Abstract
The emergence of genome-integrated molecular cytogenetic resources allows for comprehensive comparative analysis of gross karyotype architecture across related species. The identification of evolutionarily conserved chromosome segment (ECCS) boundaries provides deeper insight into the process of chromosome evolution associated with speciation. We evaluated the genome-wide distribution and relative orientation of ECCSs in three wild canid species with diverse karyotypes (red fox, Chinese raccoon dog, and gray fox). Chromosome-specific panels of dog genome-integrated bacterial artificial chromosome (BAC) clones spaced at ∼10-Mb intervals were used in fluorescence in situ hybridization analysis to construct integrated physical genome maps of these three species. Conserved evolutionary breakpoint regions (EBRs) shared between their karyotypes were refined across these and eight additional wild canid species using targeted BAC panels spaced at ∼1-Mb intervals. Our findings suggest that the EBRs associated with speciation in the Canidae are compatible with recent phylogenetic groupings and provide evidence that these breakpoints are also recurrently associated with spontaneous canine cancers. We identified several regions of domestic dog sequence that share homology with canid B chromosomes, including additional cancer-associated genes, suggesting that these supernumerary elements may represent more than inert passengers within the cell. We propose that the complex karyotype rearrangements associated with speciation of the Canidae reflect unstable chromosome regions described by the fragile breakage model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Comparative cytogenetic analysis offers a means to define regions of conserved synteny shared between genomes. Knowledge of the distribution of evolutionarily conserved chromosome segments (ECCSs) among a wide variety of species has provided key insights into karyotypic reorganization during speciation. The Canidae are believed to have undergone rapid speciation since they diverged from their last common ancestor approximately 10 Mya (Wayne 1993; Graphodatsky et al. 2008). During this time, it is thought that canid karyotypes altered via breakage–fusion events involving whole-arm ECCSs (Yang et al. 1999), which resulted in the extensive range in chromosome number and karyotypic morphology evident today among the 34 extant species of this family (Wayne 1993; Graphodatsky et al. 2008). Generally, it is agreed that the modern Canidae consists of four major groupings: fox-like canids, wolf-like canids, South American canids, and island foxes (Bardeleben et al. 2005; Lindblad-Toh et al. 2005). Early phylogenies represented species divergence from a common ancestor based on morphological and geographical features. More recently, phylogenetic trees have been constructed by DNA sequence analysis of various nuclear- and mitochondrial-encoded loci (Wayne 1993; Wayne and Ostrander 1999; Zrzavy and Ricankova 2004; Bardeleben et al. 2005; Lindblad-Toh et al. 2005; Grzes et al. 2009; Schröder et al. 2009), but these have led to a number of contradictory phylogenetic placements.
Conventional chromosome banding and molecular cytogenetic studies have enabled the construction of alternative phylogenetic trees for the Canidae, taking into account gross karyotypic changes that occurred during speciation (Wayne et al. 1987a, b, 1997; Nash et al. 2001; Graphodatsky et al. 2008). As with all wolf-like canids, the karyotype of the domestic dog (Canis familiaris, CFA) comprises 38 pairs of single-armed autosomal chromosomes and a pair of bi-armed sex chromosomes. In contrast, the more distantly related red fox (Vulpes vulpes, VVU) karyotype comprises 16 pairs of larger bi-armed (metacentric and submetacentric) autosomes (Wayne 1993) plus a variable number of supernumerary elements, referred to as B chromosomes. B chromosomes are also evident in Chinese raccoon dog (Nyctereutes procynoides procynoides, NPRp), Japanese raccoon dog (Nyctereutes procynoides viverrinus, NPRv), and maned wolf (Chrysocyon brachyurus, CBR; Mäkinen 1985; Mäkinen et al. 1986; Ward et al. 1987; Yang et al. 1999; Trifonov et al. 2002; Pieńkowska-Schelling et al. 2008) and are part of the normal chromosome complement of these species, but may vary in number across populations and even within individuals (Graphodatsky et al. 2000, 2008; Nie et al. 2003). B chromosomes are found in a diverse range of other species, including mammals, plants, and insects (Vujosević and Blagojević 2004). While most agree that they originate from A (standard) chromosomes (Camacho et al. 2000), the mechanism for B chromosome formation and persistence is unknown. Though they were originally thought to be largely inert, the proto-oncogene cKIT has been localized on B chromosomes of the red fox, and there is evidence that it may be transcribed and translated into functional KIT protein (Graphodatsky et al. 2005; Yudkin et al. 2007). Aside from these findings, however, the sequence composition and transcriptional functionality of canid B chromosomes has yet to be defined in detail, though we hypothesize that B chromosomes may contain additional cancer-associated sequences.
Karyotype evolution has long been studied by cytogenetic comparison of the constitutional chromosomes of species within and between phylogenetic groups. In a similar manner, cytogenetic evaluation has been crucial to understanding the complex chromosome changes that occur in cancer and congenital abnormalities. The fragile breakage model (FBM) was developed to explain the identification of shared chromosome breakpoints, evident in evolution and diseased karyotypes, especially mammalian cancers (Pevzner and Tesler 2003; Murphy et al. 2005; Peng et al. 2006; Alekseyev and Pevzner 2007, 2010; Alekseyev 2008).
The extensive variation in karyotype structure among canid species, coupled with expanding knowledge of genomic aberrations in spontaneous dog cancers, makes the Canidae ideally suited to the examination of the relationship between evolutionary and tumor-associated breakpoints.
Prior molecular cytogenetic studies of the Canidae used chromosome painting analysis to identify ECCSs within the karyotypes of non-domesticated canids (Yang et al. 1999; Nie et al. 2003; Graphodatsky et al. 2008). While providing a gross representation of the distribution of ECCSs, chromosome painting is restricted in resolution to 5–10 Mb and does not permit relative orientation of ECCSs. The development of a whole genome sequence assembly for the domestic dog (Lindblad-Toh et al. 2005), combined with the emergence of canine molecular cytogenetic techniques with increasing resolution, now enables a more comprehensive interrogation of ECCS to gain insight into chromosome evolution. In this study, we evaluated the genome-wide distribution and relative orientation of ECCSs in three wild canid species with diverse karyotypes—red fox, Chinese raccoon dog, and gray fox (Urocyon cinereogenteus, UCI)—using multicolor fluorescence in situ hybridization (FISH) analysis with bacterial artificial chromosome (BAC) clones spaced at ∼10-Mb intervals along each domestic dog chromosome (Thomas et al. 2007). Shared evolutionary breakpoint regions (EBRs) were investigated in detail across these and eight additional wild canid species using BAC clones spaced at ∼1-Mb intervals (Thomas et al. 2008). The distribution of dog FISH probe signals was evaluated to identify sequence conservation between the domestic dog genome and canid B chromosomes and to examine whether these supernumerary elements may harbor additional gene-coding regions. We discuss the findings from the present study in context with emerging data for recurrent DNA copy number aberrations within a range of spontaneous canine cancers to assess the relationship between evolutionary and tumor-associated chromosome breakpoints. Our findings suggest that the EBRs associated with speciation in the Canidae are compatible with recent phylogenetic groupings (Lindblad-Toh et al. 2005) and provide evidence that these breakpoints are also recurrently associated with spontaneous canine cancers. We identified several regions of domestic dog sequence that share homology with canid B chromosomes, including additional cancer-associated genes, suggesting that these supernumerary elements may represent more than inert passengers within the cell.
Materials and methods
Generation of chromosome preparations from canid species
Metaphase chromosome preparations for 11 wild canid species and the domestic dog were prepared either from mitogen stimulation of peripheral lymphocyte culture (Breen et al. 1999) or from cryopreserved cells. Cryopreserved cells were expanded in αMEM supplemented with 15% (v/v) fetal bovine serum (Cellgro, Manassas, VA), 2 mM l-analyl-l-glutamine (Glutamax™, Gibco, Carlsbad, CA), and 88 μg/mL Primocin™ (Invivogen, San Diego, CA). At 80% confluency; dividing cells were arrested at metaphase by exposure to colcemid (50 ng/mL) for 4 h and then harvested using routine hypotonic exposure and methanol/glacial acetic acid fixation. Fixed cells were dropped onto clean glass slides, air-dried, dehydrated, and stored at −80°C until required. Table 1 describes the species studied and the source of the cells examined.
Chromosome-specific, dog genome-integrated BAC panels
Two hundred seventy-five clones from the CHORI-82 BAC library (http://bacpac.chori.org, Children’s Hospital Oakland Research Institute, Oakland, CA) were selected for use as single-locus probes in multicolor FISH analysis. These genome-anchored, cytogenetically validated clones are spaced at approximately 10-Mb intervals along the length of the 38 domestic dog (CFA) autosomes and the X chromosome (Thomas et al. 2007). DNA was isolated from each BAC clone using Qiagen REAL Prep 96 kits (Qiagen, Valencia, CA). For each BAC clone, 500 ng of DNA was differentially labeled by nick translation to incorporate nucleotides conjugated with one of four fluorophores [SpectrumRed-dUTP, SpectrumGreen-dUTP, Spectrum-Orange-dUTP (Vysis, Abbott Park, IL), diethylaminomethylcoumarin-5-dUTP (Perkin Elmer Life Sciences, San Jose, CA)] or with biotin-11-dUTP (Perkin Elmer Life Sciences), detected using Cy5-Streptavidin (GE Healthcare, Piscataway, NJ), as described previously (Breen et al. 2004). The single-locus probes in each chromosome-specific panel were labeled with alternating fluorophores to produce a multicolored tiling pattern, revealing probe order and allowing relative orientation of each ECCS between species (Electronic supplementary material (ESM) Fig. 1). Additional BAC clones spaced at ∼1-Mb intervals (Thomas et al. 2008) were used to evaluate selected breakpoint regions at higher resolution.
Selection of B chromosome BAC clones
B chromosomes were identified in metaphase preparations using a dog BAC clone (524B22) containing the canine cKIT proto-oncogene sequence (Thomas et al. 2008), which has been shown previously to map to all B chromosomes in the red fox, Chinese raccoon dog, and Japanese raccoon dog (Yudkin et al. 2007; Graphodatsky et al. 2008). As part of a complementary study (Trifonov, unpublished), B chromosome-specific sequences were selected by affinity capture (Chen-Liu et al. 1995) using flow-sorted red fox B chromosomes. The resulting sequences were interrogated using the BLAST tool (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi), and sequences with 90–95% homology to the canine genome were identified. Fifteen B chromosome-derived sequences were used to identify corresponding dog BAC clones for FISH analysis (Table 2). Thirty-six additional BACs, each containing a gene known to exhibit recurrent cancer-associated copy number aberrations (Thomas et al. 2008), were selected to explore our hypothesis that canid B chromosomes may contain cancer-associated genes in addition to cKIT. Additional BAC clones were selected as required from the dog genome sequence assembly (canFam2, http://genome.ucsc.edu/) for high-resolution FISH analysis of targeted chromosome regions.
FISH analysis of chromosome preparations
Panels of five consecutive, differentially labeled BAC probes were hybridized to metaphase chromosome preparations from clinically healthy domestic dog donors to verify their unique cytogenetic location as reported previously (Thomas et al. 2007, 2008). The 10-Mb BAC panels representing each domestic dog autosome were then hybridized to metaphase chromosome preparations of three wild canids, red fox, Chinese raccoon dog, and gray fox, in the presence of sonicated CFA DNA as competitor. The presence of common breakpoints in conserved synteny, defined as those identified in at least two of these three species, was then assessed in eight additional wild canids; kit fox (Vulpes macrotis, VMA), fennec fox (Fennecus zerda, FZE), Japanese raccoon dog (N. p. viverrinus, NPRv), bat-eared fox (Otocyon megalotis, OME), crab-eating fox (Cerdocyon thous, CTH), hoarey fox (Dusicyon vetulus, DVE), maned wolf (C. brachyurus, CBR), and bush dog (Speothus venaticus, SVE). In addition, breakpoints common to the closely related Chinese and Japanese raccoon dog species were also examined in all 11 species. The 10-Mb intervals defined by whole chromosome analysis were subsequently evaluated at ∼1-Mb resolution in all 11 species to refine the common breakpoint. Chromosome preparations were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) as described previously (Breen et al. 1999). In total, this study used 320 canine BAC clones in 400 independent multicolor FISH reactions involving all 11 species. When referring to each BAC clone, we referred to the genome location of each clone by the mid-pint of the BAC sequence in the canine genome assembly.
Image capture and analysis was performed using a semi-automated multicolor FISH workstation comprising a Zeiss Axioplan 2 fluorescence microscope equipped with a motorized Ludl stage, narrow pass filter sets, and a Hamamatsu CCD camera and Smart Capture 3 software (Digital Scientific, Cambridge, UK). The distribution of BAC probe hybridization signals was defined within each species, from which ECCSs were defined and oriented relative to that in the domestic dog. Where BAC probes from a single dog chromosome hybridized to more than one chromosome in the wild canid species, the corresponding breakpoint regions were compared with data from prior chromosome painting experiments (Yang et al. 1999; Nie et al. 2003; Graphodatsky et al. 2008). The domestic dog karyotype was described using the nomenclature of Breen et al. (1999). The chromosome nomenclatures used for the red fox, Chinese raccoon dog, and gray fox were based on those of Yang et al. (1999), Nie et al. (2003), and Wayne et al. (1987a, b), respectively. Chromosome nomenclature for the gray fox was also compared with that used by Graphodatsky et al. (2008) (ESM Tables 1–4). DAPI-banded ideograms were prepared for all three wild caninds based on evaluation of 15 metaphase preparations for each species.
Results
When hybridized to metaphase preparations of the domestic dog, all BAC clones used in this study revealed each a unique cytogenetic location, as has been reported previously (Thomas et al. 2007, 2008). Metaphase preparations generated for 10 of the 11 wild canid species used in this study had karyotypes comparable to previous reports, with two exceptions; the karyotype of the crab-eating fox reported previously by Nash et al. (2001) likely had an aberrant chromosome not present in the cells used in the present study (see below for “Discussion”), and the maned wolf used in this study did not contain B chromosomes. All BAC clones had a unique cytogenetic location in the karyotypes of all 11 wild canid species, with the exception of those that also mapped to the B chromosomes (see below). FISH analysis of the 10-Mb resolution BAC panel on the karyotypes of red fox, Chinese raccoon dog, and gray fox defined the location and relative orientation of ECCSs (Figs. 1, 2, 3, and 4 and ESM Tables 1–4) and identified common breakpoint regions in conserved synteny for CFA1, 9, 13 (two breakpoint regions), 18, and 19 ECCSs (Fig. 5). These breakpoint regions were then explored in eight additional wild canid species and narrowed to ∼1- to 3-Mb intervals (Fig. 6). The B chromosomes present in red fox, Chinese raccoon dog, and Japanese raccoon dog were explored, identifying the presence of duplicate sequences shared with A chromosomes (Figs. 7, 8, 9, and 10). These findings are discussed in detail below.
Hybridization of domestic dog BAC clones to red fox chromosome preparations reveals the location and orientation of dog ECCSs. A Inverted DAPI-stained image of VVU1. B Hybridization of CFA 10-Mb probe panels to red fox chromosomes identified ECCSs shared by VVU1 and regions of CFA1, 12, and 33. C The VVU1 ideogram is annotated with the dog genome locations of the probe panels and the orientation of the dog ECCSs. The relative orientation of each ECCS is evident from the order of dog BAC clones along the length of VVU1 and is indicated by vertical arrows against the VVU1 ideogram. This defined an evolutionary breakpoint within the genomic interval CFA1;22.34–32.26 Mb in the red fox. Scale bar, 10 μm
Annotation of the red fox (VVU) ideogram to define ECCSs shared with the domestic dog. A The orientation of each dog ECCS is indicated by a vertical arrow. The red fox karyotype has four evolutionary breakpoints in dog synteny at CFA1;27.6–28.7Mb; CFA13;40.8–41.6Mb; CFA18;27.4–29.0Mb; and CFA19;21.8–25.0Mb. The breakpoints are annotated on VVU1, 2, 4, 5, and 13 in red text. Regions of each dog ECCS on either side of the EBR are indicated with a suffix [i.e., CFA1(a) and CFA1(b)]. B The probe representing CFA19;46.00Mb (yellow) hybridized both to VVU5p and all VVUB chromosomes, distal to the location marked by a BAC probe containing cKIT (red)
Annotation of the Chinese raccoon dog (NPRp) ideogram to define ECCS shared with the domestic dog. A The orientation of each dog ECCS is indicated by a vertical arrow. The Chinese raccoon dog karyotype has six evolutionary breakpoints in dog synteny at CFA1;27.6–28.7Mb; CFA9;20.2–22.4Mb; CFA13;40.8–41.6Mb; CFA13;63.5–64.9Mb; CFA19;21.8–25.0Mb; and CFA3;3.13–15.17Mb. The breakpoints on NPRp1, 5, 6, 7, 9, and 17 are noted in red text and the breakpoint in the CFA3 ECCS (on NPRp6) is italicized. While CFA3;15.17–94.31Mb hybridized to NPRp6mid-tel, CFA3;3.13Mb co-localized with the probe representing CFA13;64.87Mb on NPRp6prox. The proximal one third of NPRp2p (noted with a red asterisk) contains a region that did not hybridize to our probe panels. Several Chinese raccoon dog chromosomes contain chromatin proximal to any signal from the probe panels. These regions are noted with a closing curly bracket on NPRp6-18, and 20–26. Regions of each dog ECCS on either side of the breakpoint are indicated with a suffix [i.e., CFA1(a) and CFA1(b)]. B The probe representing CFA29;44.52Mb (blue) hybridized both to VVU1q and to the largest NPRpB chromosome (Bi), co-localizing with the distal cluster of signal from a probe containing cKIT (red)
Annotation of the gray fox (UCI) ideogram to define ECCS shared with the domestic dog. The orientation of each dog ECCS is indicated by a vertical arrow. The gray fox karyotype has nine evolutionary breakpoints in dog synteny: CFA1;22.34–32.26Mb; CFA2;3.13–13.77Mb; CFA2;33.76–43.63Mb; CFA2;43.63–53.35Mb; CFA5;32.83–43.05Mb; CFA10;13.94–23.36Mb; CFA13;33.74–44.32Mb; CFA18;23.79–33.27Mb; and CFA19;13.48–25.12Mb. Breakpoints in CFA1, 13, 18, 19 (in red text) were narrowed to CFA1;27.6–28.7Mb; CFA13;40.8–41.6Mb; CFA18;27.4–29.0Mb; and CFA19;21.8–25.0Mb on UCI1, 7, 8, 11, 20, 22, 27, and 31. Breakpoints in CFA2, 5, 10 ECCSs (not narrowed) are italicized on UCI21 and 22, 4, 14, respectively. Several gray fox chromosomes contain chromatin proximal to any signal from the probe panels. These regions are noted with a closing curly bracket on UCI3, 6, 9, 10, 14, 23, and 25. Regions of each dog ECCS on either side of the breakpoint are indicated with a suffix [i.e., CFA1(a) and CFA1(b)]. CFA7 and CFA28, 37 ECCSs are found on UCI12 and UCI15 (outlined with a box), respectively. In a previous study, Graphodatsky et al. (2008) organized these ECCSs differently, with CFA7 and 28 ECCSs together on the largest chromosome and CFA37 ECCS alone on a small chromosome. We believe that the cells used in that study had undergone a translocation relative to those described here and in Wayne et al. 1987a
Domestic dog ideogram annotated to show evolutionary breakpoints within 11 wild canid species. Probe panels representing BACs spaced ∼10 Mb apart were applied to 11 species, and the resulting breaks in dog ECCSs, evident when two adjacent probes hybridized onto different autosomes (or with intrachromosomal rearrangements, out of sequence), are noted with red bars. Regions of each dog ECCS on either side of the breakpoint are indicated with a suffix (i.e., CFA1a and CFA1b). The species possessing each breakpoint are indicated. Chromosome painting experiments identified a breakpoint in synteny in some cases (noted with symbols). Breakpoints at CFA1;22.34–32.26Mb; CFA9;13.50–23.42Mb; CFA13;33.74–44.32Mb; CFA13;54.97–64.87Mb; CFA18;23.79–33.27Mb; and CFA19;13.48–25.12Mb were narrowed to CFA1;27.6–28.7Mb; CFA9;20.2–22.4Mb; CFA13;40.8–41.6Mb; CFA13;63.5–64.9Mb; CFA18;27.4–29.0Mb; and CFA19;21.8–25.0Mb in 11 species. There were no detectable CFAX breakpoints in the 11 species studied
Six breakpoints identified using probe panels at 10-Mb resolution were refined at high resolution. A Six EBRs, located on ECCSs of CFA1, 9, 13 (two regions), 18, and 19, were narrowed to 1- to 3-Mb intervals across the 11 wild canid species. Dog chromosomes (inverted DAPI-stained) are shown first, with regions of the dog chromosome represented by separate ECCSs in the 11 non-dog canids annotated. Regions of each dog ECCS on either side of the breakpoint are indicated with a suffix (i.e., CFA1a and CFA1b). The orientation of the ECCS in each species is noted next to each corresponding chromosome. Nomenclature used in Figs. 2, 3, and 4 is shown next to red fox (VVU), Chinese raccoon dog (NPRp), and gray fox (UCI) chromosomes. Patterns of ECCSs across species generally correspond to accepted phylogenetic groupings as noted by Lindblad-Toh et al. (2005). Of the fox-like grouping, the kit fox (VMA), red fox (VVU), and fennec fox (FZE) share the same breakpoints. The bat-eared fox (OME) (fox-like canid) has one fewer breakpoint (CFA18 ECCS) than these species. The Chinese raccoon dog (NPRp) and Japanese raccoon dog (NPRv), also fox-like canids, are most closely related to each other and share the same breakpoints. An additional breakpoint at CFA13;63.5–64.9Mb is present in both Chinese and Japanese raccoon dogs (NPRp and NPRv), rearranging the bottom ∼25 Mb of the CFA13 ECCS. All species except the maned wolf (CBR) and bush dog (SVE) contained a breakpoint at CFA19;21.8–25.1Mb, splitting the ECCSs across two chromosome pairs in kit fox (VMA), red fox (VVU), fennec fox (FZE), Chinese raccoon dog (NPRp), Japanese raccoon dog (NPRv), and bat-eared fox (OME). In the crab-eating fox (CTH) and hoarey fox (DVE), a p-arm is present that contains the proximal-most CFA19 ECCS, with the distal-most CFA19 ECCS present as a q-arm. All four species containing the CFA18 breakpoint (kit fox, red fox, fennec fox, and gray fox) have the CFA38 ECCS adjacent to the CFA18a ECCS. The gray fox (UCI) is grouped as an island fox, having diverged from all other examined species at least 10 Mya (Bailey et al. 2004). Despite this, the gray fox shares CFA1, 13, 18, and 19 breakpoints with fox-like canids. B Both raccoon dogs have a breakpoint at CFA9;20.2–22.4Mb, and the resulting ECCSs have been inverted such that the proximal-most and distal-most probes are adjacent in the resulting chromosomes. The hoarey fox and crab-eating fox contain four additional breakpoints at CFA9;3.1–13.5, 23.4–44.5, 44.5–43.6, and 54.2–64.1Mb. The crab-eating fox contained three chromosome structures with CFA9 sequence, which resulted from at least seven breakpoints compared with CFA9. This includes the previously mentioned breaks and two additional breaks at CFA9;13.1–14.5 and 16.8–17.8Mb. Scale bars, 10 μm
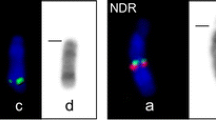
Sequence containing cKIT hybridizes to the B chromosomes of three canid species. A cKIT hybridizes to VVUB chromosomes in addition to the CFA13 ECCS on VVU2. Signal is shown on ideograms and B chromosomes of red fox (B, C), Chinese raccoon dog (D, E), and Japanese raccoon dog (F, G). VVUB chromosomes appear identical (C), but the pattern of signal and banding on the B chromosomes of the two raccoon dogs are distinct. Signal strength on NPRpBi and Bii is equal to the signal found on Chinese raccoon dog autosomes, while the single signal on NPRpBiii is much weaker (E). Signal on NPRvBi and Bii chromosomes is located near the telomeric end, while signal on NPRv Biii is more proximal (F, G). Scale bars, 10 μm (A), 2 μm (B–G)
Sequence containing LRIG1, which encodes a negative regulator of EGFR, hybridizes to NPRpB chromosomes. B chromosomes of the Chinese raccoon dog (NPRp) are marked with the cKIT probe (yellow). A BAC probe (465J12) containing LRIG1 (green) hybridized to NPRpBi and Bii. The signal for this probe is attenuated across the reaction to distinguish the probe signal on NPRpBi and Bii while still being visible on NPRp25. The signal from the LRIG1 probe is coincident and/or adjacent to signal from the cKIT probe in both Bi and Bii (inset). Scale bars, 10 μm, 2 μm (inset)
The region of CFA20 ECCS containing sequence that hybridizes to NPRpB chromosomes was identified. A Overlapping BACs were chosen from the dog genome assembly on either side of 465J12 (contains LRIG1) and hybridized sequentially to chromosomes of the Chinese raccoon dog (NPRp). BACs highlighted with blue only hybridized to NPRp25 (CFA20 ECCS location), while BACs highlighted with red hybridized both to NPRp25 and to B chromosomes. A red box outlines this region in the assembly. Colors of probe signals are noted adjacent to each BAC shown in B. B Six BACs spanning CFA20;27,568,093-28,169,083Mb (approximately 601 kb) hybridized to NPRpB chromosomes. Each probe’s hybridization pattern is different on Bi and Bii chromosomes, which suggests that each region contains different duplicated CFA20 sequence. Signal strength was reduced (465J12, marked with an asterisk) or increased (107D16, marked with a number sign) across the entire reaction to distinguish the probe signals on Bi and Bii, resulting in artificially weak or enlarged signal on NPRp25. Probes marked with a plus sign (084C23 and 459J13) were hybridized in a separate reaction; chromosomes were of similar length and, hence, represent a similar point in the cell cycle as other chromosomes. Scale bar, 2 μm
B chromosome ideograms are annotated with probe locations. The cKIT probe (red) was used to identify B chromosomes in red fox (VVU), Chinese raccoon dog (NPRp), and Japanese raccoon dog (NPRv) metaphase spreads. Additional probes also hybridized to canid B chromosomes as shown. The characteristic hybridization patterns of the cKIT probe in the Chinese raccoon dog provide a novel method for distinction between the three B chromosomes present in this species
FISH analysis of red fox (VVU) chromosomes refines four known disruptions of conserved synteny with the domestic dog genome
The autosomal karyotype of the red fox comprises 16 submetacentric autosomes, supplemented with up to eight small submetacentric B chromosomes, each similar in size to the red fox Y chromosome (VVUY). Hybridization of the 10-Mb resolution dog BAC panel to red fox chromosomes demonstrated that ECCSs shared by the domestic dog and red fox remain grossly intact, identifying the four known breaks in conserved synteny (Yang et al. 1999) and now refining these breakpoints regions to within ∼10-Mb intervals on CFA1, 13, 18, and 19, corresponding to regions of VVU1, 2, 4, 5, and 13 (Fig. 2a). The orientation of CFA ECCSs within the karyotype of red fox is noted in Fig. 2a. One dog BAC clone (287N13: CFA9;46.00Mb) hybridized to VVU5p, consistent with flanking markers from the CFA19 BAC panel, but also mapped to all B chromosomes, distal to the region containing sequence homologous to cKIT (Figs. 2b and 10).
FISH analysis of Chinese raccoon dog (NPRp) chromosomes refines three known disruptions of conserved synteny with the domestic dog genome and identifies three previously unreported breakpoint regions
The Chinese raccoon dog autosomal karyotype comprises 5 submetacentric autosomes, 21 acrocentric autosomes, and up to 4 B chromosomes. The B chromosomes range in size, comparable to NPRp6-8, and are characterized as three types: Bi, Bii, and Biii. The distribution and relative orientation of ECCSs identified by the application of the dog 10-Mb resolution BAC panel to Chinese raccoon dog are summarized in Fig. 3. Breakpoints in conserved synteny between Chinese raccoon dog chromosomes and regions of CFA1, 13, and 19 have been identified previously by chromosome painting (Nie et al. 2003). Application of the 10-Mb resolution dog BAC panel refined these breakpoints to within ∼10-Mb intervals and also revealed three additional, previously unreported breakpoint regions corresponding to regions of CFA3, 9, and 13 (Fig. 3). The newly identified breaks result in rearrangements on NPRp6, and while all of the CFA9 BAC panel hybridized exclusively to the full length of NPRp5q, it did so in two ECCSs that are inversely oriented (Fig. 3a).
One dog BAC clone (335J13: CFA29;44.52Mb) hybridized to NPRp1q in common with flanking CFA29 clones, but also mapped to the distal end of the largest B chromosome (Fig. 3b). Clone 524B22 (CFA13;50.4Mb, containing cKIT) hybridized to all Chinese raccoon dog B chromosomes, with two distinct regions of hybridization evident on the two larger B chromosomes (Figs. 3b, 7, 8, and 10). Nie et al. 2003 reported previously that the full length of NPRp2p was an ECCS shared with CFA25. In the present study, the CFA25 panel hybridized only to the distal two thirds of the p-arm of NPRp2.
FISH analysis of gray fox (UCI) chromosomes refines five known disruptions of conserved synteny with the domestic dog genome and identifies four previously unreported breakpoint regions
The autosomal complement of the gray fox comprises 32 pairs of acrocentric chromosomes and no B chromosomes. Prior chromosome painting studies identified five breaks in conserved synteny between the dog and gray fox karyotypes within regions corresponding to CFA1, 2, 13, 18, and 19 (Graphodatsky et al. 2008). These regions were confirmed and their precise location refined in the present study (Fig. 4). Two previously unknown intrachromosomal rearrangements resulting from two breaks were also identified. The first, corresponding to CFA5;32.83–43.05Mb, resulted in an inversion of the orientation of the two ECCSs on UCI4. The second, at CFA10;13.94–23.36Mb, resulted in an inversion of both CFA10 ECCSs on UCI14. Graphodatsky et al. (2008) reported a breakpoint using a CFA2 paint, placing the upper CFA2 ECCS on UCI22 with the lower CFA2 ECCS on UCI21. Our data revealed two additional breakpoints at CFA2;3.13–13.77Mb and CFA2;43.63–53.35Mb, resulting in the recombination of CFA2 ECCSs. Two BACs, 125F22 (CFA2;3.13Mb) and 126P21 (CFA2;43.63Mb), hybridized to UCI22, while CFA2;13.77–33.76 and CFA2;53.35–88.13Mb hybridized to UCI21 (Fig. 4).
Refinement of common evolutionary breakpoint regions across 11 wild canid species
Application of the 10-Mb resolution dog BAC panel to chromosome preparations of the red fox, Chinese raccoon dog, and gray fox identified four common evolutionary breakpoints shared by two or more of the species, located on CFA1 (22.34–32.26 Mb), 13 (33.74–44.32Mb), 18 (23.79–33.27Mb), and 19 (13.48–25.12Mb; Figs. 2, 3, and 4). Two additional breakpoints were common to the two species of raccoon dog, located on CFA9 (13.50–23.42Mb) and 13 (54.97–64.87Mb). The locations of the breakpoints in red fox, Chinese raccoon dog, and gray fox, relative to the dog, are shown in Fig. 5. These ∼10-Mb regions were evaluated at higher resolution in all 11 species using FISH analysis with BAC probes spaced at ∼1-Mb intervals, refining the breakpoint regions further to CFA1;27.6–28.7Mb; CFA9;20.2–22.4Mb; CFA13;40.8–41.6Mb; CFA13;63.5–64.9Mb; CFA18;27.4–29.0Mb; and CFA19;21.8–25.0Mb (Fig. 6).
Exploration of B chromosomes in red fox (VVU), Chinese raccoon dog (NPRp), and Japanese raccoon dog (NPRv)
Previously, it was shown that the cKIT sequence is present in the B chromosomes of the red fox, Chinese raccoon dog and Japanese raccoon dog (Graphodatsky et al. 2005; Yudkin et al. 2007). This is supported in the present study by FISH analysis using a dog BAC clone containing the canine cKIT gene. In addition to hybridizing to the CFA13 ECCS on VVU2p, NPRp6, and NPRv2q (the presumptive primary genomic locations of cKIT in these species), the cKIT BAC also hybridized to all B chromosomes in these three species (Fig. 7). In the red fox, the cKIT BAC produced a large signal that covered much of the q-arm of all B chromosomes (Fig. 7a–c). When hybridized to the Chinese raccoon dog, the cKIT signal was present on all three B chromosomes, but with distinct pattern of hybridization: three locations on Bi, four locations on Bii, and one location on Biii (Fig. 7d, e). This differential pattern of hybridization on NPRpBi, Bii, and Biii allows each to be identified conclusively. In the Japanese raccoon dog, the cKIT signal was smaller than in the red fox, and while two of the B chromosomes (Bi and Bii) had signal in the same location, the signal on Biii was further from the telomere (Fig. 7f, g).
Among the cancer-associated BACs used in this study, the BAC clone 521A01 contains the proto-oncogene RET (a receptor tyrosine kinase) and maps to CFA28;6.89Mb. When hybridized to the species containing B chromosomes, this clone produced signal on NPRpBi and Bii, but not to Biii and did not map to any of the B chromosomes of the red fox or Japanese raccoon dog. Similarly, a BAC containing the gene LRIG1 (which maps to CFA20;27.8Mb) did not hybridize to the B chromosomes of the red fox or Japanese raccoon dog. However, in the Chinese raccoon dog, while LRIG1 did not map NPRpBiii, it hybridized to both NPRpBi and Bii in multiple copies (Fig. 8). To explore this region further, we tiled the regions proximal and distal to LRIG1 with 15 additional BACs selected from the dog genome sequence assembly (Fig. 9). These data revealed that the size of the genome sequence shared between CFA20 and NPRp Bi and Bii was 601 kb, with the upper breakpoint located at CFA20;27,559,082-27,568,093Mb and the lower breakpoint located at CFA20;28,169,083-28,211,725Mb.
Of the 15 dog BACs specifically selected following affinity capture to target the VVUB chromosome (Table 2), all hybridized to the B chromosomes of at least one species (Fig. 10). Additional dog BAC clones that co-hybridize to the canid B chromosomes of the red fox and/or Chinese raccoon dog were located at CFA12, 15, 19, 31, and 34. Only cKIT hybridized to the B chromosomes of the Japanese raccoon dog (Fig. 10).
Discussion
Comparative molecular cytogenetic analysis using genome-anchored domestic dog (CFA) BAC probes revealed shared breakpoints among 11 wild canid species
CFA1;27.6–28.7Mb
This breakpoint was present in 7 of the 11 species studied, six of which were Fox-like species (kit fox, red fox, fennec fox, Chinese raccoon dog, Japanese raccoon dog, and bat-eared fox) and one was an Island fox (gray fox; Table 1). As a known EBR, this breakpoint has also been described in at least five other mammal genomes including the mouse, rat, human, chimpanzee, and cow (Derrien et al. 2007). It is therefore likely that this genetic sequence was organized as two ECCSs in the mammalian ancestral karyotype. As canid speciation occurred, the karyotypes of the ancestor of both wolf-like (domestic dog, maned wolf) and two of the South American (SA) canids (crab-eating fox and hoarey fox) underwent a fusion to create the modern CFA1 chromosome seen these species.
CFA9;20.2–22.4Mb
The CFA9 sequence exists as a single ECCS in 7 of the 11 wild canid species studied, four fox-like (kit fox, red fox, fennec fox, bat-eared fox), the island fox (gray fox), and two of wolf-like canids (maned wolf and bush dog). However, extensive intrachromosomal rearrangements involving the CFA9 ECCSs were observed in crab-eating fox and hoarey fox. Unlike the Chinese and Japanese raccoon dogs, which possessed only one breakpoint, the crab-eating fox and hoarey fox displayed at least four additional breakpoints in CFA9 ECCSs. The shared breakpoint at CFA9;20.2–22.4Mb, located in the Chinese and Japanese raccoon dogs, crab-eating fox, and hoarey fox, would seemingly only exist if it occurred in two separate events: one in the ancestor of the two raccoon dogs after divergence from other fox-like canids and one in the ancestor of the crab-eating fox and hoarey fox, both of which could have occurred coincidently with a CFA19 ECCS fusion. It has been proposed based on mtDNA sequences that the two raccoon dogs should be grouped basally to other fox-like canids, but separately from SA canids (Wayne et al. 1997), and the CFA9 breakpoints are consistent with this proposed grouping. Regardless, two events causing CFA9 ECCS rearrangements would explain the additional breakpoints found only in the crab-eating fox and hoarey fox.
CFA13;40.8–41.6Mb
This region is a common EBR present in the same seven canids as the EBR in CFA1 and also in and at least five mammals (Derrien et al. 2007). It is likely, therefore, that the two CFA13 ECCSs fused in the ancestor(s) of wolf-like and SA canids to create the modern CFA13 chromosome, while the other species retained two separate ECCSs on different autosomes. It is possible that the chromosome restructuring which occurred fusing the two CFA1 ECCS took place coincidently with fusions of ECCSs on either side of this EBR in the ancestor(s) of wolf-like and SA canids.
CFA13;63.5–64.9Mb
An additional break in CFA13 synteny occurred within this region in the lineage of two species of raccoon dog, rearranging the bottom ∼25 Mb of the distal ECCS. Although this breakpoint is only present in the two species of raccoon dog, this region appears to be a location prone to rearrangement in other mammals (Derrien et al. 2007) and may reflect an inherently fragile region as predicted by the fragile breakpoint model.
CFA18;27.4–29.0Mb
Four canids (kit fox, red fox, fennec fox, and gray fox) possess this EBR. In these species, the CFA38 ECCS is adjacent to one, if not both, CFA18 ECCSs (Fig. 6). Prior studies have reported that DNA segments corresponding to regions of CFA18 and 38 are associated as neighboring units on a single chromosome in several canid species (Graphodatsky et al. 2008). In three fox-like canids (kit fox, red fox, and fennec fox), the CFA38 ECCS is located between and in opposing orientation to the two CFA18 ECCSs (Fig. 2, VVU5, and Fig. 6). However, in the gray fox, the proximal CFA18 ECCS is located adjacent to the CFA38 ECCS, and both are oriented in the same direction (Fig. 4, UCI22, and Fig. 6). At least five additional mammals (human, chimpanzee, mouse, rat, and cow) contain an EBR at CFA18;27.4–29.0Mb (Derrien et al. 2007), while the Chinese raccoon dog, Japanese raccoon dog, and bat-eared fox (also fox-like canids in the accepted phylogenetic tree; Lindblad-Toh et al. 2005) organize CFA18 sequence as one single ECCS, as is the case in the wolf-like and SA canids. A more parsimonious tree would thus have two fusion events to create the CFA18 form in wolf-like and SA canids and in three fox-like canids (Chinese raccoon dog, Japanese raccoon dog, and bat-eared fox). Also, there would be two separate fusion events to associate CFA38 with the proximal CFA18 ECCS—one in the gray fox ancestor and one in the ancestor of the kit fox, red fox, and fennec fox—after divergence from the ancestor of the Chinese raccoon dog, Japanese raccoon dog, and bat-eared fox. The CFA38 ECCS would have fused to the proximal CFA18 ECCS (opposite orientations in each fusion), which in the kit fox, red fox, and fennec fox ancestor would also be fused with the distal CFA18 ECCS.
CFA19;21.8–29.0Mb
This is a known EBR in at least five other mammals (Derrien et al. 2007), and the mammalian ancestral karyotype probably organized this sequence as two ECCSs, which then fused in the ancestor(s) of the wolf-like and SA canids. The karyotypes of the domestic dog, maned wolf, and bush dog have CFA19 sequence organized as one ECCS comprising the full length of a single chromosome arm, along which the relative order of BAC probes from the 10-Mb panel remained intact in all three species. In contrast, two other SA canid karyotypes (crab-eating fox and hoarey fox) share the CFA19 breakpoint. In both of these species, the two resulting ECCSs are arranged in opposing orientation within a single bi-armed chromosome whose p-arm and q-arm correspond to the proximal and distal regions of CFA19, respectively (Fig. 6). Examination of additional SA canids should clarify whether the inverted fusion present in the crab-eating fox and hoarey fox was established in a shared ancestor after the maned wolf/bush dog ancestor diverged. It is unclear whether the fusion of two ECCSs to form CFA19 (and likewise CFA9 ECCSs) occurred separately in the ancestors of the wolf-like canids and maned wolf/bush dog SA canids or whether the phylogeny should be adjusted to reflect the shared CFA19 (and CFA9) breakpoint(s), which would contradict phylogenies based solely on mtDNA sequence.
Retention and reorientation of ECCSs
Comparative cytogenetics has demonstrated that despite the extensive variation in chromosome number and morphology that developed during speciation of the Canidae, the majority of ECCSs appear to have remained largely intact in the karyotypes of extant canid species, although the relative orientation of ECCSs is not always conserved. For example, the CFA9 ECCS is located on VVU2p (Fig. 2), but is oriented such that the sequence corresponding to the centromeric region of CFA9 is located at the VVU2p telomere. If there were no sequence deletions, VVU2p would contain interstitial telomeric repeats present because of the translocation of the CFA9 ECCS adjacent to the distal CFA13 ECCS that makes up VVU2p. In the Chinese raccoon dog, the larger CFA9 ECCS present in VVU2 is split at CFA9;20.2–22.4Mb, and the resulting ECCSs are oriented on NPRp5q (Fig. 3) so that the centromeric end is closest to the NPRp5q telomere and the telomeric end is interstitial. The CFA38 ECCS is located on NPRp5p, oriented so that the centromeric and telomeric ends are inverted. This means that the centromere on NPRp5 is new relative to CFA. Shifts in the location of centromere and telomere sequences have been studied in human and horse (Lin and Yan 2008; Marshall et al. 2008; Wade et al. 2009). In equids, centromere locations have shifted over the course of just the last 2 million years via centromere repositioning concomitant with fusion of ECCSs (Piras et al. 2010). Further study in canids would be necessary to clarify the origin of centromeres, whether they are artifacts of the ancestral centromeres or formed de novo as a consequence of translocations during speciation.
Three pairs of ECCSs (CFA19a/32, CFA13a/29, and CFA28/37) have been retained together as contiguous segments in canids and other mammalian species despite the paired segment being located adjacent to different ECCSs across speciation. CFA19a and 32 ECCSs are adjacent in all canids (Figs. 2–4, 11; Wayne et al. 1987a, b; Graphodatsky et al. 2008) and also in human and chimpanzee (Derrien et al. 2007). CFA13a and 29 ECCSs are adjacent in the Chinese raccoon dog, Japanese raccoon dog, bat-eared fox, maned wolf, bush dog, and gray fox (Figs. 3, 4 and 11; Wayne et al. 1987a, b; Graphodatsky et al. 2008), as well as in human and chimpanzee (Derrien et al. 2007). This pair are adjacent, but separated by a centromere, in kit fox, red fox, and fennec fox (Figs. 1 and 11; Wayne et al. 1987b). CFA28 and 37 ECCSs are adjacent in the Chinese raccoon dog, Japanese raccoon dog, bat-eared fox, maned wolf, bush dog, and gray wolf (Figs. 3, 4 and 11; Wayne et al. 1987a, b; Graphodatsky et al. 2008). It is possible that these associations are a consequence of the karyotype organization of the mammalian ancestor, with a centromerization event occurring between CFA13a and 29 ECCSs in the ancestor(s) to kit fox, red fox, and fennec fox after divergence from the Chinese raccoon dog, Japanese raccoon dog, and bat-eared fox. These paired associations are not present in cow, mouse, or rat (Derrien et al. 2007), suggesting that additional breakage–fusion events occurred during speciation of ruminants and rodents. Such patterns of associated ECCSs have been identified in mammalian genomes previously. One such association, human chromosomes 12 and 22 (HSA12/22), is considered ancestral for all mammals (Haig 1999), and its loss in various lineages is derived. HSA12 and 22 are each represented by two ECCSs (HSA12a, 12b and HSA22a, 22b) that are associated as HSA12a-22a and HSA12b-22b in cattle, pigs, and mice (Haig 1999). Translocations between HSA12 and 22 have been identified in a range of cancers (Chemitiganti et al. 1985; Mrózek et al. 1993), and the regions surrounding such breaks could correspond to fragile regions as predicted by the fragile breakpoint model.
Regions of wild canid chromosomes that were not detected with domestic dog BAC panels
The centromeric regions of several Chinese raccoon dog and gray fox chromosomes exhibit chromatin that lies beyond the proximal boundary of the dog BAC set used in the present study, resulting in chromosomal regions with no corresponding dog BAC assignment (noted with asterisks in Figs. 3 and 4). These apparent “CFA-vacant” regions appear to be approximately 5–15 Mb in size by comparison to known cytogenetic intervals defined by the BAC panel within the same karyotype. As such, each of these regions is more extensive than the ∼3 Mb of centromeric sequence from each dog chromosome that is absent from the genome sequence assembly due to its highly repetitive nature (Lindblad-Toh et al. 2005) and which in turn is not represented within the BAC panels used in the present study. For the CFA-vacant regions of NPRp6, 7, 8, 9, 10 and UCI9 and 23, overall agreement between chromosome painting and our data suggests that during speciation, these chromosomes likely gained sequence (or, conversely, the corresponding CFA chromosomes lost centromeric-adjacent sequence), resulting in the presence of chromatin regions in the Chinese raccoon dog and gray fox for which a homologous region is not evident in the domestic dog. As an example, the region of additional chromatin on NPRp7 is adjacent to sequence proximal to CFA1, 32.26 Mb. Since the dog BAC panels provide coverage extending, on average, from 3.34 Mb to the telomere of each dog chromosome, it is possible that the CFA-vacant regions in the wild canids share sequence with the centromeric regions of dog chromosomes. This would cause the chromosomes to appear to have additional chromosomal material observed by FISH, but not in chromosome painting, since the latter involves hybridizing probes made from entire dog chromosomes. This could explain the CFA-vacant regions seen in our studies (but not reported in earlier work) on NPRp11, 12, 13, 16, 17, 18, 20, 22, 25, 26 and UCI3, 6, 25. However, the larger CFA-vacant regions, such as the region on NPRp18, are too large to be explained by the approximately 3.17 Mb of CFA7 not represented in the BAC panel. NPRp14, 15, 21, 23, 24, and UCI10 contain dog sequence oriented so that the telomeric end of the ECCS is interstitial while the centromeric end is near the telomeric end of the Chinese raccoon dog chromosome (Fig. 3, NPRp23). This orientation means that the CFA-vacant region on the Chinese raccoon dog chromosome is proximal to the distal-most BAC (essentially the dog telomeric region) in the associated panel. This scenario could mean that the sequence represented in these regions is gained in the Chinese raccoon dog (or lost in the dog), but since this was not noted in previous work, it is unclear whether this is the case. Alternatively, there may actually be more chromatin at the telomeres of dog chromosomes not described in the genome assembly. This is very unlikely since, at least cytogenetically, our BAC panels hybridize to the ends of each dog chromosome (Thomas et al. 2008, 2009).
Duplicated sequences—dog sequences show co-hybridization to wild canid B chromosomes
Twenty-five BAC clones used in this study mapped to more than one cytogenetic location in the wild canids despite having a unique location in the dog. Intriguingly, these duplicated sequences were all located on the canid B chromosomes and in some cases represent cancer-associated genes (i.e., cKIT and LRIG1) that were present in multiple copies on the B chromosomes. This raises an interesting question about the possible role of canid B chromosomes. Could B chromosomes represent an evolutionary mechanism to sequester additional copies of genes that are generated at the breakpoints associated with speciation? It has been suggested that duplications in sequence occur at breakpoint regions (Bailey et al. 2004). Dog BAC clones identified here that were duplicated in the B chromosomes of the red fox, Chinese raccoon dog, and Japanese raccoon dog (noted in Figs. 2, 3, 5, 6, 7, 8, 9, and 10) may indicate the presence of additional breakpoint regions whose boundaries are beyond the limits of detection of this study. Further investigation of the regions using higher resolution analysis would determine the precise size of all genomic regions of the dog that are duplicated in the wild canids.
While our data show that canid B chromosomes contain numerous regions shared with A chromosomes, it is unclear at this stage whether the duplicated sequences on the B chromosomes result in a corresponding increase in functional transcripts. Previous studies attempted to address this issue by evaluating the level of cKIT transcripts in cells with varying numbers of B chromosomes (Yudkin et al. 2007), but it was unclear whether the associated increase in transcripts in animals with more B chromosomes results in an increase in functional cKIT transcripts. Further examination of the duplicated regions identified in the present study would help clarify whether the B chromosomes contain functional copies of sequence present on A chromosomes.
Discrepancies with prior studies
We identified several discrepancies between our data and those published previously. First, the proximal one third of NPRp2p, a region estimated to be ∼20 Mb (by comparison to the size of the CFA25 ECCS), did not contain the CFA25 ECCS reported by Nie et al. (2003). Since the limit of resolution for chromosome painting is approximately 5–10 Mb, it is possible that some of this region of NPRp2p erroneously appeared to contain hybridization signal in chromosome painting experiments. Although individuals may have variation in sequence repeats in this region, our data suggest that NPRp2pprox contains genetic information not represented by our domestic dog BAC panels. Secondly, two intrachromosomal breakpoints were identified in the present study, in ECCSs shared between CFA5 and UCI4, and between CFA10 and UCI14, which were not evident in previous studies, where chromosome paints for CFA5 and CFA10 each hybridized to just single gray fox chromosomes. The presence of these breakpoints, and the consequent internal sequence rearrangements, is supported by visible differences in the DAPI banding patterns of UCI4 and UCI14, relative to those of the corresponding dog chromosomes. Further discrepancies (highlighted in Fig. 4 and ESM Tables 1–4) for the gray fox are likely due to the presence of a translocation event in the individual described in Graphodatsky et al. 2008.
In previous studies of the crab-eating fox, the CFA9 ECCS was divided across two chromosomes (Nash et al. 2001). In the present study, the data show that crab-eating fox cells contained three chromosome structures with CFA9 sequence, which resulted from at least seven breakpoints compared with CFA9 (Fig. 6). This includes the previously reported breaks and two additional breaks at CFA9;13.1–14.5Mb and CFA9;16.8–17.8Mb. Regardless, both the crab-eating fox and the hoarey fox contain multiple breakpoints in CFA9 ECCSs that are specific to their branches of the SA canids, which would likely mean that these breakpoints arose after divergence from the other SA canids based on accepted phylogenetic groupings (Lindblad-Toh et al. 2005). Figure 11 illustrates an accepted phylogenetic tree of the Canidae (Lindblad-Toh et al. 2005) annotated with the ECCS patterns described in this study.
Phylogenetic tree shown with ECCSs. a The accepted phylogenetic tree of the Canidae (Lindblad-Toh et al. 2005) annotated with the ECCS patterns described in the text. We propose that the mammalian ancestor organized CFA9 ECCSs together as one ECCS, while CFA1, 13, 18, and 19 ECCSs were organized as two ECCSs each. The changes in ECCS organization during speciation are noted in bold for each branch of the tree. In addition, the CFA19a/32, CFA13a/29, and CFA28/37 associations are noted. Species within each colored region (i.e., CBR and SVE) share the same breakpoints and ECCS organization. The CFA13a/32 association was either present in the ancestral canid karyotype but then lost in the South Amercian and wolf-like canids or arose in the island fox and fox-like canid branches separately. Likewise, the CFA28/37 association could have been present in the ancestral canid karyotype but was lost in the VMA/VVU/FZE, CTH/DVE, and wolf-like branches or arose separately in the NPRp/NPRv, CBR/SVE, OME, and UCI branches. Since the CFA19a/32 association is present in primates and canids but not other mammals, this association either arose separately in the ancestral canid karyotype and ancestral primate karyotypes or was present in the mammalian ancestor and was lost in all other non-canid, non-primate modern mammals. We suggest that two centromerization events occurred, one in the VMA/VVU/FZE branch to separate CFA13a and CFA29 ECCSs and one in the CTH/DVE branch to separate the CFA19a/CFA19b ECCSs that fused in the ancestor to South American and wolf-like canids. b A more parsimonious tree (changes noted in red) may move the OME branch to the location noted with an asterisk in a, which would mean that the fusion of CFA18a/CFA18b and association of CFA28/37 common in NPRp, NPRv, and OME occurred once rather than separately in the NPRp/NPRv and OME branches
Support for the fragile breakage model
The FBM proposed by Pevzner and Tesler (2003) proposes that breakpoints in evolution or disease neither occur at random sites nor are uniformly distributed across genomes as described in the random breakage model (Ohno 1973). Instead, Pevzner and Tesler (2003) proposed that breakpoints evident in evolution and disease are reused/shared. They concluded that there must be an evolutionarily maintained feature of such regions that results in a localized increase in breakpoints. Although still controversial, numerous groups have been able to provide support for the model by comparing varying chromosome structures and sequence information of several mammals (human, dog, mouse, rat, primates) and other organisms (Murphy et al. 2005; Ruiz-Herrera et al. 2005). By performing multiple pairwise alignments between the sequences from multiple organisms, breakpoints deemed EBRs were identified and described as reused if the EBR was common across species from different clades (Murphy et al. 2005; Larkin et al. 2009). Studies examining the breakpoints observed in cancer and speciation (Murphy et al. 2005) confirm that this theory applies to mammals, and our work supports this theory in the Canidae.
Comparison of common breakpoint regions with cancer-associated genomic aberrations
The common evolutionary breakpoint regions identified in the present study were evaluated further to investigate any potential correlation with genomic aberrations identified in spontaneous canine cancers. This revealed recurrent DNA copy number imbalances that were highly localized to the genomic intervals defining the CFA9 and CFA19 breakpoint regions, within a range of canine cancers. Aberrations of the CFA9 region (20.2–22.4 Mb) were evident in non-Hodgkin’s lymphoma (Thomas et al. 2011), as well as in our ongoing studies of leukemia and transitional cell carcinoma of the bladder. Recurrent genomic imbalances within the CFA19 breakpoint region (21.8–25.1 Mb) were again evident in lymphoma and leukemia and also among cases of appendicular osteosarcoma (Angstadt et al. 2011) and hemangiosarcoma (unpublished). Osteosarcoma cases also exhibited aberrations consistent with the CFA1 (27.6–28.7 Mb) breakpoint and with the CFA18 (27.4–29.0 Mb) region (Angstadt et al. 2011), in common with lymphoma (Thomas et al. 2011). These findings provide intriguing new data supportive of an association between cancer-associated genomic instability and karyotypic rearrangement during speciation. Interestingly, the CFA9, 18, and 19 EBR intervals coincide with defined regions of natural copy number variation identified within normal domestic dog populations, providing further evidence for their inherent instability.
While there was evidence of recurrent subregional genomic aberrations in canine cancers that were highly localized to the CFA1, 9, 18, and 19 breakpoint intervals, this was not apparent for the two CFA13 breakpoint regions. Genomic gain of CFA13 is the most widely encountered copy number aberration identified across a wide range of canine tumor types, evident in approximately 50–60% of B cell lymphomas (Thomas et al. 2011), gliomas (Thomas et al. 2009), and osteosarcomas (Angstadt et al. 2011) and approximately 20–25% of leukemias and hemangiosarcomas (unpublished), and is often associated with concomitant overrepresentation of the MYC and cKIT oncogenes. This aberration typically spans the majority of the length of the chromosome and therefore may mask subregional genomic imbalances involving evolutionary breakpoint regions. The high incidence of CFA13 in canine cancers is of particular interest in light of evidence from the present study revealing that sequence consistent with regions of CFA13 is present on all B chromosomes of the red fox, Chinese raccoon dog, and Japanese raccoon dog. Since the CFA13 ECCS is also evident within the A chromosome complement of these species, their normal karyotypes therefore exhibit overrepresentation of these subchromosomal regions relative to the normal domestic dog genome. Each B chromosome harbors at least one copy of the cKIT oncogene, with tandem duplication of this locus evident on NPRp Bii. This gains greater significance with the discovery of additional cancer-associated gene sequences on supernumerary chromosomes of this species, with the RET oncogene also being detected on NPRpBi and Bii chromosomes. Furthermore, both NPRpBi and Bii exhibit multiple copies of LRIG1, a regulator of cell signaling by receptor tyrosine kinases, for which genomic dosage and transcriptional dysregulation has been implicated in a variety of human cancers (Yang et al. 2006; Ljuslinder et al. 2007, 2009). In a previous study, we identified disruption of the LRIG1 locus and neighboring regions of CFA20 in canine soft tissue sarcoma (Thomas et al. 2008), resulting in extensive reorganization of chromosome structure and amplification of the LRIG1 gene. Figure 10 shows that the B chromosomes of both raccoon dog species comprise extensive tracts of as-yet undefined sequence, which did not share homology with any probes from the 10-Mb dog BAC panel. Targeted evaluation of these regions will be required in order to define their composition and establish whether they share additional features in common with the architecture of canine cancer genomes.
Conclusions
The domestic dog is one of 34 living species comprising the family Canidae. The high-quality genome sequence assembly of the domestic dog (Lindblad-Toh et al. 2005) provides new opportunities for information transfer to other canids. We have used molecular cytogenetic reagents developed for the domestic dog to compare the genome architecture of 11 other members of the Canidae, thus facilitating the transfer of the enormous amount of domestic dog genomics data. Newly discovered breakpoints and duplicated regions were identified in the canid species studied, including 37 new regions of ECCSs present on the B chromosomes of the red fox and Chinese raccoon dog. Six breakpoint regions shared between the 11 species in relation to the domestic dog support the fragile breakage model as proposed by Pevzner and Tesler (Pevzner and Tesler 2003; Murphy et al. 2005; Ruiz-Herrera et al. 2005) and also correspond to known breakpoints in synteny between dog, human, and mouse (Derrien et al. 2007). It has been shown that the sequence is duplicated in breakpoint regions (Bailey et al. 2004), and so the 25 duplicated regions reported here could indicate additional breakpoint regions yet to be explored at an adequate resolution. Our results have enabled us to generate a set of integrated physical genome maps of three wild canid species (red fox, Chinese raccoon dog, and gray fox) and defined shared breakpoints across eight additional species. This study provides the most extensive B chromosome sequence identification to date, which strongly supports the unique characteristics of canid B chromosomes. We believe that evolutionary breakpoints associated with speciation in the Canidae also are associated with genomic instability in cancer. Taking into account these new data, we propose that the complex rearrangements in the Canidae occur due to fragile regions described by the fragile breakpoint model and, furthermore, that the B chromosomes present in three of the species studied may represent artifacts of this rearrangement and serve to sequester additional copies cancer-associated genes often found at breakpoints.
Abbreviations
- BAC:
-
Bacterial artificial chromosome
- BLAST:
-
Basic local alignment search tool
- CBR:
-
Chrysocyon brachyurus (maned wolf)
- CFA:
-
Canis familiaris (domestic dog)
- CHORI:
-
Children’s Hospital Oakland Research Institute
- cKIT :
-
Cellular homolog for feline sarcoma viral oncogene vKIT
- CTH:
-
Cerdocyon thous (crab-eating fox)
- DNA:
-
Deoxyribonucleic acid
- DVE:
-
Dusicyon vetulus (hoarey fox)
- EBR:
-
Evolutionary breakpoint region
- ECCS:
-
Evolutionarily conserved chromosomal segment
- FBM:
-
Fragile breakage model
- FISH:
-
Fluorescence in situ hybridization
- FZE:
-
Fennecus zerda (fennec fox)
- LRIG1 :
-
Leucine-rich repeats and immunoglobulin-like domain protein 1
- NPRp:
-
Nyctereutes procynoides procynoides (Chinese raccoon dog)
- NPRv:
-
Nyctereutes procynoides viverrinus (Japanese raccoon dog)
- OME:
-
Otocyon megalotis (bat-eared fox)
- RET :
-
Rearranged during transfection
- SA:
-
South American
- SVE:
-
Speothus venaticus (bush dog)
- UCI:
-
Urocyon cinereogenteus (gray fox)
- VMA:
-
Vulpes macrotis (kit fox)
- VVU:
-
Vulpes vulpes (red fox)
References
Alekseyev MA (2008) Multi-break rearrangements and breakpoint re-uses: from circular to linear genomes. J Comput Biol 15(8):1117–1131
Alekseyev MA, Pevzner PA (2007) Are there rearrangement hotspots in the human genome? PLoS Comp Biol 3(11):e209
Alekseyev M, Pevzner P (2010) Comparative genomics reveals birth and death of fragile regions in mammalian evolution. Genome Biol 11(11):R117–R117
Angstadt A, Motsinger-Reif A, Thomas R et al. (2011) Characterization of canine osteosarcoma by array comparative genomic hybridization and RT-qPCR: signatures of genomic imbalance in canine osteosarcoma parallel the human counterpart. Genes, Chromosomes, Cancer. doi:10.1002/gcc.20908
Bailey JA, Baertsch R, Kent WJ, Haussler D, Eichler EE (2004) Hotspots of mammalian chromosomal evolution. Genome Biol 5(4):R23
Bardeleben C, Moore RL, Wayne RK (2005) A molecular phylogeny of the Canidae based on six nuclear loci. Mol Phylogenet Evol 37(3):815–831
Breen M, Bullerdiek J, Langford CF (1999) The DAPI banded karyotype of the domestic dog (Canis familiaris) generated using chromosome-specific paint probes. Chromosome Res 7(5):401–406
Breen M, Hitte C, Lorentzen TD et al (2004) An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics 5(1):65
Camacho JP, Sharbel TF, Beukeboom LW (2000) B-chromosome evolution. Philos Trans R Soc Lond, B, Biol Sci 355(1394):163–178
Chemitiganti S, Verma RS, Silver RT, Coleman M, Dosik H (1985) Unusual translocations involving chromosomes 12;22 and 9;12 in a case of chronic myelogenous leukemia. Cancer Genet Cytogenet 14(1–2):61–65
Chen-Liu LW, Huang BC, Scalzi JM et al (1995) Selection of hybrids by affinity capture (SHAC): a method for the generation of cDNAs enriched in sequences from a specific chromosome region. Genomics 30(2):388–392
Derrien T, Andre C, Galibert F, Hitte C (2007) AutoGRAPH: an interactive web server for automating and visualizing comparative genome maps. Bioinformatics 23:498–499
Graphodatsky AS, Yang F, O'Brien PC et al (2000) A comparative chromosome map of the Arctic fox, red fox and dog defined by chromosome painting and high resolution G-banding. Chromosome Res 8(3):253–263
Graphodatsky AS, Kukekova AV, Yudkin DV et al (2005) The proto-oncogene C-KIT maps to canid B-chromosomes. Chromosome Res 13(2):113–122
Graphodatsky AS, Perelman PL, Sokolovskaya N (2008) Phylogenomics of the dog and fox family (Canidae, Carnivora) revealed by chromosome painting. Chromosome Res 16:129–146
Grzes M, Nowacka-Woszuk J, Szczerbal I, Czerwinska J, Gracz J, Switonski M (2009) A Comparison of coding sequence and cytogenetic localization of the myostatin gene in the dog, red fox, Arctic fox and Chinese raccoon dog. Cytogenet Genome Res 126:173–179
Haig D (1999) A brief history of human autosomes. Philos Trans R Soc Lond, B, Biol Sci 354(1388):1447–1470
Larkin DM, Pape G, Donthu R, Auvil L, Welge M, Lewin HA (2009) Breakpoint regions and homologous synteny blocks in chromosomes have different evolutionary histories. Genome Res 19(5):770–777
Lin KW, Yan J (2008) Endings in the middle: current knowledge of interstitial telomeric sequences. Mutat Res 658(1-2):95–110
Lindblad-Toh K, Wade CM, Mikkelsen TS et al (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438(7069):803–819
Ljuslinder I, Golovleva I, Palmqvist R et al (2007) LRIG1 expression in colorectal cancer. Acta Oncol 46(8):1118–1122
Ljuslinder I, Golovleva I, Henriksson R, Grankvist K, Malmer B, Hedman H (2009) Co-incidental increase in gene copy number of ERBB2 and LRIG1 in breast cancer. Breast Cancer Res 11(3):403
Mäkinen A (1985) The standard karyotype of the silver fox (Vulpes fulvus Desm.). Committee for the Standard Karyotype of Vulpes fulvus Desm. Hereditas 103(2):171–176
Mäkinen A, Kuokkanen MT, Valtonen M (1986) A chromosome-banding study in the Finnish and the Japanese raccoon dog. Hereditas 105(1):97–105
Marshall OJ, Chueh AC, Wong LH, Choo KHA (2008) Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet 82(2):261–282
Mrózek K, Karakousis CP, Perez-Mesa C, Bloomfield CD (1993) Translocation t(12;22)(q13;q12.2-12.3) in a clear cell sarcoma of tendons and aponeuroses. Genes, Chromosom Cancer 6(4):249–252
Murphy WJ, Larkin DM, Everts-van der Wind A et al (2005) Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science 309(5734):613–617
Nash WG, Menninger JC, Wienberg J, Padilla-Nash HM, O'Brien SJ (2001) The pattern of phylogenomic evolution of the Canidae. Cytogenet Cell Genet 95(3–4):210–224
Nie W, Wang J, Perelman P, Graphodatsky AS, Yang F (2003) Comparative chromosome painting defines the karyotypic relationships among the domestic dog, Chinese raccoon dog and Japanese raccoon dog. Chromosome Res 11(8):735–740
Ohno S (1973) Ancient linkage groups and frozen accidents. Nature 244(5414):259–262
Peng Q, Pevzner PA, Tesler G (2006) The fragile breakage versus random breakage models of chromosome evolution. PLoS Comp Biol 2(2):e14
Pevzner P, Tesler G (2003) Human and mouse genomic sequences reveal extensive breakpoint reuse in mammalian evolution. Proc Natl Acad Sci USA 100(13):7672–7677
Pieńkowska-Schelling A, Schelling C, Zawada M, Yang F, Bugno M, Ferguson-Smith M (2008) Cytogenetic studies and karyotype nomenclature of three wild canid species: maned wolf (Chrysocyon brachyurus), bat-eared fox (Otocyon megalotis) and fennec fox (Fennecus zerda). Cytogenet Genome Res 121(1):25–34
Piras FM, Nergadze SG, Magnani E et al (2010) Uncoupling of satellite DNA and centromeric function in the genus Equus. PLoS Genet 6(2):1–10
Ruiz-Herrera A, García F, Mora L, Egozcue J, Ponsà M, Garcia M (2005) Evolutionary conserved chromosomal segments in the human karyotype are bounded by unstable chromosome bands. Cytogenet Genome Res 108(1–3):161–174
Schröder C, Bleidorn C, Hartmann S, Tiedemann R (2009) Occurrence of Can-SINEs and intron sequence evolution supports robust phylogeny of pinniped carnivores and their terrestrial relatives. Gene 448(2):221–226
Thomas R, Duke SE, Bloom SK et al (2007) A cytogenetically characterized, genome-anchored 10-Mb BAC set and CGH array for the domestic dog. J Hered 98(5):474–484
Thomas R, Duke SE, Karlsson EK et al (2008) A genome assembly-integrated dog 1 Mb BAC microarray: a cytogenetic resource for canine cancer studies and comparative genomic analysis. Cytogenet Genome Res 122(2):110–121
Thomas R, Duke SE, Wang HJ et al (2009) 'Putting our heads together': insights into genomic conservation between human and canine intracranial tumors. J Neurooncol 94(3):333–349
Thomas R, Seiser EL, Motsinger-Reif A et al (2011) Refining tumor-associated aneuploidy through 'genomic recoding' of recurrent DNA copy number aberrations in 150 canine non-Hodgkin lymphomas. Leuk Lymphoma 52(7):1321–1335
Trifonov VA, Perelman PL, Kawada SI, Iwasa MA, Oda SI, Graphodatsky AS (2002) Complex structure of B-chromosomes in two mammalian species: Apodemus peninsulae (Rodentia) and Nyctereutes procyonoides (Carnivora). Chromosome Res 10(2):109–116
Vujosević M, Blagojević J (2004) B chromosomes in populations of mammals. Cytogenet Genome Res 106(2–4):247–256
Wade CM, Giulotto E, Sigurdsson S et al (2009) Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 326(5954):865–867
Ward OG, Wurster-Hill DH, Ratty FJ, Song Y (1987) Comparative cytogenetics of Chinese and Japanese raccoon dogs, Nyctereutes procyonoides. Cytogenet Cell Genet 45(3–4):177–186
Wayne RK (1993) Molecular evolution of the dog family. Trends Genet 9(6):218–224
Wayne RK, Ostrander EA (1999) Origin, genetic diversity, and genome structure of the domestic dog. Bioessays 21(3):247–257
Wayne RK, Nash WG, O'Brien SJ (1987a) Chromosomal evolution of the Canidae. I. Species with high diploid numbers. Cytogenet Cell Genet 44(2–3):123–133
Wayne RK, Nash WG, O'Brien SJ (1987b) Chromosomal evolution of the Canidae. II. Divergence from the primitive carnivore karyotype. Cytogenet Cell Genet 44(2–3):134–141
Wayne RK, Geffen E, Girman DJ, Koepfli KP, Lau LM, Marshall CR (1997) Molecular systematics of the Canidae. Syst Biol 46(4):622–653
Yang F, O'Brien PC, Milne BS et al (1999) A complete comparative chromosome map for the dog, red fox, and human and its integration with canine genetic maps. Genomics 62(2):189–202
Yang W-M, Yan Z-J, Ye Z-Q, Guo D-S (2006) LRIG1, a candidate tumour-suppressor gene in human bladder cancer cell line BIU87. BJU Int 98(4):898–902
Yudkin DV, Trifonov VA, Kukekova AV et al (2007) Mapping of KIT adjacent sequences on canid autosomes and B chromosomes. Cytogenet Genome Res 116(1–2):100–103
Zrzavy J, Ricankova V (2004) Phylogeny of recent Canidae (Mammalia, Carnivora): relative reliability and utility of morphological and molecular datasets. Zoologica Scripta 33:311–333
Acknowledgments
This study was supported by a grant from the Morris Animal Foundation awarded to MB (D08ZO-022). SEDB was funded in part by the Comparative Biomedical Sciences Graduate Program at NCSU. RW was supported by funds from the National Science Foundation (DEB0614585), and ASG and VAT were supported by funds from the Program on Molecular and Cellular Biology (MCB) and Russian Foundation of Basic Research (RFBR).
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Herbert Macgregor
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
a Thirteen BAC probes spaced ∼10 Mb apart along the length of CFA1 were hybridized together onto CFA chromosome spreads. The location and orientation of the panel represents the CFA1 ECCS. b The CFA1 probe panel was hybridized to VVU5 and VVU1, revealing a breakpoint between probes representing CFA1;22.3 Mb (yellow) and CFA1;32. 3Mb (purple). Regions of CFA1 ECCSs on either side of the breakpoint are indicated with a suffix (i.e., CFA1a and CFA1b). Through application of the multicolor labeling strategy, the location and orientation of each CFA1 ECCS was evident (inset). Scale bar, 10 μm (JPEG 2026 kb)
Supplemental Table 1–4
CFA regions corresponding to ECCSs are listed with the corresponding regions of red fox (VVU), Chinese raccoon dog (NPRp), and gray fox (UCI) indicated. Table 1 is sorted by dog region, Table 2 by red fox chromosome locations, Table 3 by Chinese raccoon dog chromosome locations, and Table 4 by gray fox chromosome locations. Regions with additional hybridization to B chromosomes of red fox and Chinese raccoon dog are noted with asterisks. While we used the nomenclature of Wayne et al. (1987a) for gray fox chromosomes, the final column lists the nomenclature used in Graphodatsky et al. (2008). The gray fox cells described in Graphodatsky et al. (2008) contain a translocation relative to the cells described in this text. The regions involved (CFA7, 28, 37 ECCSs on UCI12, 15) are noted with double asterisks here, shown in Fig. 4 and discussed in the text (XLS 47 kb)
Rights and permissions
About this article
Cite this article
Duke Becker, S.E., Thomas, R., Trifonov, V.A. et al. Anchoring the dog to its relatives reveals new evolutionary breakpoints across 11 species of the Canidae and provides new clues for the role of B chromosomes. Chromosome Res 19, 685–708 (2011). https://doi.org/10.1007/s10577-011-9233-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-011-9233-4