Abstract
Glioblastoma multiforme (GBM) is the most aggressive and common brain tumor in adults. Sorafenib, a multi-kinase inhibitor, has been shown to inhibit cell proliferation and induce apoptosis through inhibition of STAT3 signaling in glioblastoma cells and in intracranial gliomas. However, sorafenib also induces cell autophagy. Due to the dual roles of autophagy in tumor cell survival and death, the therapeutic effect of sorafenib on glioblastoma is uncertain. Here, we combined sorafenib treatment in GBM cells (U373 and LN229) and tumors with the autophagy inhibitor chloroquine. We found that blockage of autophagy further inhibited cell proliferation and migration and induced cell apoptosis in vitro and in vivo. These findings suggest the possibility of combination treatment with sorafenib and autophagy inhibitors for GBM.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Glioblastoma multiforme (GBM) is the most common primary brain tumor and the most aggressive glioma in adults. Despite surgery, chemotherapy, and radiation therapy, the prognosis remains poor due to the active angiogenesis, rapid progression, and the emergence of resistance toward conventional therapy. The median overall survival of GBM patients is 12–14 months (Van Meir et al. 2010; Wen and Kesari 2008).
Several molecular alterations in signaling pathways are commonly found in GBM. These include the pathways of phosphatidylinositol-3 kinase-Akt-mammalian target of rapamycin (PI3K/Akt/mTOR), Ras-Raf-mitogen-activated protein kinase (Ras/Raf/MAPK), vascular endothelial growth factor (VEGF), and platelet-derived growth factor (PDGF) (Cancer Genome Atlas Research N 2008; Fan et al. 2007; Furnari et al. 2007; Van Meir et al. 2010). Sorafenib (Nexavar, BAY43-9006), a potent small-molecule multikinase inhibitor, was originally developed as an inhibitor of Raf kinase, an essential kinase of the MAPK pathway, and inhibits cancer cell proliferation by targeting the MAPK pathway (Liu et al. 2006; Peereboom et al. 2013). Sorafenib also inhibits the receptor tyrosine kinases (RTKs), including vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR). By targeting the RTKs, sorafenib can prevent angiogenesis (Lee et al. 2012; Liu et al. 2006; Peereboom et al. 2013; Qin et al. 2013). Like the studies of sorafenib of U87, U251 cell lines and primary cultures of GBM (Carra et al. 2013; Siegelin et al. 2010), here we also found that sorafenib can inhibit the phosphorylation of STAT3 (Tyr 705) and Akt (Ser 473) and down-regulate the protein level of anti-apoptotic protein, myeloid cell leukemia 1 (Mcl-1) in U373, which can partially explain the antitumor effect of sorafenib. Sorafenib has shown clinical benefits for different types of cancers, including hepatocellular carcinoma, renal cell carcinoma, lung cancer, thyroid cancer, and breast cancer (Kruijtzer et al. 2002; Liu et al. 2006; Xu et al. 2014; Zhao et al. 2014). For GBM, sorafenib was shown to inhibit cell proliferation and induces apoptosis and autophagy in glioma cells (Hamed et al. 2014; Kim et al. 2007). However, a recent phase I/II study of sorafenib in combination with temsirolimus for recurrent glioblastoma or gliosarcoma showed no clinical advantage due to minimal activity and substantial toxicity of the dose used in the study (Galanis et al. 2013; Hottinger et al. 2014; Karajannis et al. 2014; Kuppens et al. 2007; Lee et al. 2012).
Autophagy is an evolutionarily conserved intracellular self-defense process. In general, autophagy functions to maintain cellular homeostasis by mediating the removal of dysfunctional or damaged organelles that are digested and recycled for cellular metabolic needs (Gozuacik and Kimchi 2004; Murrow and Debnath 2013; White 2012). In the context of diseases, autophagy has dual functions. On the one hand, activated autophagy can cause the death of certain types of cancer cells like breast cancer cells MCF-7 via the activation of AMP-activated protein kinase (Kim et al. 2015). On the other hand, it can protect cancer cells under therapeutic interventions and mediate resistance to anticancer therapies (Murrow and Debnath 2013; Nagelkerke et al. 2014; White 2012). The role of autophagy in tumor is complicated and may have opposite consequences depending on the circumstances (Hippert et al. 2006). The mechanism of how autophagy affects tumor development, progression, and treatment is not clear yet.
Previous studies showed that inhibition of autophagy augments the antitumor effect of sorafenib in hepatocellular carcinoma (Shi et al. 2011; Shimizu et al. 2012). Sorafenib also can induce autophagy in glioma cells (Siegelin et al. 2010). However, it is unknown whether autophagy activation induced by sorafenib can promote death of cancer cells or provide protection for cancer cells against anti-tumor function of sorafenib. In this study, we combined sorafenib treatment with the autophagy inhibitor chloroquine in GBM cells (U373 and LN229) and tumors. Chloroquine inhibits autophagy because it raises the lysosomal pH, which leads to inhibition of both fusion of autophagosome with lysosome and lysosomal protein degradation (Shintani and Klionsky 2004). We found that chloroquine can further inhibit cell proliferation and induce cell apoptosis together with sorafenib compared to the treatment with sorafenib alone. The co-administration of sorafenib and chloroquine inhibited tumor growth in mouse implanted with U373 xenografts in a prolonged and significant fashion and promoted survival benefit. All the evidence indicated that sorafenib induced protective autophagy in GBM. To strengthen our findings, we also used another method to inhibit autophagy, which is ATG5 siRNA technique. ATG5 is an essential protein in the formation of the autophagosome. Knock-down of ATG5 with specific siRNA can block autophagy (Gonzalez et al. 2012).We found that blockage of autophagy with ATG5 siRNA enhanced the inhibitory effect of sorafenib on cell viability of sorafenib. In summary, inhibition of autophagy enhances the antitumor efficacy of sorafenib in GBM. This combination of chloroquine and sorafenib provides a prospective therapeutic option for GBM patients.
Experimental Procedure
Ethics Statement
Animal experiments were performed following the international guidelines for the care and use of laboratory animals and with Nanjing University Ethical Committee approval.
Animal Study
Six-week-old male RAG-KO mice were maintained and kept in the animal center of Nanjing University. The mice were housed in a temperature-controlled and light-controlled environment. U373 cells (1 × 105 cells in 5 μL of DMEM medium) stably transfected with a luciferase expression plasmid (U373-Luc) were injected into the brains of the mice at 3 mm deep into the brain parenchyma and 3 mm to the right of the midline behind the bregma using a Hamilton syringe under anesthesia with chloralic hydras (4 %, 2 mL/kg, i.p.). Three days after inoculation, the mice were randomly divided into four groups (six mice per group). Treatment was initiated after 7 days of tumor cell implantation, according to the following regimens: (1) control groups receive appropriate vehicles (0.5 % DMSO in PBS); (2) sorafenib (100 mg/kg, daily i.p.); (3) chloroquine (50 mg/kg, daily oral gavage); and (4) sorafenib plus chloroquine. In the survival study, mice were allowed access to food and water ad libitum. Tumor growth was assessed twice a week by bioluminescence imaging using a Xenogen in vivo imaging system. We weighed the mice daily, checked survival and recorded the time until they reached their humane end-points. Humane end-points were defined as the loss of more than 20 % of maximum body weight or hunched appearance (in accordance with the Ethical Committee). Moribund mice were humanely sacrificed by using CO2 followed by cervical dislocation. At the end of the experiment, the mice were euthanized, and the brains were fixed in 10 % buffered formalin, embedded in paraffin and then stained with Ki67 and cleaved caspase-3. The slides were photographed using a phase-contrast microscope.
Cell Lines and Culture
LN229 and U373 established human GBM cell lines (originally purchased from The American Type Culture Collection, Manasas, VA) were cultivated in DMEM medium supplemented with 10 % fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin and 1 % l-glutamine (Invitrogen).Culture flasks were kept at 37 °C and 5 % CO2 in a humidified atmosphere.
Reagents and Antibodies
Chloroquine and dimethyl sulfoxide (DMSO) were purchased from Sigma (St Louis, MO); sorafenib was purchased from Santa Cruz Biotechnology Inc (Santa Cruz, CA) and dissolved in DMSO for in vitro studies. A maximum dose of 0.1 % DMSO was never exceeded. The antibodies to pSTAT3 (Tyr705), STAT3, pAkt (Ser473), Akt, Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), p44/42 MAPK (Erk1/2), Mcl-1, cleaved caspase-3, Ki-67, p62 and GAPDH were obtained from Cell Signaling Technology (Danvers, MA); LC3I/II antibody was purchased from Novus Biologicals (Littleton, CO); the Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies, Inc. (Rockville, MA); the Annexin V-FITC apoptosis detection kit and Matrigel were purchased from BD Biosciences (San Diego, CA); the Pierce BCA Protein Assay Kit was obtained from Thermo Scientific Corp (Hudson, New Hampshire); the HRP-linked secondary antibodies were from GE Healthcare Bio-Sciences Corp (Piscataway, NJ); other Western blot reagents were obtained from Bio-Rad Laboratories (Hercules, CA). All cell culture products were purchased from Invitrogen Corp. Cells were transiently transfected with the pSELECT-GFP-LC3 plasmid (Invitrogen) using Lipofectamine 2000 reagent (Sigma) according to the manufacturer’s protocol.
Cell Viability Assay
Cell viability was measured by CCK-8 assay. Briefly, LN229 and U373 cells were plated in 96-well plates (5 × 103 cells/well) and subjected to different treatments. We used DMSO as a vehicle for drug treatment. After a 48 h incubation at 37 °C in a humidified atmosphere containing 5 % CO2/95 % air, 5 μl of CCK-8 reagent was added to the cells. The plates were read at 450 nm on a multi-scan plate reader after a 2 h incubation.
Wound Healing Assay
Cellular ability to migrate was measured by a wound migration assay. Briefly, LN229 and U373 cells were plated onto 6-well tissue culture plates (5 × 105/well) and cultured in medium containing 10 % FBS to nearly confluent monolayers. A scratch was made on a uniform layer of cells using a sterile micropipette tip and cells were washed to remove debris. The wounded monolayers were then subjected to the drug treatments. At the end of treatments, the cells were photographed using a phase-contrast microscope (Nikon) and analyzed for the distance migrated by the leading edge of the wound at 0 and 24 h. The experiments were performed in triplicate wells and repeated at least three times.
Transwell Invasion Assay
Cell ability to invade was measured by a transwell invasion assay in modified Boyden chambers with filter inserts with 8-μm pores in 24-well plates. Cells (1 × 105/well) were resuspended in serum-free DMEM and seeded into the insert well of the plate (8-μm pores, BD biosciences, Franklin Lakes, NJ, USA) for 24 h. Remaining cells in the upper chamber were removed with a cotton swab. Migrated cells to the bottom of the membrane were fixed in paraformaldehyde (4 %), stained with DAPI and counted. Cell counts were expressed as the mean number of cells per field of view. The experiments were performed in triplicate wells and repeated at least three times.
Western Blot Analysis
Total protein was extracted from cells with M-PER Mammalian Protein Extraction Reagent (Thermo, Fremont, CA) supplied with Complete Protease Inhibitor Cocktails (Roche, Lewes, UK), according to manufactures’ protocols. The protein concentrations were measured with Pierce BCA Protein Assay Kit (Thermo, Fremont, CA). Equal amounts of protein diluted in NuPAGE-sample buffer containing reducing reagents were denatured at 95 °C for 5 min and electrophoretically separated by 16 % sodiumdodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred onto nitrocellulose membranes and the membranes were blocked in 5 % BSA/TBST for an hour at room temperature. The blots were probed with the respective primary antibodies following by their dilution factors (pSTAT3 (Tyr705) 1:1000, STAT3 1:1000, pAkt (Ser473) 1:1000, Akt 1:1000, Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) 1:1000, p44/42 MAPK (Erk1/2) 1:1000, Mcl-1 1:1000, cleaved caspase-3 1:1000, Ki-67 1:1000, p62 1:1000, LC3 1:1000 and GAPDH 1:2000) at 4 °C overnight. The membranes were rinsed with PBS and incubated with horseradish secondary antibodies. Protein bands were detected using the ECL detection system (BeyoECL Plus, Beyotime) and quantitatively analyzed using ImageJ software. The values of each protein were normalized to that of GAPDH. The experiments were performed in triplicate wells and repeated at least three times.
Small Interfering RNA Transfection
Two different sequences targeted to two different sites in Atg5 and a negative-control mismatch sequence were designed by GenePharma (Shanghai GenePharmaCo, Ltd). The sequences of siRNA were presented as follows: human Atg5 Sequence 1: CUGAAAUGGCAUUAUCCAATT; UUGGAUAAUGCCAUUUCAGTG, Sequence 2: GAAGUUUGUCCUUCUGCUATT; UAGCAGAAGGACAAACUUCTT, negative control sequence UUCUCCGAACGUGUCACGUTT; ACGUGACACGUUCGGAGAATT. The siRNAs were transfected with Lipofectamine 2000 (Invitrogen, 11668019) for 48 h in LN229 and U373 cells according to the protocol of the manufacturer. After 12 h, cells were treated with different concentration of sorafenib. Cells were lysed 48 h after siRNA transfection and protein (20 μg) was assayed using immunoblot analysis with primary antihuman antibody against ATG5 (1:1000 Cell Signaling, 2630).
Annexin V-FITC Staining
The percentage of apoptotic cells in the different treatment groups was determined with the Annexin V staining (Sigma, St Louis, MO), according to the manufacturer’s instructions. The cells positive for Annexin V-FITC and/or PI were analyzed using a BD FACS flow cytometer (San Diego, CA).
Immunohistochemistry
Solid tumors from sacrificed mice were formalin-fixed and paraffin-embedded. 1 mM EDTA (pH 8.0) was used for antigen retrieval and methanol containing 3 % hydrogen peroxide was used to quench endogenous peroxidase activity. After 2 h incubation at room temperature with normal goat serum, the slides were incubated at 4 °C overnight with primary antibodies (1:50 Ki-67 and 1:50 cleaved-caspase 3). Next, the sections were rinsed with PBS and incubated with horseradish peroxidase-linked goat anti-rabbit antibodies, followed by reaction with diaminobenzidine. The immunostained sections were counterstained using hematoxylin.
Statistical Analysis
Data are presented as mean ± standard deviation (SD) from three independent experiments. The comparisons between control and treated groups were done by using Student’s t test. Significance in relation to tumor growth in mice with drug treatment was analyzed using two-way ANOVA compared to control mice group. The combination treatment differed from sorafenib alone treatment was analyzed using two-way ANOVA after Bonferroni post test. Significance in relation to survival was analyzed using the Kaplan–Meier survival curves and the log-rank test. In all experiments, differences were considered statistically significant when P < 0.05.
Results
Sorafenib and Chloroquine Interact to Suppress Gliomas Cell Viability
To systematically address whether chloroquine would enhance sorafenib-induced inhibitory activity on GBM cell growth, we evaluated gliomas cell viability under different drug treatment by CCK-8 assay in vitro. Two gliomas cell lines LN229 and U373 tested showed that combined treatment with 0–20 μM sorafenib and 5 μM chloroquine profoundly inhibited the cell viability, as compared with 0–20 μM sorafenib treatment without chloroquine (Fig. 1) after 48 h treatment. Sorafenib and chloroquine interacted in a greater than additive fashion in both cell lines (Fig. 1). We used 10 μM sorafenib and 5 μM chloroquine as a combined treatment in future experiments except for specification. For chloroquine, 5 μM concentration had minimal toxicity (5.4 % inhibition) but was still enough to inhibit autophagy. To check the inhibitory effect of the drug treatment besides 48 h, we also measured the cell viability at 24 and 72 h (Fig. S1). Comparing the cells treated with 10 μM sorafenib and 5 μM chloroquine to the cell treated with 10 μM sorafenib alone, we found that longer time treatment further inhibited cell viability after 24 h and the combination drug treatment always had a bigger inhibitory effect at different time points (Fig. S1). To strength our findings, we also treated LN229 and U373 with ATG5 siRNA (siATG5) to block autophagy and treated the cells with different concentrations of sorafenib. Knockdown of ATG5 without any drug treatment in U373 and LN229 slightly decreased cell viability to 93 and 90 %, respectively, regarding to 100 % cell viability in their respective controls. When treated each group with different concentration of sorafenib, cell viability was more inhibited when autophagy was blocked in both LN229 and U373 by knockdown of ATG5 compared to cell viability of cells treated with control siRNA (Fig. S2).
Sorafenib and chloroquine interact to suppress glioma cell viability. Human glioma cell lines LN229 and U373 were plated in 96-well plates (5 × 103/well), and treated with sorafenib alone (black column) and in combination (grey column) with 5 µM chloroquine at the concentrations indicated for 48 h. Each group used cell viability at 0 µM sorafenib without or with chloroquine as controls respectively. Cell viability was examined by CCK-8 assay. Relative viability showed that sorafenib and chloroquine cooperate to inhibit cell viability. Each bar represents mean ± SD of triplicate determinations; results shown are representative of three identical experiments. *p < 0.05
STAT3, AKT, p44/42 MAPK (Erk1/2) and Mcl-1 are Targets of Sorafenib in U373 and LN229 Glioma Cell
To define the intracellular mechanism mediating sorafenib inhibitory effects on cell viability, we analyzed three pathways including STAT3, MAPK and Akt pathways, which are often activated in GBM. We found that the total protein levels of STAT3, MAPK and AKT were not significantly changed after 24 h sorafenib treatment. However, phosphorylation STAT3 at Tyr705, MAPK at Thr202/Tyr204 and AKT at Ser473 in U373 cells and LN229 cells were reduced in a sorafenib concentration-dependent manner (Fig. 2). We also found that the protein level of anti-apoptotic protein Mcl-1 was decreased after sorafenib treatment in U373 and LN229 (Fig. 2).
STAT3, AKT, MAPK (ERK1/2) and Mcl-1 are targets of sorafenib in U373 and LN229 glioma cell. Human glioma cell line U373 and LN229 were plated in 6-well plates (1 × 106/well) and treated with 0, 5, 10 or 20 µM sorafenib for 48 h. 10 µg of cell lysate was analyzed by Western blot using pSTAT3 (Tyr705), STAT3, pAkt (Ser473), Akt, Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), p44/42 MAPK (Erk1/2), Mcl-1 and GAPDH antibodies
Sorafenib and Chloroquine Treatment Inhibits the Migration and Invasion of Glioma Cells
We compared the wound-healing migration ability and matrigel invasion of the combined treatment with sorafenib and chloroquine to those of sorafenib, chloroquine or vehicle alone. To avoid interference of reduced cell migration and invasion due to the cell death in the presence of high concentration drugs, we treated the cells with 2.5 µM sorafenib with or without 5 µM chloroquine. The cell viability of U373 and LN229 under different treatment did not show statistical difference (Fig. S3). The combined treatment showed reduced cell mobility as determined by scratch wound-healing migration assay. Microscopic examination showed that the control cells migrated prominently toward each other after 24 h of scratch. In contrast, the migration of the cells treated with sorafenib and chloroquine combined were significantly impaired (Fig. 3a). The percentage of migrated cells in LN229 and U373 treated with sorafenib and chloroquine combined were 32.3 and 26.4 % respectively. The percentage of migrated cells in LN229 and U373 treated with sorafenib alone were 68.5 and 70.6 % respectively, while the percentage of migrated cells in LN229 and U373 treated with chloroquine alone were 63.4 and 64.0 % respectively. So quantification of the wound-healing migration assay indicated that migration of LN229 and U373 cells treated with sorafenib and chloroquine combined was greatly decreased, when compared with sorafenib or chloroquine alone (Fig. 3b). Apart from this remarkable inhibition in migration ability, sorafenib and chloroquine significantly suppressed the ability of glioma cells to invade the matrigel-coated filters when compared with sorafenib alone (Fig. 4a). Quantitative analysis demonstrated similar results both in LN229 and U373 cell lines (Fig. 4b). The percentage of invaded cells in LN229 and U373 treated with sorafenib and chloroquine combined were 57.7 and 57.2 % compared to the percentage of migrated cells in LN229 (83.4 %) and U373 (86.0 %) treated with sorafenib alone. The percentage of migrated cells in LN229 and U373 treated with chloroquine alone were 84.5 and 83.9 % respectively (Fig. 4b).
Co-treatment with sorafenib and chloroquine inhibits the migration of glioma cells in scratch wound assays in vitro. LN229 and U373 cells treated with 2.5 µM sorafenib and 5 µM chloroquine alone or in combination were scratched with a pipette tip and imaged at 0 and 24 h (a, b). Width of the wound was measured and the mean ± SD from three independent experiments were shown. *p < 0.05 versus vehicle control (no sorafenib or chloroquine treatment). Additional comparison between group treated with sorafenib alone and group treated with sorafenib and chloroquine combination was performed and p value was labeled on the figure
Sorafenib and chloroquine in combination inhibit glioma cell invasion. LN229 and U373 cells were plated onto 6-well plates (1 × 106/well) and treated with 2.5 µM sorafenib in the presence or absence of 5 µM chloroquine. At the end of treatment, the same amount of cells (1 × 105/well) were resuspended and seeded on the upper chamber of a 24-well plate coated with Matrigel. Twenty-four hours later, the cells that migrated into the lower chambers were fixed with ice-cold methanol, stained with DAPI, and then imaged and counted under a microscope (a, b). The transwell chambers were fixed with 33 % ice-cold acetic acid, vibrated and read in a microplate reader at a wavelength of 570 nm *p < 0.05 versus vehicle control (no sorafenib or chloroquine treatment). Additional comparison between group treated with sorafenib alone and group treated with sorafenib and chloroquine combination was performed and p value was labeled on the figure
Chloroquine Blocks Autophagic Flux Induced by Sorafenib in Glioma Cells
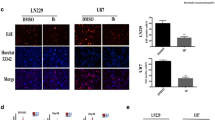
Previous studies showed that sorafenib could modulate the levels of autophagic flux (Siegelin et al. 2010). We investigated the induction of autophagy through use of LC3-GFP vesicle formation (Fig. 5a, b) and Western blot analyses (Fig. 5c, d). LC3-GFP vesicle formation assay showed a significant increase in the formation of autophagosomes after treatment with sorafenib alone (15 % in LN229 and 19 % in U373) compared to control (6 % in LN229 and 9 % in U373), as assessed by the number of cells with punctuate green staining. The percentages of cells showing LC3 puncta (mean ± SD) are indicated (Fig. 5a). This indicated that sorafenib induced autophagy in LN229 and U373. Chloroquine is a lysosome inhibitor, which can prevent the degradation of autophagosome resulting in autophagosome accumulation that could not be cleared. If we treated cells with chloroquine, the basal LC3-II level should increase and we could see more positive cells after chloroquine treatment (Klionsky et al. 2012). As expected, we observed more GFP-puncta in single cell and overall higher percentage of positive cells after chloroquine treatment (26 % in LN229 and 27 % in U373) compared to control (6 % in LN229 and 9 % in U373). Combination treatment the cells with both chloroquine and sorafenib resulted in even higher percent of positive cells (41 % in LN229 and 47 % in U373) compared to treatment with chloroquine alone (26 % in LN229 and 27 % in U373). This double confirmed that sorafenib induced autophagy in LN229 and U373. Western blot analysis demonstrated that sorafenib alone increased LC3 II expression and reduced p62 expression, indicating that sorafenib stimulated autophagic flux. Addition of chloroquine resulted in increased conversion of LC3-I to LC3-II and increased p62 protein level due to accumulation of autophagosome that cannot be cleared. Combined exposure to sorafenib with chloroquine resulted in further increased LC3 II conversion, suggesting that chloroquine suppressed the sorafenib-induced autophagy (Fig. 5c, d).
Chloroquine blocks the autophagic flux induced by sorafenib. a GFP-LC3 punctum formation in glioma cell lines was observed by microscopy. Tumor cells were seeded on coverslips and treated with 10 µM sorafenib and 5 µM of chloroquine, alone and in combination for 48 h. Autophagosomes were visualized by the presence of LC3 puncta. The drug combination treatment showed more autophagosome accumulation than either single agent alone. The percentages of cells showing LC3 puncta (mean ± SD) are indicated. Scale bar is 50 µm. b The percentages and statistical analysis were further performed in column format. c Western blot demonstrated the protein levels of LC3-II and p62. d Quantitation of the ratio of LC3-II/LC3-I after single and combination treatment in comparison to the untreated control. Data show that the combination treatment resulted in considerable increases in LC3-II ratio compared to treatment with chloroquine alone, indicating the blockade of autophagic flux. *p < 0.05 versus vehicle control (no sorafenib or chloroquine treatment). Additional comparison between group treated with sorafenib alone and group treated with sorafenib and chloroquine combination was performed and p value was labeled on the figure
Inhibition of Autophagy by Chloroquine Enhanced the Apoptosis Induced by Sorafenib in Glioma Cells
An increasing number of studies have shown that autophagy may serve as a protective mechanism in tumor cells and that therapy-induced cell death can be potentiated through autophagy inhibition (Nagelkerke et al. 2014; White 2012).We further investigated apoptosis in glioma cells treated with sorafenib alone or combined with chloroquine. As clearly demonstrated by the increases in the amounts of cleaved caspase-3 and in Annexin V staining (LN229 and U373), inhibition of autophagy by chloroquine enhanced the apoptosis induced by sorafenib in glioma cells (Fig. 6a, b).
Effect of inhibiting autophagy on the sorafenib-induced apoptosis in glioma cells. LN229 and U373 cells were treated with the indicated concentration of sorafenib for 48 h in the presence or absence of chloroquine. At the end of treatment, apoptosis was determined by Western blot of cleaved caspase-3 (a) and by cytometric analysis of Annexin V staining (b) GAPDH was used as a loading control. *p < 0.05 versus vehicle control (no sorafenib or chloroquine treatment). Additional comparison between group treated with sorafenib alone and group treated with sorafenib and chloroquine combination was performed and p value was labeled on the figure
Combined Treatment of Sorafenib with Chloroquine Exerts a Potent Antitumor Effect In Vivo
To further determine whether autophagy blockade can augment the effects of sorafenib in vivo, we monitored, measured and compared the tumor growth inhibition and the survival-promoting effects of combined treatment with chloroquine and sorafenib versus either agent alone in U373 xenografts. Mice treated with vehicle or chloroquine exhibited rapid increases in bioluminescence (Fig. 7a, b). Sorafenib treatment initially slowed tumor growth, but this effect was disappeared after 24 days. Mice treated with combination therapy showed longer stable bioluminescence through 32 days and tumor continue to grow after that. Two way ANOVA analysis indicated that the drug treatment (chloroquine, sorafenib or combination therapy) is effective compared to vehicle treatment (p < 0.05). At the meanwhile, combination therapy is more effective than sorafenib treatment alone (p < 0.05). From the survival data, we observed that chloroquine and sorafenib together inhibited tumor growth in a prolonged and significant fashion and promoted survival (Fig. 7c. The remarkable decrease in tumor cell proliferation (Ki67) and increase in apoptosis (cleaved caspase-3) noted in combination-treated xenografts showed that autophagy blockade enhanced the antitumor activity of sorafenib in glioblastoma (Fig. 7d–g).
Anti-glioma activity of combinatorial treatment with sorafenib and chloroquine in vivo. a Bioluminescence during 28 days of treatment of U373_Luc tumors (n = 6 mice per group). Vehicle, sorafenib, chloroquine or combination therapy was given daily. b Bioluminescence during 49 days of treatment in mice implanted with U373_Luc cells. The treatment groups differed by two-way ANOVA (p < 0.05), and the combination treatment differed from sorafenib alone treatment after Bonferroni post (p < 0.05). c RAG-KO mice stereotactically implanted with U373 cells in the right cerebral striatum were treated systemically with different treatments: (1) control groups receive appropriate vehicles (0.5 % DMSO in PBS); (2) sorafenib (100 mg/kg, daily i.p.); (3) chloroquine (50 mg/kg, daily i.p.); and (4) sorafenib plus chloroquine. Treatment began after 7 days of tumor cell implantation. Survival curves per group are shown. p = 0.0223 (Log-rank test) for difference between control and sorafenib; p = 0.1542 (Log-rank test) for difference between control and chloroquine; p = 0.0007 (Log-rank test) for difference between control and sorafenib + chloroquine; p = 0.0403 (Log-rank test) for difference between sorafenib and sorafenib + chloroquine; differences were considered statistically significant when p < 0.05. Histopathology of sorafenib plus chloroquine shows anti-glioma activity, in vivo. Representative brain sections from mice treated with vehicle, sorafenib, chloroquine and sorafenib plus chloroquine were harvested after 28 days, and cell proliferation (Ki67) (d) and apoptosis (cleaved caspase-3) (f) were analyzed. Magnification, ×400. Quantification of proliferation (e) and apoptosis (g). Labeled cells were counted in an average of 10-15 high-power fields. Data are the mean ± SD *p < 0.05 versus vehicle control (no sorafenib or chloroquine treatment). Additional comparison between group treated with sorafenib alone and group treated with sorafenib and chloroquine combination was performed and p value was labeled on the figure
Discussion
Despite recent advances in surgery, chemotherapy, and radiation therapy for the treatment of GBM, the outcome is still poor. New agents and novel therapeutic strategies targeting GBM are urgently needed (Mischel and Cloughesy 2003; Wen and Kesari 2008; Wurth et al. 2014). In GBM, aberrant activation of Ras-Raf-MAPK signaling leads to proliferation, and the dysregulation of VEGFR2 and PDGFR2 drive tumor-associated angiogenesis. Sorafenib as a multikinase inhibitor which can inhibit Raf, a critical enzyme in the Ras signaling cascade, as well as VEGFR and PDGFR at the same time, appears particularly promising in glioma given the robust activity of both pathways in the disease (Liu et al. 2006; Siegelin et al. 2010; Xu et al. 2014).
Akt pathway, STAT3 and MAPK pathway are relevant for the biology and development of GBM (de la Iglesia et al. 2009; Holland et al. 2000; Molina et al. 2010; Robinson et al. 2011; Zhang et al. 2013). Dephosphorylation of STAT3, AKT and MAPK (ERK1/2) by sorafenib were found in other tumor entities (Blechacz et al. 2009; Geng et al. 2014; Yang et al. 2008) like cholangiocarcinoma, hepatocellular carcinoma and medulloblastomas. Here we showed that sorafenib inhibited STAT3, AKT and MAPK phosphorylation at Tyr705, Ser473 and Thr202/Tyr204, respectively, in glioma cell line U373 and LN229. Expression of a pro-apoptotic protein Mcl-1 was regulated by STAT3 signaling (Yu and Jove 2004). Sorafenib down-regulated the expression of Mcl-1 in several kinds of tumor cells (Rahmani et al. 2007; Yu et al. 2005). Here we showed that sorafenib decreased Mcl-1 expression in established cell line U373. Therefore inhibition of phosphorylation of STAT3 and Akt may be important mechanisms of anti-tumor effect of sorafenib in GBM.
In this study, we also showed that sorafenib inhibited GBM cell proliferation through caspase 3 apoptosis in a dose-dependent manner. In mice implanted with glioblastoma cells, systemic administration of sorafenib significantly suppressed tumor growth and prolonged survival, as was shown previously (Fan et al. 2010). Our wound healing and migration assays also showed that sorafenib inhibited GBM cell migration and invasion in vitro. Collectively, this evidence provides a clinical rationale for testing sorafenib for GBM treatment.
However, phase II clinical trials that used sorafenib as a combinational reagent have all had disappointing results (Nabors et al. 2011). Most studies blamed this failure on the drug–drug interaction causing an insufficient dose, the overlapping toxicity, and the limited passage of sorafenib through the brain–blood barrier because sorafenib is a substrate for the breast cancer resistance protein (ABCG2/BRCP), blood–brain barrier efflux pumps (Galanis et al. 2013). However, these studies did not consider that sorafenib could also cause autophagy in addition to its anti-proliferation and anti-angiogenesis effects. Autophagy may either stimulate or inhibit tumor growth depending on tumor type, stage and genetic background.
Our study therefore examined the function of autophagy induced by sorafenib in GBM. We clearly showed that in vitro, sorafenib decreased p62 level, increased the level of a faster migration form of LC3, and induced a punctate pattern of LC3-GFP labeling, which were all biochemical markers of autophagy. When we added chloroquine, a clinically applicable autophagy inhibitor, to the sorafenib treatment, we found that chloroquine further inhibited the GBM cell viability, migration and invasion and enhanced caspase-3 dependent cell apoptosis. We also used ATG5 siRNA technique to inhibit autophagy. We found that blockage of autophagy with ATG5 siRNA enhanced the inhibitory effect of sorafenib on cell viability of sorafenib in U373 and LN229. One recent study found controversial siATG5 effect on sorafenib induced cell death. They found siATG5 increased GBM5 cell death but decreased GBM12 cell death with sorafenib treatment. The significance of siATG5 on sorafenib was not analyzed in that paper. So the effect of siATG5 together with sorafenib on GBM cell lines may be cell type dependent. Moreover, our in vivo experiments indicated that chloroquine could further suppress tumor growth and prolong animal survival. This is the first time showing that sorafenib-induced autophagy provided cyto-protection for GBM, and that inhibition of autophagy can enhance the anti-tumor effects of sorafenib treatment. These results therefore suggest another possible reason for why clinical trials of sorafenib for glioma treatment have failed. A current clinical trial that includes chloroquine in the conventional treatment and FET-PET-based reirradiation on glioblastoma has shown promising early results (Sotelo et al. 2006).
In our study, chloroquine itself could suppress tumor growth and prolong animal survival to some extent. This result is consistent with the clinical trial using chloroquine which showed that chloroquine triggered apoptosis in primary stem-like glioma cells by blocking autophagy (Munshi 2009; Sotelo et al. 2006; Wurth et al. 2014). Recent studies also showed that high levels of oxidative stress were present in high-grade gliomas and this oxidative stress was associated with autophagy activation (Murrow and Debnath 2013; Wurth et al. 2014). Together, these evidences provide a rationale for including chloroquine in future clinical trials of sorafenib. As for the insufficient passage of sorafenib through the blood–brain barrier, it may be worth including elacrida, a dual inhibitor of ABCG2/BRCP, together with sorafenib (Kuppens et al. 2007).
In conclusion, our study showed that sorafenib had anti-tumor effects on GBM both in vitro and in vivo and also induced autophagy. Moreover, the sorafenib-induced autophagy provided cyto-protection for tumor cell survival. Future studies of sorafenib treatment on glioblastoma should include an anti-autophagy drug.
References
Blechacz BR et al (2009) Sorafenib inhibits signal transducer and activator of transcription-3 signaling in cholangiocarcinoma cells by activating the phosphatase shatterproof 2. Hepatology 50:1861–1870
Cancer Genome Atlas Research, N (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455:1061–1068
Carra E et al (2013) Sorafenib selectively depletes human glioblastoma tumor-initiating cells from primary cultures. Cell Cycle 12:491–500
de la Iglesia N, Puram SV, Bonni A (2009) STAT3 regulation of glioblastoma pathogenesis. Curr Mol Med 9:580–590
Fan QW et al (2007) A dual phosphoinositide-3-kinase alpha/mTOR inhibitor cooperates with blockade of epidermal growth factor receptor in PTEN-mutant glioma. Cancer Res 67:7960–7965
Fan QW et al (2010) Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal 3:ra81
Furnari FB et al (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21:2683–2710
Galanis E et al (2013) Phase II study of bevacizumab in combination with sorafenib in recurrent glioblastoma (N0776): a north central cancer treatment group trial. Clin Cancer Res 19:4816–4823
Geng ZM et al (2014) Sorafenib inhibition of hepatic stellate cell proliferation in tumor microenvironment of hepatocellular carcinoma: a study of the sorafenib mechanisms. Cell Biochem Biophys 69:717–724
Gonzalez P et al (2012) Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ 19:1337–1346
Gozuacik D, Kimchi A (2004) Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23:2891–2906
Hamed HA et al (2014) Sorafenib/Regorafenib and Lapatinib interact to kill CNS tumor cells. J Cell Physiol 230:131–139
Hippert MM, O’Toole PS, Thorburn A (2006) Autophagy in cancer: good, bad, or both? Cancer Res 66:9349–9351
Holland EC et al (2000) Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25:55–57
Hottinger AF et al (2014) Phase I study of sorafenib combined with radiation therapy and temozolomide as first-line treatment of high-grade glioma. Br J Cancer 110:2655–2661
Karajannis MA et al (2014) Phase II study of sorafenib in children with recurrent or progressive low-grade astrocytomas. Neuro Oncol. doi:10.1093/neuonc/nou059
Kim S et al (2007) Sorafenib inhibits the angiogenesis and growth of orthotopic anaplastic thyroid carcinoma xenografts in nude mice. Mol Cancer Ther 6:1785–1792
Kim DE et al (2015) Raloxifene induces autophagy-dependent cell death in breast cancer cells via the activation of AMP-activated protein kinase. Mol Cells 38:138–144
Klionsky DJ et al (2012) Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8:445–544
Kruijtzer CM et al (2002) Increased oral bioavailability of topotecan in combination with the breast cancer resistance protein and P-glycoprotein inhibitor GF120918. J Clin Oncol 20:2943–2950
Kuppens IE et al (2007) A phase I, randomized, open-label, parallel-cohort, dose-finding study of elacridar (GF120918) and oral topotecan in cancer patients. Clin Cancer Res 13:3276–3285
Lee EQ et al (2012) Phase I/II study of sorafenib in combination with temsirolimus for recurrent glioblastoma or gliosarcoma: North American Brain Tumor Consortium study 05-02. Neuro Oncol 14:1511–1518
Liu L et al (2006) Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis, and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res 66:11851–11858
Mischel PS, Cloughesy TF (2003) Targeted molecular therapy of GBM. Brain Pathol 13:52–61
Molina JR et al (2010) Invasive glioblastoma cells acquire stemness and increased Akt activation. Neoplasia 12:453–463
Munshi A (2009) Chloroquine in glioblastoma–new horizons for an old drug. Cancer 115:2380–2383
Murrow L, Debnath J (2013) Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annu Rev Pathol 8:105–137
Nabors LB et al (2011) Phase I trial of sorafenib in patients with recurrent or progressive malignant glioma. Neuro Oncol 13:1324–1330
Nagelkerke A et al (2014) Therapeutic targeting of autophagy in cancer. Part I: Molecular pathways controlling autophagy. Seminars cancer biology. Academic Press, London
Peereboom DM et al (2013) NABTT 0502: a phase II and pharmacokinetic study of erlotinib and sorafenib for patients with progressive or recurrent glioblastoma multiforme. Neuro Oncol 15:490–496
Qin Y et al (2013) SL1122-37, a novel derivative of sorafenib, has greater effects than sorafenib on the inhibition of human hepatocellular carcinoma (HCC) growth and prevention of angiogenesis. Biosci Trends 7:237–244
Rahmani M et al (2007) The multikinase inhibitor sorafenib induces apoptosis in highly imatinib mesylate-resistant bcr/abl + human leukemia cells in association with signal transducer and activator of transcription 5 inhibition and myeloid cell leukemia-1 down-regulation. Mol Pharmacol 72:788–795
Robinson JP et al (2011) Akt signaling is required for glioblastoma maintenance in vivo. Am J Cancer Res 1:155–167
Shi YH et al (2011) Targeting autophagy enhances sorafenib lethality for hepatocellular carcinoma via ER stress-related apoptosis. Autophagy 7:1159–1172
Shimizu S et al (2012) Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer 131:548–557
Shintani T, Klionsky DJ (2004) Autophagy in health and disease: a double-edged sword. Science 306:990–995
Siegelin MD et al (2010) Sorafenib exerts anti-glioma activity in vitro and in vivo. Neurosci Lett 478:165–170
Sotelo J, Briceno E, Lopez-Gonzalez MA (2006) Adding chloroquine to conventional treatment for glioblastoma multiforme: a randomized, double-blind, placebo-controlled trial. Ann Intern Med 144:337–343
Van Meir EG et al (2010) Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 60:166–193
Wen PY, Kesari S (2008) Malignant gliomas in adults. N Engl J Med 359:492–507
White E (2012) Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer 12:401–410
Wurth R, Barbieri F, Florio T (2014) New molecules and old drugs as emerging approaches to selectively target human glioblastoma cancer stem cells. Biomed Res Int 2014:126586
Xu M et al (2014) Sorafenib blocks the HIF-1alpha/VEGFA pathway, inhibits tumor invasion, and induces apoptosis in hepatoma cells. DNA Cell Biol 33:275–281
Yang F et al (2008) Sorafenib inhibits signal transducer and activator of transcription 3 signaling associated with growth arrest and apoptosis of medulloblastomas. Mol Cancer Ther 7:3519–3526
Yu H, Jove R (2004) The STATs of cancer–new molecular targets come of age. Nat Rev Cancer 4:97–105
Yu C et al (2005) The role of Mcl-1 downregulation in the proapoptotic activity of the multikinase inhibitor BAY 43-9006. Oncogene 24:6861–6869
Zhang J et al (2013) Whole-genome sequencing identifies genetic alterations in pediatric low-grade gliomas. Nat Genet 45:602–612
Zhao D et al (2014) Upregulation of HIF-2alpha induced by sorafenib contributes to the resistance by activating the TGF-alpha/EGFR pathway in hepatocellular carcinoma cells. Cell Signal 26:1030–1039
Acknowledgments
This work was supported by Grants from Nanjing Jinling Hospital, Nanjing, China (2011Q047).
Author information
Authors and Affiliations
Corresponding author
Additional information
Xiangyu Liu and Kangjian Sun have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Sorafenib and chloroquine cooperated to inhibit the glioma cell viability. Human glioma cell lines LN229 (black line) and U373 (grey line) were plated in 96-well plates (5×103/well), and treated with sorafenib alone (10 µM) and in combination with 5 µM chloroquine for 24, 48 and 72 hours. Cell viability was examined by CCK-8 assay. Relative viability showed that sorafenib and chloroquine cooperate to promote cell death. Each bar represents mean ± SD of triplicate determinations; results shown are representative of three identical experiments.* p <0.05 vs. cell viability at 0 µM sorafenib in each groups (TIFF 438 kb)
Fig. S2
Inhibition of autophagy with ATG5 siRNA enhanced the inhibitory activity of sorafenib on gliomas cell viability. A. Human glioma cell line LN229 and U373 were plated in 6-well plates (1×106/well), and treated with control siRNA, siATG5 sequence 1 and sequence 2 for 48 hours. 10 µg of cell lysate was analyzed by Western blot using ATG5 and β-actin antibodies. B. Human glioma cell lines LN229 and U373 were plated in 6-well plates (1×106/well), and treated with control siRNA (siControl) and siRNA (siATG5) sequence 2 for 12 hours. Then the cells were trypsin digested and were plated in 96-well plates (5×103/well), and treated with 0-20 µM sorafenib for 48 hours. Cell viability was examined by CCK-8 assay. Relative viability showed that sorafenib and chloroquine cooperate to promote cell death. Each bar represents mean ± SD of triplicate determinations; results shown are representative of three identical experiments.* p <0.05 vs. cell viability at 0 µM sorafenib in each groups (TIFF 10,231 kb)
Fig. S3
Cell viability of LN229 and U373 treated with sorafenib alone (2.5 µM) and in combination with 5 µM chloroquine for 24 hours. 0.1% DMSO was used as control. Each bar represents mean ± SD of triplicate determinations; results shown are representative of three identical experiments. No statistical difference were found between different groups (TIFF 9,186 kb)
Rights and permissions
About this article
Cite this article
Liu, X., Sun, K., Wang, H. et al. Inhibition of Autophagy by Chloroquine Enhances the Antitumor Efficacy of Sorafenib in Glioblastoma. Cell Mol Neurobiol 36, 1197–1208 (2016). https://doi.org/10.1007/s10571-015-0318-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-015-0318-z