turtle chromosomes are precise counterparts of each other. Chromosome Res 13:601–615
Nishida-Umehara C, Tsuda Y, Ishijima J, Ando J, Fujiwara A, Matsuda Y, Griffin DK (2007) The molecular basis of chromosome orthologies and sex chromosomal differentiation in palaeognathous birds. Chromosome Res 15:721–734
Olmo E, Signorino G (2005) Chromorep: a reptile chromosomes database. Internet references. Available from http://chromorep.univpm.it. Accessed 06/04/2013
Shedlock AM, Edwards SV (2009) Amniota. In: Kumar S, Hedges SB (eds) The timetree of life. Oxford University Press, New York, pp 375–379
Srikulnath K, Matsubara K, Uno Y et al (2009a) Karyological characterization of the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Squamata) by molecular cytogenetic approach. Cytogenet Genome Res 125:213–223
Srikulnath K, Nishida C, Matsubara K et al (2009b) Karyotypic evolution in squamate reptiles: comparative gene mapping revealed highly conserved linkage homology between the butterfly lizard (Leiolepis reevesii rubritaeniata, Agamidae, Lacertilia) and the Japanese four-striped rat snake (Elaphe quadrivirgata, Colubridae, Serpentes). Chromosome Res 17:975–986
Srikulnath K, Matsubara K, Uno Y et al (2010) Genetic relationship of three butterfly lizard species (Leiolepis reveesii rubritaeniata, Leiolepis belliana belliana, Leiolepis boehmei, Agamidae Squamata) inferred from nuclear gene sequence analysis. Kasetsart J (Nat Sci) 44:424–435
Townsend TM, Larson A, Louis E, Macey JR (2004) Molecular phylogenetics of Squamata: the position of snakes, amphisbaenians, and dibamids, and the root of the squamate tree. Syst Biol 53:735–757
Uetz P (2013) The TIGR reptile database. The EMBL reptile database. Available from http://www.reptile-database.org/5/4/2013
Uno Y, Nishida C, Tarui H et al (2012) Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS ONE 7:e53027
Vidal N, Hedges SB (2005) The phylogeny of squamate reptiles (lizards, snakes, and amphisbaenians) inferred from nine nuclear protein-coding genes. C R Soc Biol 328:1000–1008
Wiens JJ, Hutter CR, Mulcahy DG et al (2012) Resolving the phylogeny of lizards and snakes (Squamata) with extensive sampling of genes and species. Biol Lett 8:1043–1046
Young MJ, O’Meally D, Sarre SD, Georges A, Ezaz T (2013) Molecular cytogenetic map of the central bearded dragon, Pogona vitticeps (Squamata: Agamidae). Chromosome Res 21:361–374
Acknowledgments
This work was financially supported by Grants-in-Aid for Scientific Research on Innovative Areas (no. 23113004) and Scientific Research (B) (no. 22370081) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Fengtang Yang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Table 1
Species and their accession numbers of the mitochondrial ND2 gene fragments used for molecular identification of V. exanthematicus(XLS 20 kb)
Supplementary Table 2
The cDNA fragments of L. reevesii rubritaeniata (LRE), G. hokouensis (GHO), L. agilis (LAG), and E. quadrivirgata (EQU) homologues of chicken genes, and nucleotide sequence identities between chicken and these squamate reptile cDNA fragments(XLS 39.5 kb)
Rights and permissions
About this article
Cite this article
Srikulnath, K., Uno, Y., Nishida, C. et al. Karyotype evolution in monitor lizards: cross-species chromosome mapping of cDNA reveals highly conserved synteny and gene order in the Toxicofera clade. Chromosome Res 21, 805–819 (2013). https://doi.org/10.1007/s10577-013-9398-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-013-9398-0