Abstract
Epigenetic alterations induced by ionizing radiation may contribute to radiation carcinogenesis. To detect relative accumulations or losses of constitutive post-translational histone modifications in chromatin regions surrounding DNA double-strand breaks (DSB), we developed a method based on ion microirradiation and correlation of the signal intensities after immunofluorescence detection of the histone modification in question and the DSB marker γ-H2AX. We observed after ionizing irradiation markers for transcriptional silencing, such as accumulation of H3K27me3 and loss of active RNA polymerase II, at chromatin regions labeled by γ-H2AX. Confocal microscopy of whole nuclei and of ultrathin nuclear sections revealed that the histone modification H3K4me3, which labels transcriptionally active regions, is underrepresented in γ-H2AX foci. While some exclusion of H3K4me3 is already evident at the earliest time amenable to this kind of analysis, the anti-correlation apparently increases with time after irradiation, suggesting an active removal process. Focal accumulation of the H3K4me3 demethylase, JARID1A, was observed at damaged regions inflicted by laser irradiation, suggesting involvement of this enzyme in the DNA damage response. Since no accumulation of the repressive mark H3K9me2 was found at damaged sites, we suggest that DSB-induced transcriptional silencing resembles polycomb-mediated silencing rather than heterochromatic silencing.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ionizing radiation is a well-characterized carcinogenic agent (Barcellos-Hoff and Nguyen 2009; Streffer 2010), but how radiation induces cancer is still a matter of debate. Since epigenetic alterations, such as altered DNA methylation status, histone modification patterns, or miRNA expression, are a hallmark of cancer cells, it has been hypothesized that irradiation may induce epigenetic alterations which then may be causally linked to the carcinogenic process (Mothersill and Seymour 2003; Loree et al. 2006; Kovalchuk and Baulch 2008).
Histone proteins (H2A, H2B, H3, H4, and variants thereof) form the nucleosomes around which the DNA molecule is wrapped (Campos and Reinberg 2009). Post-translational modifications of the histone proteins, such as methylation, acetylation, or phosphorylation, play a pivotal role for chromatin structure and transcriptional regulation by determining the charge of the nucleosome or by providing binding sites for or inhibiting binding of other proteins. Histone modification is also a major player in the cellular response to DNA damage (Altaf et al. 2007; van Attikum and Gasser 2009). Phosphatidyl inositol kinase-like kinases, such as ATM, ATR, or DNA-PK, phosphorylate the histone H2A variant, H2AX, at serine139 rapidly after induction of a DNA double-strand break (DSB), in a 1–2-Mbp large region surrounding the break. Phosphorylated H2AX is called γ-H2AX and its accumulations can be visualized by immunofluorescence as γ-H2AX foci. By providing a binding platform for mediator proteins which then recruit other proteins, phosphorylation of H2AX is thought to constitute an early and important step in the recruitment of signaling, mediator, and effector proteins to DSB sites.
DNA damage may not only induce the formation of specific histone modifications, but also alter the pattern of constitutive modifications. It is generally observed that irradiation of cells leads at early times after damage induction to a decondensation of chromatin in the vicinity of damaged sites (Kruhlak et al. 2006; Ziv et al. 2006), which may at least in part be explained by local histone acetylation (Murr et al. 2006; Stante et al. 2009). This effect is proposed to facilitate repair processes by improving the access of repair proteins to damaged sites (Sun et al. 2005; Murr et al. 2006). However, increasing evidence also suggests the installation of transcriptionally repressive chromatin patterns in the vicinity of damage sites. O’Hagan et al. (2008) observed the appearance of silencing histone modifications, including tri-methylation at lysine27 of histone H3 (H3K27me3), in the region surrounding an enzyme-mediated DSB. This was accompanied by the accumulation of several key proteins involved in establishing and maintaining transcriptional repression, including the polycomb group protein, EZH2, which is the histone methyltransferase responsible for the majority of cellular H3K27me3 marks. Chou et al. (2010) observed recruitment of repressive polycomb and NuRD complexes at damage sites induced by laser microirradiation. Recruitment of the NuRD components, CHD4 and MTA2, and a function in DNA damage response were also shown by others (Polo et al. 2010; Smeenk et al. 2010; Larsen et al. 2010). The installation of a repressive chromatin pattern correlates with a loss of active RNA polymerase II and of nascent RNA in the vicinity of the damage sites (Solovjeva et al. 2007; Shanbhag et al. 2010; Chou et al. 2010).
We questioned whether the accumulation of repressive complexes and histone modifications at damage sites is accompanied by a loss of active marks. In the case of transcriptional silencing associated with X chromosome inactivation or transgene inactivation, loss of tri-methylation at H3K4 is one of the earliest events detected (O’Neill et al. 2008). H3K4me3 is a mark for actively transcribed chromatin, which is preferentially found in the promoter and 5′ regions of genes (Barski et al. 2007). To clarify whether a loss of active histone modifications occurs in chromatin regions decorated by γ-H2AX, we planned to use immunofluorescence-based methods, as had already been done by others to study histone modifications in damaged chromatin (e.g., Solovjeva et al. 2007; Ayoub et al. 2008; Luijsterburg et al. 2009). Since antibodies detecting histone modifications generally produce a pan-nuclear staining pattern modulated by local alterations in chromatin state, the unequivocal detection of co-localization, and even more so, mutual exclusion, is not trivial. Nevertheless, most studies published so far have not relied on stringent quantification procedures.
These technical limitations have prompted a need for an improved method to detect the sites of relative accumulations or losses of histone modifications in damaged chromatin. We reasoned that targeted application of DNA damage achieved by microirradiation facilitates the detection of small alterations since irradiated regions can be differentiated from, for example, spurious accumulations of the damage markers. In recent years, laser microirradiation has been used to elucidate the characteristics of recruitment of many protein types at damaged chromatin (Grigaravicius et al. 2009). However, laser irradiation experiments in general suffer from the fact that (a) laser-induced damage types are poorly characterized, (b) high laser energy densities may lead to unspecific chromatin damage, and (c) concomitant treatment with photosensitizing agents may be necessary, which may affect histone modifications. To overcome these difficulties, we have built an ion microbeam to irradiate cells in predefined geometric patterns (Hauptner et al. 2004; Greubel et al. 2008). Using this tool, accumulation of a variety of DNA damage signaling and repair proteins, among them 53BP1, Mdc1, Rad51, phospho-ATM(Ser1981), has been shown by immunofluorescence or live cell imaging in the recent years (Hauptner et al. 2004; Greubel et al. 2008; Hable et al. 2009). Here, we use ion microirradiation in combination with state-of-the-art microscopy and image analysis methods. Besides common 3D widefield and confocal microscopy, we also introduced immunofluorescence imaging of cells after ultramicrotome slicing of cells into 150-nm thin sections, a method which is commonly used for electron microscopy (Solimando et al. 2009; Rouquette et al. 2009). With this method, the axial resolution of conventional fluorescence microscopy is greatly improved.
Various methods, e.g., Pearson’s correlation coefficient and Manders’ coefficient, are described in the literature to investigate co-localization of proteins (reviewed in Ronneberger et al. 2008). As already discussed by others (Costes et al. 2004; Bolte and Cordelières 2006; Ronneberger et al. 2008; French et al. 2008), these coefficients are suitable for the analysis of co-localization, but of limited usefulness in the analysis of anti-correlation (Barlow et al. 2010). Therefore, we here use intensity correlation analysis (Li et al. 2004) to visualize correlation and anti-correlation and Spearman’s coefficient (French et al. 2008) for quantification of correlation.
We here report that markers for transcriptional silencing, such as accumulation of H3K27me3 and loss of active RNA polymerase II, are evident after ionizing irradiation at chromatin regions labeled by γ-H2AX. We demonstrate an anti-correlation of H3K4me3 and γ-H2AX which increases with time after ionizing irradiation, suggesting an active loss of H3K4me3 at damage sites. Focal accumulation of the H3K4me3/me2 demethylase, JARID1A (Christensen et al. 2007), was observed at damaged regions inflicted by laser irradiation, suggesting involvement of this enzyme in the DNA damage response. Since there was no accumulation of the repressive mark H3K9me2 at damaged sites, we conclude that DNA double-strand break-induced transcriptional silencing resembles polycomb-mediated silencing rather than heterochromatic silencing.
Materials and methods
Cell culture and irradiation
HeLa and U2OS cells were cultivated (37°C, 5% CO2) in RPMI 1640 medium, and hTERT-immortalized BJ1 primary fibroblasts were cultivated in DMEM and 199 medium (4:1). Media were supplemented with 10% FCS, 100 U/ml penicillin, and 50 μg streptomycin. For ion irradiation, cells were seeded 24 h prior to irradiation on 6-μm mylar foil (pre-coated with Cell-TAK, BD Bioscience) in stainless steel chambers designed as described (Hauptner et al. 2004). At the time of irradiation, cells formed a confluent layer. Cells were irradiated with accelerated 55-MeV carbon ions at the ion microirradiation facility SNAKE (Hauptner et al. 2004) at the 14MV Munich tandem accelerator. With this technique, the ions were applied in a linear pattern of 1 μm × 5 μm in a perpendicular angle to the cell layer which results in a dose of 10 Gy. After irradiation at room temperature, the medium was replaced with fresh, pre-warmed medium and cells were incubated (37°C, 5% CO2) for different periods of time, as indicated. Samples denoted “5 min” were fixed immediately after irradiation (which takes about 7 min) and transfer (on ice) to the cell lab (which takes about 2 min); this is the earliest point of time amenable to analysis with this type of experimental setup. Similarly, samples denoted 20 min were fixed 15 min after having been returned to the incubator, i.e., about 17–24 min after irradiation of the individual cells, depending on their position on the cell container.
Since beam time at SNAKE is limited, some experiments were performed after α-irradiation with a 241Americium source located at the Institute of Radiation Biology of the Helmholtz Center Munich. For irradiation, HeLa cells were grown on coated mylar foil as described above. Each sample was irradiated for 4 min, which results in a dose of 0.4 Gy and on average about four particle transversals per cell nucleus. Samples denoted “2 min” were fixed immediately after irradiation.
For laser microirradiation, cells were seeded 24 h before irradiation in 7-mm MaTek chambers or ibidi μ-Dish containing a grid for relocation. Irradiation was performed with an UltraView Vox spinning disc microscope (PerkinElmer) equipped with a 63×/1.4 Plan-Apochromat oil objective and Volocity software 5.3. Irradiation was performed at 37°C and 5% CO2. For irradiation, RPMI medium was replaced with phenol-red free DMEM (10% FCS) medium containing 25 mM HEPES. Pre-selected areas of 1-μm diameter (spot irradiation) or 18-μm × 1-μm rectangles (line irradiation) were irradiated with a 405-nm diode laser in the FRAP preview mode of the Volocity software by repeated scanning (3,000 or 800 iterations, respectively).
Antibodies and immunofluorescence detection
Cells grown on a 6-μm mylar foil were fixed for 15 min in 2% para-formaldehyde. Immunofluorescence staining was performed as described previously (Hauptner et al. 2004). Briefly, following fixation, cells were washed with PBS; permeabilized with PBS, 0.1% Triton-X-100; and blocked with PBS containing 1% BSA and 0.15% glycine. The irradiated area was covered by a mixture of primary antibodies and detected with appropriate secondary antibodies, which were labeled with anti mouse IgG-Alexa488 (Invitrogen; A11029) or anti-rabbit IgG Alexa488 (Invitrogen; A11034) or with mouse anti-Cy3 F(ab)2 (Jackson Immunoresearch; 115-165-072) or with anti-rabbit Cy3 IgG (Amersham; PA43004). Primary antibodies used for immunofluorescence were mouse anti-γ-H2AX (Millipore; 05-636), rabbit anti-H3K4me1, H3K4me2, H3K4me3 (Abcam; ab3825, ab7766, ab8580 and Cell Signaling; 9727), rabbit anti-H3K9me2 (Millipore; 04-768), rabbit anti-H3K27me3 (Millipore; 07-449), and rabbit anti-JARID1A/RBP2 (Cell Signaling; 3878). Rabbit anti-JARID1A/RBP2, recognizing the large JARID1A isoform (Beshiri et al. 2010), was a kind gift from E. Benevolenskaya. Monoclonal antibodies recognizing RNA polymerase II phosphorylated either at Ser5 (IgG2a) or Ser2 (IgG1) and mouse monoclonal antibody Pol3/3 recognizing an epitope outside the CTD were kindly provided by D. Eick and were generated as described in Chapman et al. (2007). All primary antibodies were diluted in PBS containing 1% BSA and 0.15% glycine. The DNA was counterstained with DAPI. Cells were mounted in Vectashield for microscopy. H3K4me3 (ab8580) and H3K9me2 (Millipore 04-768) antibodies were tested for specificity with competing peptides in an immunofluorescence assay as described by Zinner et al. (2007). Results are shown in Fig. S9.
Ultrathin sections
Following immunodetection and DNA counterstaining with DAPI, the cell monolayer attached to the mylar foil was prepared for ultrathin sectioning. Cells were dehydrated by several washing steps in an increasing concentration of ethanol (twice with ice-cold 30% and 50% ethanol for 5 min each, twice with ice-cold 70% and 90% ethanol for 10 min each, and finally three times with 100% ethanol for 10 min each). Subsequently, cells were incubated in liquid resin (LR White) for 30 min at room temperature. Resin was replaced by fresh resin and cells were incubated overnight at 4°C. Next, the foil with the attached cells was placed on a resin-filled capsule and incubated for 48 h at 60°C to allow polymerization. Prior to sectioning, the foil was removed. Embedded cells were cut into ultrathin sections of approximately 150-nm thickness, using the Ultracut UCT ultramicrotome (Leica) and a diamond knife. Sections were placed on cover slips and embedded in anti-fade mounting solution (Vectashield) for microscopy.
Image acquisition
Image acquisition was either performed with an inverse epifluorescence microscope (Zeiss AxioObserver Z1, Germany) using a Zeiss Plan Apochromat 63×/1.4 glycerin objective, the software AxioVision 4.6 and a AxioCam Mrm camera (Zeiss), or with a confocal laser scanning microscope (SP5 DMI 6000 CS, Leica, Germany) equipped with an 63×/1.4 Plan Apochromat oil objective and the software Leica Application Suite 1.7.0. Acquisition was performed at room temperature. Scanning was done with a pixel size of 50 nm × 50 nm. In confocal microscopy, the axial distance between the optical sections was 200 nm. Scanning of optical sections was performed sequentially for the different fluorochromes. Images of the ultrathin sections were collected with a x,y-pixel size of 50 nm × 50 nm. Z-stacks of epifluorescence images were collected sequentially with 250-nm distance between optical sections.
Image processing
All Z-stack images were deconvolved with Huygens deconvolution software (Scientific Volume Imaging). The confocal images were corrected for z-shift. 2D images of ultrathin sections were not deconvolved. All images were further processed using the software ImageJ 1.37c (www.uhnresearch.ca/wcif). For correlation analysis, the images were processed according to the instructions for the ImageJ-based ICA (intensity correlation analysis) module (Khanna et al. 2006), with some variations described in the following: Shortly, for analysis, the segmented cell nucleus was cut out from the whole image and about three to seven mid-sections of the Z-stack with clear staining were selected. Gray values in each channel were normalized to 8 bit. For background subtraction, background was measured in areas where no true signals were expected, e.g., nucleoli or outside irradiated regions, and mean intensity of background signal plus 1× standard deviation across the measured area was subtracted from the whole image. After background subtraction and prior to correlation analysis, gray values were again normalized to 8 bit. Intensity correlation analysis was performed for pixel pairs of red (Cy3) and green (Alexa488) channel for all selected optical sections (three to seven slices). Only pixels which are neither zero in red nor in green channel are included in the analysis. For illustration, representative single slices from mid-sections were selected, and brightness and contrast were enhanced for the whole image where necessary. Images were assembled with Photoshop 7.0.1.
Statistical analysis
Maps of product of the differences from the mean (PDM) were used to visualize the co-localization of proteins via the correlation of intensities (Li et al. 2004). The PDM is calculated from (A − a)(B − b), where A and B represent the pixel pair intensities for two channels, and a and b are the respective means of pixel intensities for the cell analyzed in the two channels. PDM analysis was performed by using an automated graphic plug-in running with ImageJ 1.37c software (www.uhnresearch.ca/wcif). Three to seven mid-sections of a segmented cell nucleus were analyzed.
To quantify the bias introduced by the intensity correlation quotient (ICQ), we used a bootstrap technique to compute the value of ICQ under independence for a given distribution of intensities for both channels (Efron and Tibshirani 1993). For this, each pixel intensity of one channel was paired with a random pixel intensity of the other channel and the ICQ was computed. This was reiterated 1,000 times and the mean value ICQ0 was computed as the value of ICQ under assumption of no correlation. The ICQ0 value is typically not equal to zero. From this, a corrected ICQ value ICQ* = ICQ − ICQ0 can be calculated. To statistically evaluate correlation of intensities over an image, Spearman’s rank correlation coefficient ρ was used as proposed by French et al. (2008). For this, the intensities of each channel were ranked. Spearman’s coefficient is defined as ρ = 1−6 Σd i 2 / (n(n 2 − 1)), where d i is the difference of the ranks of both channels in voxel i and n is the number of foreground pixel. To test for correlation, p values for a test on the null hypothesis ρ = 0 were computed. To compare images at different points of time after radiation, we used a Mann–Whitney U test on the rank correlation coefficients to test differences in co-localization between groups. All statistical computations were performed in the statistical language R, version 2.11.1 (R Development Core Team 2010).
Immunoblotting and quantitative Western analysis
HeLa cells were grown in cell culture flasks and X-irradiated at room temperature with 0.5 or 2.0 Gy, incubated at 37°C, 5% CO2 for the indicated periods of time, and proteins were extracted with RIPA-buffer (150 mM NaCl, 1% NP-40, 10 mM MDOC, 0.1% SDS, 50 mM Tris pH 8.0) on ice. After denaturation for 10 min at 104°C, the proteins were separated with Laemmli loading dye on 10% Bis-Tris NuPAGE-gels (Invitrogen). After immunoblotting, membranes were cut in halves, blocked with Roti-Block (Roth) and incubated with the primary antibodies mouse anti-tubulin-α (Labvision; MS-581-P0) or rabbit anti-H3K4me3 (Abcam; ab8580), respectively, in Roti-Block before detection with appropriate secondary antibodies m-HRP and r-HRP (Santa Cruz; sc2004 and sc2005, respectively). Blots were developed with Amersham ECL Advance (GE Healthcare). Chemiluminescence images were acquired with a CHEMISMART documentation system (peqlab, Vilber Lourmat) and the Chemi-Capt 5000 software. Quantitative analysis was realized with the Bio-1D software (Vilber Lourmat). In order to ascertain detectability of small differences between samples, only blots were evaluated where equal amounts of protein extracts had been loaded, as determined by equal intensity of the tubulin-α signals (max. 20% variation over all lanes). H3K4me3 signals were normalized with respect to the tubulin-α signals and then with respect to the unirradiated samples.
Results
Detection of altered histone modification patterns at DSB sites
To investigate local alterations of histone modifications at sites of DNA damage, we used microirradiation with 55-MeV carbon ions applied in defined patterns (horizontal distance between single ion hits, 1 μm; vertical distance between the “lines” thus formed, 5 μm). Because of the limited range of the ions in matter, the cells were grown on 6-μm mylar foil rather than glass or plastic slides. The histone modification of interest was visualized in fixed cells by indirect immunofluorescence, concomitantly with γ-H2AX, which was used to mark chromatin regions surrounding DSB sites. The secondary antibodies used to detect the two primary antibodies were labeled with Cy3 (red) or Alexa488 (green), respectively. To visualize the correlation of pixel pairs in both color channels, the PDM was determined as PDM = (A − a)(B − b), with A and B being the fluorescence intensity of the pixels in question in the respective channels, and a and b being the mean intensity of all pixels in the respective cell nucleus. In areas where the signals in both channels are positively correlated, both intensities should be higher or lower than their means, leading to a positive PDM value. If, however, they are anti-correlated, the PDM should be negative (Li et al. 2004; Ronneberger et al. 2008). The analysis is done for all pixels in all optic slices (three to seven mid-sections of the 3D microscopic image). For visualization in a PDM map, PDM values are color-coded for each pixel in a slice. In addition, a plot of fluorescence intensity vs. PDM is produced for all pixels of the selected optic slices in the 3D microscopic image for each channel. In our analysis, we are interested in histone modification patterns in the chromatin regions labeled by γ-H2AX. Therefore, we consider the PDM values ascribed to pixels with high-intensity γ-H2AX signal as most important. In the PDM plots, the relationship between pixel intensity and PDM value can easily be seen and it can be determined if it is preferentially positive, negative, or randomly distributed.
For quantification of the correlation between channels, Li et al. (2004) proposed the ICQ, which assumes a symmetric intensity distribution for both channels. As this is unrealistic for most microscopic images, we use Spearman’s rank correlation coefficient ρ, as proposed by French et al. (2008), for quantitative evaluations.
To establish intensity correlation analysis for our system, we verified positive correlation of H3K9me2 and DAPI intensity (reviewed in Shilatifard 2008). A strong positive correlation is evident from the scatter plot of pixel intensities in both channels against each other (Fig. S1A). As expected, PDM values are clearly positive as seen from a preponderance of pink regions in color-coded PDM maps and from the PDM plots, which show positive PDM values at high pixel intensities in both channels (Fig. S1A). As a well-characterized example for anti-correlation (Zinner et al. 2006), the distribution of DAPI and H3K4me3 was analyzed (Fig. S1B). In the PDM map, a great part of the nucleus is colored in green, indicating negative PDM values. The evaluation shows that even in the case of very prominent anti-correlation, the negative PDM values tend to be close to zero for high DAPI intensity pixels (Fig. S1B). This fact is easily explained by the asymmetric distribution of fluorescence intensities, where the mean intensities (which are indicated by the waistline in the PDM sandglass distributions) are rather low and thus the absolute value of (A − a) for A < a is small. Some pink staining in the PDM maps, especially at the edge of nucleoli, can be explained by the fact that both signal intensities are below the respective mean intensities in these areas, thus appearing to be positively correlated (Fig. S1B). In spite of these caveats, we conclude that by calculation and visual display of PDM values, both positive and negative correlations can robustly be determined for the regions of interests. The Spearman’s rank correlation coefficients determined for the images shown in Fig. S1A and B are ρ = 0.86 and ρ = −0.04, respectively, demonstrating that visual impression and quantitative evaluation agree.
Using the same methods, accumulation of the repressive histone modification, H3K27me3, at regions decorated by γ-H2AX after ion irradiation was verified (Fig. 1a), similar to what has been described after laser microirradiation (Chou et al. 2010). While early (on average 5 min; see “Materials and methods”) after damage induction regions with positive and negative correlation are present, after 2 h, a strong positive correlation is seen. The respective Spearman’s coefficients are ρ = 0.051 and ρ = 0.43. Next, we addressed the question whether loss of active RNA polymerase II is detectable at sites of ion-induced damage. After laser microirradiation, relative loss of the initiating form of RNA polymerase II (RNAPII) was demonstrated (Miller et al. 2010). Here, we extend this observation to ion-induced damage (Fig. 1b), where first indications of a relative loss of the initiating form of RNAPII, which contains phosphorylated serine5 in the CTD domain (Egloff and Murphy 2008), are evident at about 5 min after irradiation (ρ = −0.02) and a prominent anti-correlation is seen after 2 h (ρ = −0.14). Similarly, we observe a relative loss of RNAPII serine5-P at 2 h after irradiation with α-particles (Fig. S2A), whereas at the earliest point in time tested (on average 2–3 min, see “Materials and methods”), anti-correlation was not yet evident. Since others had reported a loss of the elongating form of RNAPII, which is phosphorylated at serine2 in the CTD region, at laser-induced damage (Chou et al. 2010), we also tested for this form after α-irradiation and observed a similar underrepresentation as in the case of RNAPII serine5-P (Fig. S2B). No underrepresentation of total RNAPII at damage sites 2 h after damage induction is evident after use of an antibody detecting an epitope outside the CTD domain (Fig. S2C), arguing against a major damage-induced degradation, as had been described for RNAPII blocked by UV damage (Brookes and Pombo 2009). We conclude that after ionizing, irradiation increased H3K27me3 and transcriptional repression is seen at damaged chromatin regions, similar to what was observed after damage induction by laser microirradiation.
a Accumulation of H3K27me3 at γ-H2AX foci and b exclusion of initiating RNA polymerase II serine5-P at γ-H2AX foci in HeLa cells about 5 min (left) and 2 h (right) after ion microirradiation. Shown in the upper rows are microscopic images of single slices after epifluorescence 3D microscopy. The middle rows show the respective red–green merges and the PDM maps, with pink pixels representing positive PDM values and green pixels representing negative PDM values. Plots of signal intensity in the respective channels vs. PDM and the respective intensity scatter plots are shown in the bottom rows. Spearman’s rank correlation coefficients of the cell shown are a 5 min ρ = 0.051, 2 h ρ = 0.43, b 5 min ρ = −0.02, 2 h ρ = −0.14. Scale bars are 5 μm
Euchromatin markers H3K4me3 and γ-H2AX are mutually exclusive
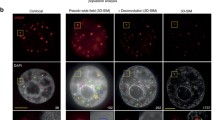
Given that loss of H3K4me3 is an early event in many silencing scenarios, we reasoned that transcriptional silencing in the vicinity of DSB may be associated with loss of H3K4me3. This hypothesis is corroborated by the fact that increased tri-methylation of H3K27 at damage sites most probably is due to recruitment of the polycomb repressive complex PRC2 (Chou et al. 2010), the activity of which is inhibited by active chromatin markers, including H3K4me3 (Schmitges et al. 2011). So far, distribution of H3K4me3 has not been studied at damaged chromatin sites in detail. Immunofluorescence with antibodies recognizing H3K4me3 gave heterogeneous, “speckle-like” signals in the cell nuclei (Fig. 2a, b). After ion microirradiation, we almost never detected an overlap or co-localization of H3K4me3 and γ-H2AX in 3D confocal microscopic images (Fig. 2a, b). Scatter plots show that high-intensity γ-H2AX pixels tend to have a low intensity in the H3K4me3 channel and vice versa, and this effect appears more pronounced at 2 h after irradiation than in cells fixed at 5 min after irradiation. Anti-correlation for high-intensity pixels is also evident from PDM plots and the PDM map (Fig. 2a, b).
Underrepresentation of H3K4me3 at γ-H2AX foci in HeLa cells. a–c Microscopic images. Shown in the upper rows are microscopic images (a, b) of single slices after confocal 3D microscopy and a 2D confocal microscopic image (c) of an ultrathin (150 nm) section (red channel γ-H2AX, green channel H3K4me3, blue channel DAPI). The middle rows show the respective red–green merges and the PDM maps, with pink pixels representing positive PDM values and green pixels representing negative PDM values. Plots of γ-H2AX or H3K4me3 signal intensity vs. PDM and the respective intensity scatter plots are shown in the bottom rows. At 5 min (a) and 2 h (b, c) after ion microirradiation in a line pattern (1 μm × 5 μm), PDM plots are skewed to negative values. Spearman’s rank correlation coefficient for c is ρ = –0.13. Scale bars are 5 μm. d Spearman’s rank correlation coefficients for γ-H2AX and H3K4me3 pixel intensities in HeLa cells at 5 min and 2 h after ion microirradiation. Indicated are the mean and standard error of the mean (SEM) of Spearman’s rank coefficients at 5 min (n = 15 cell nuclei) and 2 h (n = 19 cell nuclei) from confocal microscopy analyses. Both coefficients are significantly different from each other (Mann–Whitney U test; p = 0.0003) and significantly different from zero (p = 0.015 at 5 min; p = 7.63 × 10−6 at 2 h). e Decreased levels of H3K4me3 in whole cell protein extracts of HeLa cells after X-irradiation. Western blot images show levels of H3K4me3 in unirradiated control samples and in cells 1 and 3 h after X-irradiation with 0.5 and 2 Gy, respectively. Tubulin-α was used to verify equal loading. Graph represents levels of H3K4me3 after quantitative analysis of Western blots for protein amount normalized to the loading control and the control sample. Shown are mean values and standard deviation (SD) of two gels. Levels of H3K4me3 were significantly decreased in comparison to the respective unirradiated control at 1 and 3 h following irradiation with 2 Gy (Student’s t test; p = 0.033 at 1 h after 2 Gy; p = 0.0032 at 3 h after 2 Gy)
Because of the low axial microscopic resolution and the potential danger of contributions from out-of-focus fluorescence signals to co-localization analyses, we repeated the above-described analyses after ultrathin sectioning of cells. Cells attached to the mylar foil were microirradiated as described above. Following fixation and immunofluorescence staining, cells were embedded in resin and sequential ultrathin sections of ∼150-nm thickness were produced using an ultramicrotome. Sections were subsequently imaged by 2D confocal microscopy. Figure 2c shows a section of a cell nucleus stained for γ-H2AX, H3K4me3, and DNA (DAPI) 2 h after ion microirradiation. Compared to microscopic light-optical sections of normal samples, ultrathin sections show sharper and more defined H3K4me3 signals. This is due to their small thickness of approx. 150 nm which is far below the axial resolution limit of light microscopes of about 600 nm (Rouquette et al. 2010) and therefore reduces signal superposition. Since Faucher and Wellinger (2010) report a transient increase of H3K4me3 in a region adjacent to an endonuclease-mediated DSB in Saccharomyces cerevisiae at 10 to 40 min after induction of the endonuclease, in additional sets of experiments, we investigated H3K4me3 at about 20 min after ion irradiation. Evaluation of epifluorescence micrographs demonstrated also anti-correlation at this point of time in HeLa cells (Fig. S3). Anti-correlation of γ-H2AX and H3K4me3 is also seen in hTERT-immortalized BJ1 cells and U2OS osteosarcoma, at 20 min and 2 or 5 h after ion irradiation, respectively (Fig. S4A–D), and in HeLa cells at 2 min and 2 h after irradiation with alpha-particles (Fig. S4E, F).
Anti-correlation of H3K4me3 and γ-H2AX increases with time after irradiation
To determine whether the anti-correlation between H3K4me3 and γ-H2AX increases with time, a quantification of the correlation between both channels is needed. We rely on Spearman’s rank correlation coefficient ρ rather than the ICQ proposed by Li et al. (2004). In order to demonstrate the bias introduced by the ICQ, we used a bootstrap technique to compute the value of ICQ under independence for a given distribution of intensities for both channels (Efron and Tibshirani 1993). Figure S5A depicts a scatter plot of corrected and uncorrected ICQ values for all images of H3K4me3 analyzed and shows that for a large proportion of images the ICQ is positive, i.e., suggests correlation, where the corrected ICQ* is negative, i.e., suggests anti-correlation. While the Spearman’s coefficient ρ and ICQ disagree on the direction of correlation in many cases (Fig. S5B), ρ and the corrected ICQ* agree on the direction of correlation in nearly all cases (Fig. S5C), demonstrating the suitability of the Spearman’s rank correlation coefficient, which is easier to compute than the ICQ*, for quantitative evaluations.
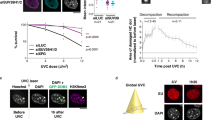
We determined the Spearman’s correlation coefficients for H3K4me3 and γ-H2AX pixel pairs at 5 min (n = 15 cells) and 2 h (n = 19 cells) after irradiation of HeLa cells and analysis of confocal images. They were ρ = −0.019 ± 0.0059 (SEM) and ρ = −0.061 ± 0.008 (SEM), respectively (Fig. 2d). For both points of time, the coefficients are significantly different from zero (p = 0.015 at 5 min and p = 7.63 × 10−6 at 2 h), indicating that exclusion is already evident at the earliest point of time amenable to this kind of analysis. Anti-correlation apparently increases with time after irradiation, since the coefficients at both points of time are significantly different from each other (Mann–Whitney U test; p = 0.0003), suggesting some active removal of H3K4me3 from the γ-H2AX-decorated chromatin after irradiation. In an independent set of experiments, a Spearman’s rank correlation coefficient of ρ = −0.059 ± 0.026 (SEM) was determined for HeLa cells (n = 10) fixed about 20 min after ion irradiation and evaluated by epifluorescence microscopy (Fig. S3). While not statistically significantly different from zero, probably due to the lower image quality of epifluorescence micrographs compared to confocal micrographs, this value ranks well between the values determined for fixation times of 5 min and 2 h, thus being consistent with the assumption of active removal of H3K4me3. This assumption was corroborated by quantitative Western analysis where a radiation-induced loss of H3K4me3 signal was observed in whole cell extracts at 1 and 3 h after X-irradiation with 2 Gy (Fig. 2e).
An increasingly negative correlation between γ-H2AX and H3K4me3 with increasing time after irradiation would be compatible with the activity of a demethylating enzyme. Several H3K4me3 demethylases are known to be present. The JARID1 family, which has four members (JARID1A–D), is able to demethylate H3K4me2 and H3K4me3 (Cloos et al. 2008). A H3K4me3-specific demethylase activity was shown for FBXL10 (KDM2B) (Frescas et al. 2007). We therefore tested the correlation between γ-H2AX and the different methylation states of H3K4 at 5 h after ion irradiation and observed anti-correlation with H3K4me2 similar to that seen with H3K4me3, while in the case of H3K4me1 at least for some high-intensity γ-H2AX pixels, a positive correlation was seen (Fig. S6). We conclude that the methylation patterns seen are compatible with the activity of a histone H3K4 demethylase of the JARID1 family.
The H3K4me3/me2 histone demethylase, JARID1A/RBP2 (KDM5A), was indirectly implicated in DSB response mechanisms (Hayakawa et al. 2007). In addition, it co-purifies with the EZH2-containing PRC2 complex and the PRC2 complex recruits JARID1A to target genes in mouse ES cells (Pasini et al. 2008). Recruitment of the PRC2 subunit EZH2 to the sites of laser-induced damage could be visualized by immunofluorescence (Chou et al. 2010). Although it has been suggested that JARID1A–PRC2 interactions may be transient, since JARID1A was not consistently detected as a component of the PRC2 complex (Simon and Kingston 2009), we set out to demonstrate recruitment of JARID1A by immunofluorescence. No recruitment of JARID1A or EZH2 was detectable after ion irradiation, even if the local dose was increased by applying ten ions to each point in the line pattern (data not shown). Therefore, we analyzed JARID1A recruitment after laser irradiation. Here, prominent γ-H2AX foci representing the irradiated line pattern of 18 μm × 1 μm size were detectable at 20, 40, and 90 min after irradiation (Fig. 3). Furthermore, we observed accumulation of JARID1A to these laser-induced γ-H2AX stripes at 20, 40, and 90 min after laser irradiation (Fig. 3). In contrast to the DNA damage response detected after ionizing irradiation, we did not detect γ-H2AX stripes at earlier times (5–10 min) after laser irradiation in our system and thus could not analyze earlier times for JARID1A recruitment.
Accumulation of JARID1A at laser-induced γ-H2AX sites after irradiation in a line pattern of 18 μm × 1 μm size at a 20, b 40, and c 90 min following irradiation. Microscopic images show single slices after 3D epifluorescence microscopy with γ-H2AX (red channel), JARID1A (green channel), and chromatin (DAPI; blue channel) in the upper rows, merges and PDM maps in the middle rows, and PDM plots of γ-H2AX and the respective enzyme, as well as the respective scatter plots in the bottom rows. An antibody against JARID1A was used which recognizes the large JARID1A isoform (Beshiri et al. 2010). JARID1A accumulates at laser-induced DNA damage at all times investigated. Evaluation with ICA confirms the positive correlation of JARID1A and γ-H2AX as indicated by the pink color in the PDM map for positive correlation, as well as positive PDM values shown in the PDM plots. Note that in the cell shown in b, a region with apparent anti-correlation in the middle of the track is due to reduced JARID1A signal in the nucleolus. Spearman’s rank correlation coefficients are a 20 min ρ = 0.29, b 40 min ρ = 0.16, c 90 min ρ = 0.16. Scale bars are 5 μm
The antibody used here for the detection of JARID1A at laser damage specifically recognizes a C terminal motif of JARID1A that is only present in the large isoform of JARID1A (Beshiri et al. 2010). This isoform contains the third plant homeo domain (PHD3), which binds H3K4me3 (Wang et al. 2009). We tested a second antibody (by Cell Signaling) and, in addition to some spot-like signals presumably representing constitutive JARID1A containing structures, observed small, but intense signals correlating with the γ-H2AX signal 30 min following laser spot irradiation with 1-μm diameter (Fig. S7A). At the same time after laser irradiation, a loss of H3K4me3 at laser-induced damage is clearly evident (Fig. S7B). For comparison, recruitment of EZH2 to laser damage is shown (Fig. S7C), which, under the conditions used, is comparable to the JARID1A recruitment.
Following exposure to ionizing radiation, H3K9me2 is partially overlapping with γ-H2AX foci
Since H3K9 methyltransferases are inhibited by tri-methylation at H3K4, a loss of H3K4me3 may result in hypermethylation at H3K9. For example, Binda et al. (2010) demonstrate that a loss of H3K4me2/me3 after knock-down of WDR82 or RBBP5, which are components of the SET1 methyltransferase complex, results in an increase of H3K9me2.
We tested the impact of ionizing irradiation on local distribution of H3K9me2 directly at γ-H2AX foci. Immunofluorescence with antibodies recognizing H3K9me2 yields a heterogeneous, “speckle”-like staining pattern (Fig. S8A, B). Radiation-induced γ-H2AX foci were accompanied by and partially overlapped with a number of H3K9me2 spots. Pixels with high γ-H2AX signal intensity show both positive and negative PDM values, hinting at a random distribution of H3K9me2 at γ-H2AX foci both at 20 min and 2 h after damage induction.
PDM mapping indicates co-localization of γ-H2AX and H3K9me2 in some small areas within the γ-H2AX foci (highlighted in pink in the PDM maps in Fig. S8A, B), but also larger areas within the γ-H2AX foci where the H3K9me2 signal is underrepresented (green in Fig. S8A, B). We performed a statistical evaluation of the correlation between H3K9me2 and γ-H2AX pixel intensities for four to five cells per data point. Mean values of Spearman’s coefficients calculated for H3K9me2 and γ-H2AX pixel pairs were ρ = 0.075 ± 0.031 (SEM) at 15 min (n = 5 cells) and ρ = 0.034 ± 0.014 (SEM) at 2 h (n = 4 cells) after ion microirradiation, respectively. Both values are not significantly different from each other (Mann–Whitney U test; p = 0.093) and also not significantly different from ρ = 0 (p = 0.13 for 15 min and p = 0.88 for 2 h), suggesting a random distribution of H3K9me2 at the γ-H2AX foci.
We repeated the above-described analyses after ultrathin sectioning of cells. Figure S8C shows a section of a cell nucleus stained for γ-H2AX, H3K9me2, and DNA (DAPI) 2 h after ion microirradiation. Again, H3K9me2 spots were found to overlap with some γ-H2AX foci or to locate at their borders, while other γ-H2AX foci were devoid of H3K9me2 signals. We conclude from 3D confocal microscopy of whole cells and from ultrathin sectioning of cells with subsequent 2D confocal microscopy that the post-translational histone modification H3K9me2 is not specifically enriched at the γ-H2AX foci, nor excluded from the γ-H2AX decorated chromatin.
Discussion
In the present work, we used elaborate microscopy techniques, including ultrathin sectioning of cells, combined with quantitative image analysis methods, to investigate how patterns of post-translational histone modifications alter in the vicinity of DSB. We clearly demonstrate that the topographies of the active histone mark H3K4me3 and γ-H2AX are negatively correlated. We observed this effect after different types of ionizing radiation and in different cell lines (HeLa, U2OS, and hTERT-immortalized BJ1). In contrast to our observations, Faucher and Wellinger (2010) report in S. cerevisiae a transient accumulation of H3K4me3 after ChIP analysis of a ∼700-bp fragment adjacent to an HO endonuclease-mediated DSB. It remains to be investigated whether the apparent discrepancy is due to differences in the organism or damage type tested, in the size of the region analyzed (about 1,000 bp vs. Mbp domain) or in the time frames investigated.
We observed a slight, but statistically significant loss of H3K4me3 also in Western analysis of whole cell extracts after X-irradiation. In our laboratory, this was independently reproduced in HeLa and other cell lines (B. Mazurek and A.A. Friedl, unpublished). The fact that Tjeertes et al. (2009) did not observe an alteration in H3K4me3 in their extensive screen for histone alterations after treatment of cells with hydroxyurea or phleomycin does not necessarily contradict our observations, as these authors did not rely on a thorough quantification of bands in the initial screening phase of their project and probably would have detected only larger alterations.
An extensive statistical analysis of the microscopic data obtained at 5 min and 2 h after ion irradiation suggests that the H3K4me3 exclusion effect increases with time after irradiation. Thus, the effect cannot entirely be explained by a model claiming that γ-H2AX formation does not occur in active promoter regions characterized by the presence of H3K4me3 and RNA polymerase II, as has been proposed after induction of restriction enzyme-mediated breaks and ChIP analysis (Iacovoni et al. 2010).
One possibility to explain increasing exclusion of H3K4me3 at the damage sites would be H3 eviction. However, the fact that patterns of H3K9me2 do not alter after irradiation argues against a general and large-scale H3 eviction, since “new” histones generally are not methylated (Loyola and Almouzni 2007). Our observations are thus compatible with the activity of a H3K4 demethylase in the vicinity of DSB sites. Since we observe not only a loss of H3K4me3, but also of H3K4me2, JHMD1B/FBXL10, which is able to demethylate H3K4me3, but not H3K4me2 (Frescas et al. 2007), is not a likely candidate. Among the members of the JARID1 family, which are able to demethylate di- and tri-methylated H3K4, JARID1A/RBP2 (KDM5A) appears most interesting because of a suggested involvement in the DSB response process (Hayakawa et al. 2007) and because it interacts with the PRC2 complex (Pasini et al. 2008). Indeed, we detect accumulation of JARID1A after laser microirradiation. The fact that we could detect recruitment of JARID1A or EZH2 after laser, but not after ion irradiation is not unprecedented, since also other chromatin/histone-modifying factors that are detectable at laser-induced damage, such as ACF1 (Sánchez-Molina et al. 2011), were not detectable at ion-induced damage in our hands (D. M. Seiler, S. Auer and A. A. Friedl, unpublished). Presumably, this means that these proteins are not foci-forming proteins in a sense that a high number of proteins accumulate in the vicinity of a single DNA lesion (as is, for example, the case with γ-H2AX, 53BP1, or Mdc1, just to name a few), but rather that accumulation is only detectable by microscopy if the local damage load is extremely high, as is often the case with laser-induced damage (Suzuki et al. 2011; Nagy and Soutoglou 2009). Similar discrepancies for protein accumulation at laser-induced vs. ion-induced damage have been reported by Splinter et al. (2010). Given the importance of risks associated with exposure to ionizing radiation, for example by medical applications and by natural sources, these discrepancies underline the importance of testing protein accumulation not only after laser treatment, but also after ionizing irradiation.
In spite of several attempts to knock-down JARID1A, using a variety of siRNAs that were successfully used by others (Benevolenskaya et al. 2005), we have so far not succeeded in the efficient inactivation of JARID1A. Therefore, at present, we cannot exclude a role for other H3K4me3/me2 demethylases in the removal of methyl marks in the vicinity of DSB. In this context, it is interesting that during revision of our manuscript, Nottke et al. (2011) reported that SPR-5, the Caenorhabditis elegans homolog of the H3K4me2-specific demethylase LSD1, has a role for repair of meiotic DSB, presumably by removing H3K4me2.
Our observations provide further evidence for the hypothesis that DSB-induced transcriptional repression resembles polycomb-mediated silencing rather than heterochromatic silencing (Tang and Greenberg 2010). In addition to H3K27 tri-methylation by the PRC2 core component EZH2, also H2A monoubiquitination by the PRC1 complex has recently been involved in transcriptional repression at DSB sites (Chagraoui et al. 2011; Ginjala et al. 2011). In addition, a loss of H3K56ac in response to DNA damage was described (Tjeertes et al. 2009; Yang et al. 2009; Miller et al. 2010), although controversial data exist (Das et al. 2009). Miller et al. (2010) demonstrate that histone deacetylases HDAC1 and HDAC2 are responsible for damage-associated H3K56 deacetylation. Since these histone deacetylases interact with the PRC2 complex via EED, it is reasonable to assume that H3K27 tri-methylation and H3K56 deacetylation occur in a concerted action. Demethylation of H3K4me3/me2 may be part of the same concerted action.
In contrast, DSB-induced alterations of histone modification patterns do not involve an increase of the repressive mark H3K9me2, at least not in the period of time investigated. Similarly, others investigated H3K9me3 patterns and did not detect DSB-induced alterations (Luijsterburg et al. 2009; Ayoub et al. 2008). The fact that the H3K9me3/me2-binding HP1 proteins do accumulate in the vicinity of DSB some time after damage induction (Ayoub et al. 2009; Luijsterburg et al. 2009; Zarebski et al. 2009) does not contradict this interpretation, since it has been shown that this accumulation occurs independently of H3K9me3/me2 binding (Luijsterburg et al. 2009; Baldeyron et al. 2011).
Since it is a hallmark of polycomb-mediated silencing that it persists through cell division (Sawarkar and Paro 2010) and since aberrant silencing may be associated with a carcinogenic development (reviewed in Chi et al. 2010), it will be important to elucidate how DSB-mediated silencing is reversed to reconstitute the gene expression status as it was before damage induction.
Abbreviations
- 2D:
-
two-dimensional
- 3D:
-
three-dimensional
- DSB:
-
double-strand break
- ICA:
-
intensity correlation analysis
- ICQ:
-
intensity correlation quotient
- MeV:
-
Mega electron volts
- PDM:
-
product of the differences from the mean
References
Altaf M, Saksouk N, Côté J (2007) Histone modifications in response to DNA damage. Mutat Res 618:81–90
Ayoub N, Jeyasekharan AD, Bernal JA, Venkitaraman AR (2008) HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature 453:682–686
Ayoub N, Jeyasekharan AD, Venkitaraman AR (2009) Mobilization and recruitment of HP1: a bimodal response to DNA breakage. Cell Cycle 8:2945–2950
Baldeyron C, Soria G, Roche D, Cook AJL, Almouzni G (2011) HP1alpha recruitment to DNA damage by p150CAF-1 promotes homologous recombination repair. J Cell Biol 193:81–95
Barcellos-Hoff MH, Nguyen DH (2009) Radiation carcinogenesis in context: how do irradiated tissues become tumors? Health Phys 97:446–457
Barlow AL, Macleod A, Noppen S, Sanderson J, Guérin CJ (2010) Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of Pearson’s correlation coefficient. Microsc Microanal 16:710–724
Barski A, Cuddapah S, Cui K et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG (2005) Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell 18:623–635
Beshiri ML, Islam A, DeWaal DC, Richter WF, Love J, Lopez-Bigas N, Benevolenskaya EV (2010) Genome-wide analysis using ChIP to identify isoform-specific gene targets. J Vis Exp 41:2101
Binda O, LeRoy G, Bua DJ, Garcia BA, Gozani O, Richard S (2010) Trimethylation of histone H3 lysine 4 impairs methylation of histone H3 lysine 9: regulation of lysine methyltransferases by physical interaction with their substrates. Epigenetics 5:767–775
Bolte S, Cordelières FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232
Brookes E, Pombo A (2009) Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 10:1213–1219
Campos EI, Reinberg D (2009) Histones: annotating chromatin. Annu Rev Genet 43:559–599
Chagraoui J, Hebert J, Girard S, Sauvageau G (2011) An anticlastogenic function for the Polycomb Group gene Bmi1. Proc Natl Acad Sci USA 108:5284–5289
Chapman RD, Heidemann M, Albert TK et al (2007) Transcribing RNA polymerase II is phosphorylated at CTD residue serine-7. Science 318:1780–1782
Chi P, Allis CD, Wang GG (2010) Covalent histone modifications—miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer 10:457–469
Chou DM, Adamson B, Dephoure NE et al (2010) A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc Natl Acad Sci USA 107:18475–18480
Christensen J, Agger K, Cloos PAC et al (2007) RBP2 belongs to a family of demethylases, specific for tri- and dimethylated lysine 4 on histone 3. Cell 128:1063–1076
Cloos PAC, Christensen J, Agger K, Helin K (2008) Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev 22:1115–1140
Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G, Lockett S (2004) Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86:3993–4003
Das C, Lucia MS, Hansen KC, Tyler JK (2009) CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459:113–117
Efron B, Tibshirani R (1993) An introduction to the bootstrap. Chapman & Hall, New York
Egloff S, Murphy S (2008) Cracking the RNA polymerase II CTD code. Trends Genet 24:280–288
Faucher D, Wellinger RJ (2010) Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet 6:e1001082
French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc 3:619–628
Frescas D, Guardavaccaro D, Bassermann F, Koyama-Nasu R, Pagano M (2007) JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450:309–313
Ginjala V, Nacerddine K, Kulkarni A et al (2011) BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol Cell Biol 31:1972–1982
Greubel C, Hable V, Drexler GA et al (2008) Competition effect in DNA damage response. Radiat Environ Biophys 47:423–429
Grigaravicius P, Greulich KO, Monajembashi S (2009) Laser microbeams and optical tweezers in ageing research. Chemphyschem 10:79–85
Hable V, Greubel C, Bergmaier A et al (2009) The live cell irradiation and observation setup at SNAKE. Nucl Instr and Meth in Phys Res B 267:2090–2097
Hauptner A, Dietzel S, Drexler GA et al (2004) Microirradiation of cells with energetic heavy ions. Radiat Environ Biophys 42:237–245
Hayakawa T, Ohtani Y, Hayakawa N et al (2007) RBP2 is an MRG15 complex component and down-regulates intragenic histone H3 lysine 4 methylation. Genes Cells 12:811–826
Iacovoni JS, Caron P, Lassadi I et al (2010) High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J 29:1446–1457
Khanna R, Li Q, Sun L, Collins TJ, Stanley EF (2006) N type Ca2+ channels and RIM scaffold protein covary at the presynaptic transmitter release face but are components of independent protein complexes. J Neurosci 140:1201–1208
Kovalchuk O, Baulch JE (2008) Epigenetic changes and nontargeted radiation effects—is there a link? Environ Mol Mutagen 49:16–25
Kruhlak MJ, Celeste A, Dellaire G et al (2006) Changes in chromatin structure and mobility in living cells at sites of DNA double-strand breaks. J Cell Biol 172:823–834
Larsen DH, Poinsignon C, Gudjonsson T et al (2010) The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. J Cell Biol 190:731–740
Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF (2004) A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci 24:4070–4081
Loree J, Koturbash I, Kutanzi K, Baker M, Pogribny I, Kovalchuk O (2006) Radiation-induced molecular changes in rat mammary tissue: possible implications for radiation-induced carcinogenesis. Int J Radiat Biol 82:805–815
Loyola A, Almouzni G (2007) Marking histone H3 variants: how, when and why? Trends Biochem Sci 32:425–433
Luijsterburg MS, Dinant C, Lans H et al (2009) Heterochromatin protein 1 is recruited to various types of DNA damage. J Cell Biol 185:577–586
Miller KM, Tjeertes JV, Coates J et al (2010) Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol 17:1144–1151
Mothersill C, Seymour C (2003) Radiation-induced bystander effects, carcinogenesis and models. Oncogene 22:7028–7033
Murr R, Loizou JI, Yang Y-G et al (2006) Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 8:91–99
Nagy Z, Soutoglou E (2009) DNA repair: easy to visualize, difficult to elucidate. Trends Cell Biol 19:617–629
Nottke AC, Beese-Sims SE, Pantalena LF, Reinke V, Shi Y, Colaiácovo MP (2011) SPR-5 is a histone H3K4 demethylase with a role in meiotic double-strand break repair. Proc Natl Acad Sci USA 108:12805–12810
O’Hagan HM, Mohammad HP, Baylin SB (2008) Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet 4(8):e1000155
O’Neill LP, Spotswood HT, Fernando M, Turner BM (2008) Differential loss of histone H3 isoforms mono-, di- and tri-methylated at lysine 4 during X-inactivation in female embryonic stem cells. Biol Chem 389:365–370
Pasini D, Hansen KH, Christensen J, Agger K, Cloos PAC, Helin K (2008) Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 22:1345–1355
Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP (2010) Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. EMBO J 29:3130–3139
R Development Core Team (2010) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. http://www.R-project.org
Ronneberger O, Baddeley D, Scheipl F et al (2008) Spatial quantitative analysis of fluorescently labeled nuclear structures: problems, methods, pitfalls. Chromosome Res 16:523–562
Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S (2009) Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: a novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res 17:801–810
Rouquette J, Cremer C, Cremer T, Fakan S (2010) Functional nuclear architecture studied by microscopy: present and future. Int Rev Cell Mol Biol 282:1–90
Sánchez-Molina S, Mortusewicz O, Bieber B, Auer S, Eckey M, Leonhardt H, Friedl AA, Becker PB (2011) Role for hACF1 in the G2/M damage checkpoint. Nucl Acids Res. doi:10.1093/nar/gkr435
Sawarkar R, Paro R (2010) Interpretation of developmental signaling at chromatin: the Polycomb perspective. Dev Cell 19:651–661
Schmitges FW, Prusty AB, Faty M et al (2011) Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell 42:330–341
Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA (2010) ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141:970–981
Shilatifard A (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20:341–348
Simon JA, Kingston RE (2009) Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 10:697–708
Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H (2010) The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J Cell Biol 190:741–749
Solimando L, Luijsterburg MS, Vecchio L, Vermeulen W, van Driel R, Fakan S (2009) Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J Cell Sci 122:83–91
Solovjeva LV, Svetlova MP, Chagin VO, Tomilin NV (2007) Inhibition of transcription at radiation-induced nuclear foci of phosphorylated histone H2AX in mammalian cells. Chromosome Res 15:787–797
Splinter J, Jakob B, Lang M, Yano K, Engelhardt J, Hell SW, Chen DJ, Durante M, Taucher-Scholz G (2010) Biological dose estimation of UVA laser microirradiation utilizing charged particle-induced protein foci. Mutagenesis 25:289–297
Stante M, Minopoli G, Passaro F, Raia M, Vecchio LD, Russo T (2009) Fe65 is required for Tip60-directed histone H4 acetylation at DNA strand breaks. Proc Natl Acad Sci USA 106:5093–5098
Streffer C (2010) Strong association between cancer and genomic instability. Radiat Environ Biophys 49:125–131
Sun Y, Jiang X, Chen S, Fernandes N, Price BD (2005) A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci USA 102:13182–13187
Suzuki K, Yamauchi M, Oka Y, Suzuki M, Yamashita S (2011) Creating localized DNA double-strand breaks with microirradiation. Nat Protoc 6:134–139
Tang J-B, Greenberg RA (2010) Connecting the dots: interplay between ubiquitylation and SUMOylation at DNA double strand breaks. Genes Cancer 1:787–796
Tjeertes JV, Miller K, Jackson SP (2009) Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J 28:1878–1889
van Attikum H, Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19:207–217
Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD (2009) Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459:847–851
Yang B, Zwaans BMM, Eckersdorff M, Lombard DB (2009) The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle 8:2662–2663
Zarebski M, Wiernasz E, Dobrucki JW (2009) Recruitment of heterochromatin protein 1 to DNA repair sites. Cytometry A 75:619–625
Zinner R, Albiez H, Walter J, Peters AHFM, Cremer T, Cremer M (2006) Histone lysine methylation patterns in human cell types are arranged in distinct three-dimensional nuclear zones. Histochem Cell Biol 125:3–19
Zinner R, Teller K, Versteeg R, Cremer T, Cremer M (2007) Biochemistry meets nuclear architecture: multicolor immuno-FISH for co-localization analysis of chromosome segments and differentially expressed gene loci with various histone methylations. Adv Enzyme Regul 47:223–241
Ziv Y, Bielopolski D, Galanty Y et al (2006) Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 8:870–876
Acknowledgments
We thank the staff of the Maier-Leibnitz-Laboratorium for operating the accelerator facility; K. Pfleghaar and K. Schneider for their help with laser microirradiation; B. Scholz for the help with RT-PCR; H. Hofmann, D. Wieland, and S. Lindemaier for excellent technical assistance, and S. Auer for managing the beam time. Anti-polymerase II Ser5-P and Ser2-P monoclonal antibodies as well as antibody Pol3/3 were a kind gift of D. Eick, anti-JARID1A a kind gift of E. Benevolenskaya, and anti-H3K27me3 a kind gift of T.E. Schmid. Parts of this work were supported by grants of the Bundesministerium für Bildung und Forschung (02S8457 and 03NUK007), the DFG Cluster of Excellence Munich Centre for Advanced Photonics, and the LMU innovative project Biomed-S.
Conflicts of interest
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Beth A. Sullivan.
Electronic supplementary material
Below is the link to the electronic supplementary material.
MOESM 1
(PDF 5231 kb)
Rights and permissions
About this article
Cite this article
Seiler, D.M., Rouquette, J., Schmid, V.J. et al. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res 19, 883–899 (2011). https://doi.org/10.1007/s10577-011-9244-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9244-1