Abstract
Convincing evidence has shown that microRNAs (miRNAs) are involved in the pathogenesis of stroke. This study aimed to examine whether miRNA biogenesis genes polymorphisms are associated with risk of large artery atherosclerosis (LAA) stroke. Three polymorphisms (DROSHA rs10719 T>C, RAN rs3803012 A>G, and PIWIL1 rs10773771 C>T) were screened by certain criteria. A total of 1,785 (710 cases and 1,075 controls) study subjects were included in this study. We found that rs10773771 CC genotype was associated with a decreased risk of LAA stroke (CC vs. TT/CT: OR 0.63, 95% CI 0.46–0.86, P = 3 × 10–3). In silico analysis suggested that rs10773771 can change the mRNA secondary structure of PIWIL1 and affect the binding of the miRNAs and regulatory motifs to the 3′-UTR of PIWIL1. Expression quantitative trait loci analysis showed that rs10773771 could change the expression of PIWIL1 in human skin (P = 1.534 × 10–10) and thyroid tissues (P = 4.869 × 10–6). These findings suggested that PIWIL1 rs10773771 may be associated with a decreased risk of LAA stroke.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Stroke is a complex disease caused by environmental and genetic factors. Existing studies have shown that genetic factors play a substantial role in the risk of stroke (Bak et al. 2002; Jerrard-Dunne et al. 2003). About 80% of stroke is ischemic stroke (IS). The most common subtypes of IS are large artery atherosclerosis (LAA), cardioembolic stroke (CES), and small vessel disease (SVD) (Jerrard-Dunne et al. 2003). The genetic heritability varies by different subtypes of IS, of which LAA has the highest heritability of all IS subtypes (40.3%) (Bevan et al. 2012).
MicroRNAs (miRNAs) are a class of small noncoding RNAs that can bind to the 3′-untranslated regions (3′-UTRs) in mRNAs regulating their expression (Ambros 2004). Increasing evidences suggest that miRNAs have been involved in the pathogenesis of many diseases, such as cancer (Lu et al. 2005), schizophrenia (Beveridge et al. 2010), Parkinson's diseases (Kim et al. 2007), and stroke (Khoshnam et al. 2017). The biosynthesis of miRNAs is a complex process involving multiple miRNA biogenesis genes (Ambros 2004). Briefly, primary miRNA is produced in the nucleus by RNA polymerase II. Then, the primary miRNA is further processed into the precursor miRNA (pre-miRNA) by RNase III DROSHA and RNA binding protein DGCR8. The pre-miRNA is exported to the cytoplasm via Ran-GTPase (RAN) and Exporti-5 (XPO5). At last, the pre-miRNAs are processed to produce the mature miRNAs through a multiprotein complex that includes DICER, PIWIL1, GEMIN, and RAN (Bartel 2004).
We conducted this case–control study to investigate whether there is an association between three known miRNA biogenesis genes polymorphisms (DROSHA rs10719 T > C, RAN rs3803012 A > G, and PIWIL1 rs10773771 C > T) and LAA stroke risk.
Materials and Methods
Study Subjects
This case-control study was approved by the institutional review boards of the local participating hospitals (2017NZGKJ-041). Written informed consent was obtained from all participants. This is a case–control study designed with samples from the Chinese Han population. Our study comprised 710 LAA patients and 1,075 controls. All LAA patients had a focal neurologic deficit lasting > 24 h, which were confirmed by computed tomography (CT) or magnetic resonance imaging (MRI). All LAA patients were diagnosed according to the Trial of Org 10172 in Acute Stroke Treatment (TOAST) criteria (Adams et al. 1993). All patients with tumor, autoimmune diseases or hemorrhagic diseases were excluded. The healthy controls were recruited from those presenting at local hospitals for health physical examinations during the same period. The controls did not have neurological diseases and cerebrovascular disease as well as cardiovascular diseases according to clinical examinations or history taken. Risk factors of LAA stroke patients and controls were determined according to established criteria. Hypertension was defined as systolic pressure ≥ 140 mmHg and diastolic pressure ≥ 90 mmHg on at least three separate occasions or currently taking anti-hypertensive medications or has a history of hypertension. Diabetes mellitus was defined as fasting glucose ≥ 126 mg/dl or taking anti-diabetic medications or has a history of diabetes.
SNP Selection and Genotyping
The single-nucleotide polymorphisms (SNPs) were selected from eight known miRNA biogenesis genes (DROSHA, DGCR8, RAN, XPO5, DICER, PIWIL1, GEMIN3, GEMIN4). The selection criteria were as follows: (1) in the 3′-UTR region; (2) the minor allele frequency of each SNP ≥ 0.05 in 1000 Genome CHB (Han Chinese in Beijing) data on the GRCh37 reference assembly (www.1000genomes.org/); (3) each SNP can influence the miRNA binding predicted by miRanda (John et al. 2004) and miRbase (Griffiths-Jones et al. 2008) database using the SNPinfo Web Server (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html) (Xu and Taylor 2009). A total of three SNPs (DROSHA rs10719, RAN rs3803012, and PIWIL1 rs10773771) met the selection criteria described above and were included in this study. Genomic DNA was isolated from whole blood. Genotyping for DROSHA rs10719, RAN rs3803012, and PIWIL1 rs10773771 was conducted using SNPscan technology, with technical assistance from Center for Genetic and Genomic Analysis, Genesky Biotechnologies Inc. (Shanghai, China) (Supplementary Figs. 1–3). For quality control and validation purposes, a random 5% of sample was repeated genotyping to check for consistency, and the results were 100% consistent.
Statistical Analysis
Categorical variables were presented as number (percentage) and continuous variables as mean (SD, standard deviation). Categorical variables were compared with Chi-square test and continuous variables with Student’s t test. The association between microRNA biogenesis genes polymorphisms and LAA stroke was estimated with the odds ratio (OR) and 95% confidence interval (CI) by multivariate logistic regression model, adjusting for age, sex, hypertension and diabetes. The Hardy–Weinberg equilibrium, the association between these polymorphisms and LAA stroke were calculated by SNPStats web tool (http://bioinfo.iconcologia.net/SNPstats) (Sole et al. 2006). Goodness of fit was evaluated using Akaike information criterion (AIC) for selecting the best genetic model to the SNPs (Sole et al. 2006).
All reported P values are two-sided. P < 0.05 was deemed as statistical significance unless otherwise specified. All analyses were performed with IBM SPSS Statistics version 23.0 (Armonk, NY: IBM Corp.).
In Silico Analysis
To evaluate the potential function of the 3′-UTR SNP rs10773771, we conducted in silico analysis using RNAfold Web server (http://rna.tbi.univie.ac.at/) (Hofacker 2003), SNPinfo Web server (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html) (Xu and Taylor 2009) and HaploReg v4.1 (https://pubs.broadinstitute.org/mammals/haploreg/haploreg.php) (Ward and Kellis 2016; Kheradpour and Kellis 2014).
Expression Quantitative Trait Loci (eQTL)
For SNP rs10773771, we queried publicly available eQTL database GTEx V7 (https://www.gtexportal.org/home/) (The Genotype-Tissue Expression (GTEx) project 2013).
Results
miRNA Biogenesis Genes Polymorphisms and the Risk of LAA Stroke
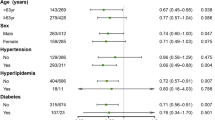
The clinical characteristics of LAA patients and controls in the study are summarized in Table 1. Distribution of the genotypes in the control groups was not deviated from Hardy–Weinberg equilibrium (P > 0.05 for all 3 SNPs). As shown in Table 2, no significant associations between rs10719, rs3803012 and LAA stroke were detected in the multivariate logistic regression analysis. However, the rs10773771 CC genotype was significantly associated with a decreased risk of LAA stroke in both codominant (P = 0.014) and recessive model (P = 3 × 10–3). According to the AIC values, recessive model is the best-fit model for rs10773771. In the recessive model, compared with TT/CT genotype, rs10773771 CC genotype was associated with significantly decreased risk of LAA stroke (OR 0.63, 95% CI 0.46–0.86, P = 3 × 10–3). Further stratification analysis suggested that this decreased risk was only observed in males (OR 0.63, 95% CI 0.42–0.96, P = 0.032), those aged above 66 (OR 0.52, 95% CI 0.33–0.82, P = 0.005), those without hypertension (OR 0.49, 95% CI 0.28–0.89, P = 0.018), and those without diabetes (OR 0.63, 95% CI 0.46–0.87, P = 0.005). (Supplementary Table 1).
In Silico Analysis
The mRNA secondary structure can affect the interactions between mRNA and miRNA. Therefore, we examined whether mRNA structure of PIWIL1 could be altered by rs10773771. After inputting a 200 bp fragment of the 3′-UTR of PIWIL1 including rs10773771 and changing the rs10773771 allele from C to T in the RNAfold, we found the change of the mRNA secondary structure, suggesting that it may affect the stability of mRNA, with an change of minimum free energy (MFE) from ∆G = − 35.5 kcal/mol to − 36.6 kcal/mol (Fig. 1a). We further investigated whether the rs10773771 could influence the combination of miRNAs to the 3′-UTR of PIWIL1 using the SNPinfo web server. The in-silico results suggested that the rs10773771 could affect the combination of three miRNAs (hsa-miR-1264, hsa-miR-340, hsa-miR-590-3p; Table 3) to the 3′-UTR of PIWIL1. In addition, based on the HaploReg v4.1, we identified that rs10773771 is likely to alter the binding affinity of regulatory motifs HNF1, Irx, Pou2f2, Sp100 and TATA (Table 4).
a Bioinformatics analysis for prediction of rs10773771 influence on RNA folding structures; MFE minimum free energy. b Association between rs10773771 genotypes and PIWIL1 expression in human skin tissues based on GTEx V7. c Association between rs10773771 genotypes and PIWIL1 expression in human thyroid tissues based on GTEx V7. d Allele frequency of rs10773771 among different populations from the 1000 genome project. Population descriptions: AMR American, AFR African, EUR European, EAS East Asian, SAS South Asian
eQTL Analysis
Based on gene expression data extracted from GTEx, rs10773771 was identified to be an eQTL for PIWIL1 in human skin (P = 1.534 × 10–10; Fig. 1b) and thyroid tissues (P = 4.869 × 10–6; Fig. 1c).
Discussion
In this study, we evaluated the associations between three miRNA biogenesis genes polymorphisms (DROSHA rs10719 T>C, RAN rs3803012 A>G, and PIWIL1 rs10773771 C>T) and the risk of LAA stroke. To the best of our knowledge, this is the first study to report the association between rs10773771 and LAA stroke. These findings indicated that genetic variant at PIWIL1 contribute to the development of LAA stroke.
There is mounting evidence that miRNAs have been involved in the pathogenesis of stroke (Khoshnam et al. 2017; Rink and Khanna 2011; Mirzaei et al. 2018). PIWIL1 plays an important role in stem cell renewal, division and RNA silencing (Hutvagner and Simard 2008). Moreover, it can also take part in the process of microRNA biogenesis that pre-miRNAs are processed to produce the mature miRNAs (Bartel 2004). Given the function of PIWIL1 in miRNA biogenesis and the involvement of miRNA in the pathogenesis of stroke, SNPs in PIWIL1 may affect the risk of stroke through regulating its function. The SNP rs10773771, which reached statistical significance in this study, is located in the 3′-UTR of PIWIL1. Furthermore, we performed a number of complementary in silico analysis to predict the potential function of the 3′-UTR SNP rs10773771. The SNP rs10773771 can change the mRNA secondary structure of PIWIL1. Additionally, the rs10773771 can influence the combination of three miRNAs (hsa-miR-1264, hsa-miR-340, hsa-miR-590-3p) to the 3′-UTR of PIWIL1. Reporter gene assays indicated that rs10773771 can change the binding ability of hsa-miR-1264 to the 3′-UTR of PIWIL1 in a previous study (Liu et al. 2013). The miR-1264/1298/448 cluster has been identified as a biomarker to monitor reperfusion after cerebral ischemia (Uhlmann et al. 2017). Based on the HaploReg v4.1, we also identified that rs10773771 is likely to alter the binding affinity of regulatory motifs HNF1, Irx, Pou2f2, Sp100 and TATA. Based on gene expression data extracted from GTEx, rs10773771 was identified to be eQTL for PIWIL1 in human skin (P = 1.534 × 10–10) and thyroid tissues (P = 4.869 × 10–6). The elucidation of its mechanism on the development of stroke needs further research. The allele frequency of rs10773771 from different population was different according to the 1000 genomes project (Fig. 1d), suggesting that ethnic differences may exert different influences on the association observed in our study.
In addition, we also found no significant association between DROSHA rs10719 T>C, RAN rs3803012 A>G and LAA stroke. DROSHA and RNA are essential for miRNA biogenesis. A Korean study has found that DROSHA rs10719 was associated with the risk of IS (Kim et al. 2018). However, this association was not detected in the LAA stroke, which was consistent with the results in our study. This is the first study investigating the association between RAN rs3803012 and LAA stroke. In the current study, we found that DROSHA rs10719 and RAN rs3803012 were not associated with the risk of LAA stroke, suggesting that these SNPs might not be associated with the susceptibility of LAA stroke in Chinese population.
The strengths of this study included the large sample size and in silico analyses. However, several limitations in this study should also be considered: (1) some risk factors of stroke, such as smoking and hyperlipoidemia were not included in the study, which may introduce additional biases. (2) Our study was a hospital-based case–control study, and the selection bias cannot be eliminated.
In summary, we have identified an association between rs10773771 and LAA stroke. The underlying mechanisms remain to be elucidated in the future.
References
Adams HP, Bendixen BH, Kappelle LJ, Biller J, Love BB, Gordon DL, Marsh EE (1993) Classification of subtype of acute ischemic stroke. Definitions for use in a multicenter clinical trial. TOAST. Trial of Org 10172 in Acute Stroke Treatment. Stroke 24(1):35–41
Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355. https://doi.org/10.1038/nature02871
Bak S, Gaist D, Sindrup SH, Skytthe A, Christensen K (2002) Genetic liability in stroke: a long-term follow-up study of Danish twins. Stroke 33(3):769–774
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Bevan S, Traylor M, Adib-Samii P, Malik R, Paul NL, Jackson C, Farrall M, Rothwell PM, Sudlow C, Dichgans M, Markus HS (2012) Genetic heritability of ischemic stroke and the contribution of previously reported candidate gene and genomewide associations. Stroke 43(12):3161–3167. https://doi.org/10.1161/STROKEAHA.112.665760
Beveridge NJ, Gardiner E, Carroll AP, Tooney PA, Cairns MJ (2010) Schizophrenia is associated with an increase in cortical microRNA biogenesis. Mol Psychiatry 15(12):1176–1189. https://doi.org/10.1038/mp.2009.84
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154-158. https://doi.org/10.1093/nar/gkm952
Hofacker IL (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31(13):3429–3431. https://doi.org/10.1093/nar/gkg599
Hutvagner G, Simard MJ (2008) Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 9(1):22–32. https://doi.org/10.1038/nrm2321
Jerrard-Dunne P, Cloud G, Hassan A, Markus HS (2003) Evaluating the genetic component of ischemic stroke subtypes: a family history study. Stroke 34(6):1364–1369. https://doi.org/10.1161/01.str.0000069723.17984.fd
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human MicroRNA targets. PLoS Biol 2(11):e363. https://doi.org/10.1371/journal.pbio.0020363
Kheradpour P, Kellis M (2014) Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res 42(5):2976–2987. https://doi.org/10.1093/nar/gkt1249
Khoshnam SE, Winlow W, Farbood Y, Moghaddam HF, Farzaneh M (2017) Emerging roles of microRNAs in ischemic stroke: as possible therapeutic agents. J Stroke 19(2):166–187. https://doi.org/10.5853/jos.2016.01368
Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A (2007) A MicroRNA feedback circuit in midbrain dopamine neurons. Science (New York, NY) 317(5842):1220–1224. https://doi.org/10.1126/science.1140481
Kim JO, Bae J, Kim J, Oh SH, An HJ, Han IB, Oh D, Kim OJ, Kim NK (2018) Association of MicroRNA biogenesis genes polymorphisms with ischemic stroke susceptibility and post-stroke mortality. J Stroke 20(1):110–121. https://doi.org/10.5853/jos.2017.02586
Liu L, An J, Liu J, Wen J, Zhai X, Liu Y, Pan S, Jiang J, Wen Y, Liu Z, Zhang Y, Chen J, Xing J, Ji G, Shen H, Hu Z, Fan Z (2013) Potentially functional genetic variants in microRNA processing genes and risk of HBV-related hepatocellular carcinoma. Mol Carcinog 52(Suppl 1):E148-154. https://doi.org/10.1002/mc.22062
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838. https://doi.org/10.1038/nature03702
Mirzaei H, Momeni F, Saadatpour L, Sahebkar A, Goodarzi M, Masoudifar A, Kouhpayeh S, Salehi H, Mirzaei HR, Jaafari MR (2018) MicroRNA: relevance to stroke diagnosis, prognosis, and therapy. J Cell Physiol 233(2):856–865. https://doi.org/10.1002/jcp.25787
Rink C, Khanna S (2011) MicroRNA in ischemic stroke etiology and pathology. Physiol Genomics 43(10):521–528. https://doi.org/10.1152/physiolgenomics.00158.2010
Sole X, Guino E, Valls J, Iniesta R, Moreno V (2006) SNPStats: a web tool for the analysis of association studies. Bioinformatics (Oxford, England) 22(15):1928–1929. https://doi.org/10.1093/bioinformatics/btl268
The Genotype-Tissue Expression (GTEx) project (2013). Nature genetics 45(6):580–585. https://doi.org/10.1038/ng.2653
Uhlmann S, Mracsko E, Javidi E, Lamble S, Teixeira A, Hotz-Wagenblatt A, Glatting KH, Veltkamp R (2017) Genome-wide analysis of the circulating miRNome after cerebral ischemia reveals a reperfusion-induced MicroRNA cluster. Stroke 48(3):762–769. https://doi.org/10.1161/strokeaha.116.013942
Ward LD, Kellis M (2016) HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res 44(D1):D877-881. https://doi.org/10.1093/nar/gkv1340
Xu Z, Taylor JA (2009) SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res 37:W600-605. https://doi.org/10.1093/nar/gkp290
Acknowledgements
This study was supported by National Natural Science Foundation of China (Grant Nos. 81771285 and 81530038).
Author information
Authors and Affiliations
Contributions
MW: study design, statistical analysis, and manuscript drafting. JG: study design, statistical analysis, manuscript drafting. CS: data collection. WT: statistical analysis. XX: statistical analysis. ZZ: study design, interpretation of results, and critical revision of manuscript. XL: study design, interpretation of results, and critical revision of manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
This study was approved by the institutional review boards of the local participating hospitals.
Informed consent
Informed consent was obtained from all participants.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (TIF 42 KB)
Genotyping of rs10773771 was undertaken using SNPscan technology (Genesky Biotechnologies Inc. Shanghai, China).
Supplementary file2 (PDF 68 KB)
Genotyping of rs10719 was undertaken using SNPscan technology (Genesky Biotechnologies Inc. Shanghai, China).
Supplementary file3 (PDF 71 KB)
Genotyping of rs3803012 was undertaken using SNPscan technology (Genesky Biotechnologies Inc. Shanghai, China).
Rights and permissions
About this article
Cite this article
Wang, M., Gu, J., Shen, C. et al. Association of MicroRNA Biogenesis Genes Polymorphisms with Risk of Large Artery Atherosclerosis Stroke. Cell Mol Neurobiol 42, 1801–1807 (2022). https://doi.org/10.1007/s10571-021-01057-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-021-01057-8