Abstract
Neuronal activity regulates the expression of brain-derived neurotrophic factor (BDNF) in brain. In darkness, reduced neuronal activity in the visual cortex markedly decreases total BDNF transcription level in adult rats. Epigenetic mechanisms are crucially involved in the regulation of gene expression in response to environmental stimuli. In this study, we examined the effect of 1 week of light deprivation (LD) on the activity-dependent changes in BDNF expression from different promoters in the visual cortex and hippocampus. We analyzed the correlation between the chromatin state of Bdnf promoters, exon-specific transcripts levels, and total protein levels in light-deprived rats and in rats reared under normal light–dark cycle. We found that 1 week of LD significantly reduced Bdnf mRNA and protein in the visual cortex but not in the hippocampus. However, epigenetic analysis revealed that LD increased histone-3 methylation and DNA methylation at the Bdnf promoter IV in both the visual cortex and hippocampus. These data highlight the spatial differences in signaling pathways that lead to the BDNF expression in response to diminished ambient light.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Experience reorganizes neuronal networks via activity-dependent regulation of gene expression. Neuronal activity differentially affects the brain-derived neurotrophic factor (BDNF) transcription driven from eight promoters (Timmusk and Metsis 1994; Aid et al. 2007), and different signaling pathways mediate the transport of exon-specific BDNF transcripts into dendrites (An et al. 2008; Chiaruttini et al. 2008, 2009). Light deprivation (LD) for a week has been shown to markedly decrease BDNF transcription level in the visual cortex or adult rats (Castren et al. 1992; Schoups et al. 1995). Reduced neuronal activity in the visual cortex in darkness may also affect the synthesis, transport, and secretion of BDNF protein to other brain regions, such as hippocampus and cerebellum, which receive indirect visual input from the visual cortex (Pollock et al. 2001). It has been shown that rearing animals in the dark from birth (dark rearing, DR) produced changes in BDNF mRNA and protein expression in several brain regions (Pollock et al. 2001; Tropea et al. 2001), while phosphorylation but not expression of the BDNF receptor TrkB in the visual cortex was reduced by DR (Viegi et al. 2002).
A recent study showed that LD in adult rats induces structural and behavioral alterations associated with depression and sensitizes rats to stress (Gonzalez and Aston-Jones 2008), which is reminiscent of the vulnerability of humans to seasonal depression when the ambient light is reduced. Activity-dependent expression of BDNF in hippocampus is implicated in depression in experimental animals and in humans (for review, see Duman and Monteggia 2006; Castren et al. 2007; Krishnan and Nestler 2008). In this study, we examined the activity-dependent changes in BDNF expression from different promoters and the expression of TrkB in the visual cortex and hippocampus induced by 1 week of LD of adult rats. We analyzed the correlation between the chromatin state of Bdnf promoters, levels of exon-specific transcripts, and total protein levels in LD rats and in rats reared under normal 12/12 h light–dark cycle. We show that 1 week of LD reduces the Bdnf mRNA levels in promoter-specific manner and BDNF protein levels only in the visual cortex. Interestingly, we found that LD promotes significant repressive alterations of the chromatin structure at Bdnf promoter regions not only in the visual cortex but also in the hippocampus of adult rats.
Materials and Methods
Animals and Housing
All animal experiments were approved by the County Administrative Board of Southern Finland. Adult male Wistar rats (Harlan, The Netherlands) were housed under 12/12 h light–dark cycle (lights on between 7:00 a.m. and 7:00 p.m.; control group, 6 rats) or in constant darkness (light deprived; LD group, 6 rats) for 7 days. On the final day of housing, rats were stunned with CO2 (LD rats in light-impermeable box) and killed with cervical dislocation between 10:00 a.m. and 11:00 a.m. Bilateral visual cortex and hippocampus were rapidly dissected on a cooled dish and the samples divided for western blot/ELISA (left) and mRNA/chromatin (right) analyses.
BDNF ELISA Assay
Left-side brain samples were homogenized in NP++ buffer (137 mM NaCl, 20 mM Tris, 1% NP-40, 10% glycerol, 48 mM NaF, 2× complete inhibitor mix (Sigma–Aldrich, St. Louis, MO) and 2 mM Na3VO4), incubated on ice for at least 15 min, centrifuged (16,000×g, 15 min, +4°C), and the supernatants were collected for measures. The NP++ lysed brain samples were diluted (visual cortex: 1:7, hippocampus: 1:14) in Hanks buffer (125 mM NaCl, 5 mM KCl, 1.2 mM NaH2PO4, 1 mM CaCl2, 1.2 mM MgCl2, 1 μM ZnCl2, 10 mM glucose, 25 mM HEPES, 0.25% BSA; pH 7.4); samples were acidified at RT to pH 3 with 1 N HCl and neutralized within 15 min with 1 N NaOH. Next, BDNF standards (7.8–1,000 pg/ml in Hanks; Promega/Alomone), samples (170 μl) (all run in duplicate), and POD-conjugated second BDNF antibody (30 μl, 1:1,900 in Hanks containing 6.66% BSA and 0.66% Triton X-100) were transferred to pre-blocked (300 μl; Hanks buffer, 2% BSA, 0.1% Triton X-100, 2 h, RT) Maxisorb® ELISA plates that were previously coated with the 1st BDNF antibody (200 μl 1:4,000 in carbonate buffer, overnight, +4°C). Next morning, the plates were washed with PBS-T (300 μl × 4), and the POD substrate was added to the wells (200 μl; BM Blue; Roche). The colorimetric reaction was stopped within 20 min with 1 N H2SO4 (50 μl), and absorbance was immediately measured at 490 nm. The assay has significantly higher (~30×) affinity to the mature-BDNF than to the pro-BDNF (Fig. S1a). The specificity and validity of the assay was further tested using brain tissues obtained from adult conditional BDNF −/− mice (kindly provided by Dr. M. Rios) (Fig. S1b). BDNF content was calculated according to the standard curve. The r 2 for standard curve was ≥0.98 in all the measurements.
Quantification of TrkB Phosphorylation
TrkB phosphorylation at the PLCγ1 binding site of TrkB (Y816; kind gift from Dr. Moses Chao) and the catalytic domain (Y706/707; Cell Signalling Technology, Inc., USA) were analyzed from glycosylated pool of proteins (~0.5 mg) as previously described (Rantamaki et al. 2007). In order to analyze the levels of full-length and truncated TrkB, total protein samples (~50 μg) were separated in an SDS-PAGE, blotted on PVDF membrane, and the content of specific proteins visualized with conventional chemiluminescence methods using an antibody directed against the extracellular part of TrkB (1:1,000, BD Biosciences Cat. 610101). Band intensities were analyzed with ImageJ program (National Institute of Health). Phospho-TrkB levels were normalized against the total level of TrkB within the lane. Equal loading was controlled with Ponceau staining.
Messenger RNA Quantification
Total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. One microgram of total RNA was treated with DNAse I mix (Fermentas GmbH, Helsinki, Finland) and was reverse transcribed using oligo(dT) primer and SuperScript III Reverse Transcriptase mix (Invitrogen). The control reactions without Reverse Transcriptase were also performed. The amount of cDNA was quantified using RT-PCR. The primers used to amplify specific cDNA regions of the transcripts are listed in the Table S I. Control PCRs were performed with the primers for the housekeeping genes GAPDH and β-actin. DNA amplification reactions were run in triplicate at least two independent times in the presence of SYBR-Green (Applied Biosystems). Ct values from each sample were obtained using the Sequence Detector 1.1 software. Relative quantification of template was performed as described previously (Chakrabarti et al. 2002) and using the Applied Biosystems manual using ΔΔCt method, with cDNA data being normalized to the control GAPDH level.
Chromatin Immunoprecipitation Assay
A recently published protocol was used (Onishchenko et al. 2008). Specifically, the chromatin was subjected to immunoprecipitation using antibodies against trimethyl-Histone H3 (K27) and acetyl-Histone H3 (K9 + K14) (Upstate, Temecula, CA, USA). The specificity of these antibodies in the chromatin immunoprecipitation (ChIP) assay has been established previously (Kumar et al. 2005; Bernstein et al. 2007). To control for the specificity of antibody binding in our conditions, non-immune rabbit IgG (Chemicon, Temecula, CA, USA) and no-antibody immunoprecipitations were performed for each chromatin sample. The primers used to selectively amplify the portion of Bdnf promoters are listed in the Table S I. Control PCR with β-actin promoter-specific primers was performed to ensure the validity of the ChIP assay. The ChIP data were analyzed by RT-PCR identically to the mRNA data, using the ΔΔCt method (Chakrabarti et al. 2002), except that ChIP data were normalized to the “input.”
DNA Methylation Assay
To analyze the methylation status of DNA at the Bdnf promoters, genomic DNA from brain samples was first converted by sodium bisulfite (Frommer et al. 1992; Clark et al. 1994). Then Bdnf promoter IV region was amplified (Table S I), PCR product was purified from nucleotides and primers using the Qiaquick gel extraction kit (Qiagen, Valencia, CA, USA), and directly subjected to sequencing analysis. Sequencing traces at individual CpG sites at the Bdnf promoter IV were analyzed using the Sequence Scanner Software v1.0 (Applied Biosystems), and the signal of methylated C over the signal sum of both methylated and unmethylated C [Cm/(C + Cm)] represented percentage methylation.
Statistical Analysis
Data are presented as mean + SEM and as percentage of control group values unless otherwise stated. Data were statistically evaluated for significance by using an unpaired Student’s t test with a two-tailed distribution. A P-value < 0.05 was considered statistically significant.
Results
Darkness Differentially Affects the Expression of BDNF Exons in the Visual Cortex and Hippocampus
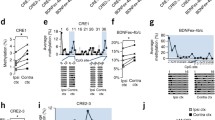
Light deprivation has been shown to markedly decrease the Bdnf transcription in the developing and adult visual cortex (Castren et al. 1992; Pollock et al. 2001). In our experiment, both total Bdnf mRNA (Fig. 1a) and BDNF protein (Fig. 1b) levels were significantly decreased in the visual cortex of adult LD rats as compared to controls. However, no changes in the Bdnf mRNA or protein levels were evident in the hippocampus of the LD rats.
Differential effect of 1 week of LD on BDNF expression in the visual cortex, VC, and the hippocampus, HC, of adult rats. LD significantly decreased the total Bdnf mRNA (a) and BDNF protein levels (b) in the VC but not in the HC. c Measurement of the Bdnf exons expression in the VC revealed strong down-regulation of the most Bdnf transcripts and Bdnf IV transcript specifically. d Levels of Bdnf exons mRNA were not changed by LD in the HC (n = 6 per group), * P < 0.05, ** P < 0.01, *** P < 0.001
Brain-derived neurotrophic factor promoter IV mediates activity-dependent BDNF transcription both in vivo and in vitro (Tao et al. 1998; Hong et al. 2008; Sakata et al. 2009). To test whether prevention of visual input differentially altered the expression of distinct Bdnf exons, we analyzed the levels of Bdnf exons I–VIII (Aid et al. 2007) in the visual cortex and hippocampus by RT-PCR. The levels of Bdnf exons V, VII, and VIII were undetectable by RT-PCR in both brain regions and were not included in further analysis. For the remaining Bdnf transcripts in the visual cortex, LD down-regulated all, except Bdnf III, and the most dramatic decrease was observed for the Bdnf IV transcript (Fig. 1c). In contrast to the visual cortex, expression levels of Bdnf exons in the hippocampus remained unchanged (Fig. 1d). Thus, LD down-regulated BDNF synthesis in the visual cortex by affecting the expression from the multiple Bdnf promoters, but these changes were absent in the hippocampus.
Disruption of the Bdnf promoter IV function specifically affects the development of cortical inhibition and reduces the expression of GABAergic markers (Hong et al. 2008; Sakata et al. 2009). We therefore investigated whether LD regulates the expression of GABAergic synaptic markers GABAA1-receptor and the neuronal transcription factor Npas4 (Lin et al. 2008). We found that LD significantly decreased the mRNA levels of both GABAA1-receptor and Npas4 in the visual cortex, but not in the hippocampus (Fig. 2). Moreover, the members of the CREB-transcription activating complex (Tao et al. 1998) containing the histone acetyltransferase CREB-binding protein (CBP) directly bind to the Bdnf promoter IV and specifically induce Bdnf IV transcription (Alarcon et al. 2004; Hong et al. 2008). We observed that LD reduced the expression of CBP in the visual cortex but not in the hippocampus (Fig. 2).
LD Induces Chromatin Remodeling at BDNF Promoters
Chromatin remodeling, such as histone modifications and DNA methylation, regulates gene expression by altering chromatin structure at promoter regions (Felsenfeld and Groudine 2003; Kouzarides 2007). To assess the impact of chromatin remodeling on the LD-induced down-regulation of the Bdnf exons, we performed a ChIP assay with the antibodies against modified histones. ChIP analysis of the visual cortical tissues revealed that LD significantly increased the level of a repressive chromatin mark H3K27me3 only at the Bdnf IV region, while an activating mark, acetylation of histone H3 (acH3), was not changed at any of the studied promoters (Fig. 3a).
One week of LD induced chromatin remodeling at the Bdnf promoters in the VC and the HC of adult rats. a, b Results of the ChIP analysis in LD animals are presented as % over control group ±SEM, whereas control group’s means equal 0% ±SEM are not shown. P I–VI Bdnf promoters I–VI. a In the VC of the LD animals, level of H3 methylated K27 was significantly increased only at the Bdnf promoter IV. Acetylated form of histone H3 was not changed at any studied promoter. b In the HC, LD-induced H3K27 tri-methylation at all studied Bdnf promoters. Moreover, simultaneous increase in acH3 was observed only at the Bdnf promoter II. c, d DNA methylation analysis at the activity-dependent Bdnf promoter IV. 1 week of LD induced CG methylation at the specific sites in the VC (c) and the HC (d) (n = 6 per group), * P < 0.05, ** P < 0.01, *** P < 0.001
Unexpectedly, chromatin remodeling by LD was not restricted to the visual cortex. In the hippocampus, LD increased the level of a repressive chromatin mark H3K27me3 at all Bdnf promoters studied (Fig. 3b). Moreover, we found that the acetylation of H3 was up-regulated at the Bdnf promoter II with a similar trend for Bdnf promoter I (P = 0.1). No changes were detectable by RT-PCR at the promoter of housekeeping β-actin gene, which was used as a control, in both studied tissues (data not shown).
We next analyzed the DNA methylation pattern at the Bdnf IV promoter region. We found that 1-week LD induced CG methylation at specific and different sites in adult rat visual cortex and hippocampus (Fig. 3c, d). Together, these results suggest that light-dependent neuronal activity remodels chromatin structure at the Bdnf promoters both in the visual cortex and hippocampus of adult rats in a brain region specific manner.
LD Does Not Affect the TrkB Signaling
Decreased neuronal activity in the visual cortex of the LD rats down-regulated the BDNF protein level. We examined whether LD affected the BDNF signaling pathways through its high-affinity receptor TrkB. First, there were no detectable alterations in the levels of full-length TrkB mRNA in the LD rats when compared to controls (Fig. 4a). Next, we found that the total TrkB protein levels were not affected by LD neither in the visual cortex, nor in hippocampus (data not shown). Moreover, TrkB phosphorylation at the PLCγ1 binding site of TrkB (Y816) remained unchanged after 1 week of LD in both studied brain areas (Fig. 4b). Comparable results were obtained with the antibody directed against the phosphorylated TrkB catalytic domain (Y706/707) (data not shown).
Discussion
This study shows that in adult rats, 1 week of LD decreased BDNF synthesis only in the visual cortex in the Bdnf promoter-specific manner. However, repressive chromatin changes at Bdnf promoter regions were induced both in the visual cortex and hippocampus and, importantly, these changes were more pronounced in the hippocampus.
Signaling pathways that lead to the BDNF expression may be different between the visual cortex and hippocampus. Visual cortex rapidly responds to the external light input via activity-regulated gene expression. Synaptic activation initiates calcium-dependent signaling cascade that induces both transcriptional and neurotrophic factors (Tao et al. 1998; West et al. 2001; Hong et al. 2008; Lin et al. 2008). Chromatin structure of the Bdnf promoter IV is highly responsive to activating stimuli because of multiple binding sites for calcium-responsive transcription factors, including Npas4 and the members of CREB-transcriptional complex (West et al. 2001; Martinowich et al. 2003; Lin et al. 2008). In line with these studies, we found that Bdnf exon IV expression was down-regulated in the visual cortex of the LD rats. An additional mechanism that may mediate the reduction in BDNF expression in the LD visual cortex is the decrease in the expression of Npas4 which has been previously been shown to directly bind to the Bdnf promoters I and IV region (Lin et al. 2008).
Curiously, we found that within 1 week of LD, alterations in the chromatin structure and DNA methylation pattern at Bdnf promoter regions took place not only in the visual cortex but also in the hippocampus. Moreover, repressive H3K27 methylation was spread along all studied Bdnf promoters in the hippocampus. A recent study showed that prolonged LD in adult rats induces depression-like behavior and sensitizes rats to stressful factors (Gonzalez and Aston-Jones 2008). The repressive epigenetic mark at the BDNF promoter region might be at least one of the factors that predisposes LD animals to stress.
Why did the repressive chromatin remodeling not bring about a decreased BDNF synthesis in the hippocampus? First of all, the abundance of modifications on the histone tail raises the possibility that different histone modifications may cancel out the effect of each other (Kouzarides 2007). For example, increased H3 acetylation observed at the Bdnf promoter II in the hippocampus of the LD rats might have overcome the repressive chromatin state and prevented the reduction in the Bdnf exon II mRNA levels. Alternatively, it is possible that additional decrease in activity-dependent transcription factors, such as Npas4 and CBP, might be required for Bdnf mRNA down-regulation in the hippocampus.
We did not detect any influence of LD on the TrkB expression. This finding is in agreement with the previous study on the neurotrophins signaling in the rats reared in complete darkness from the birth (Viegi et al. 2002). Moreover, TrkB phosphorylation was also unaffected in the LD rats in our study, while Viegi and collaborators observed the decrease in Trk signaling in the dark-reared rats. This mismatch may reflect the differences in the mechanisms through which LD of adult rats and DR from birth affect neurotrophin signaling.
There is accumulating evidence that highlights the role of epigenetic mechanisms in the gene–environment interactions (Sweatt 2009). We have here shown that chromatin remodeling underlies the effects of ambient light on the expression of BDNF in the visual cortex. We further showed that LD induces a repressive epigenetic mark in the BDNF promoter regions in the hippocampus, although no changes in the expression of BDNF mRNA or protein were observed. This finding suggests that sensory deprivation may modify the epigenetic status of critical activity-regulated genes not only in the primary sensory areas but more widespread in brain. To which extent LD affects other brain areas apart from the visual cortex and hippocampus remains unclear, and therefore, the results obtained here cannot be generalized to other brain areas without further studies. Such an epigenetic mark might be one of the factors explaining why diminished ambient light predisposes both humans and experimental animals to depression (Magnusson and Partonen 2005; Gonzalez and Aston-Jones 2008).
References
Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T (2007) Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res 85:525–535
Alarcon JM, Malleret G, Touzani K, Vronskaya S, Ishii S, Kandel ER, Barco A (2004) Chromatin acetylation, memory, and LTP are impaired in CBP+/− mice: a model for the cognitive deficit in Rubinstein–Taybi syndrome and its amelioration. Neuron 42:947–959
An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, Xu B (2008) Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 134:175–187
Bernstein BE, Meissner A, Lander ES (2007) The mammalian epigenome. Cell 128:669–681
Castren E, Zafra F, Thoenen H, Lindholm D (1992) Light regulates expression of brain-derived neurotrophic factor mRNA in rat visual cortex. Proc Natl Acad Sci USA 89:9444–9448
Castren E, Voikar V, Rantamaki T (2007) Role of neurotrophic factors in depression. Curr Opin Pharmacol 7:18–21
Chakrabarti SK, James JC, Mirmira RG (2002) Quantitative assessment of gene targeting in vitro and in vivo by the pancreatic transcription factor, Pdx1. Importance of chromatin structure in directing promoter binding. J Biol Chem 277:13286–13293
Chiaruttini C, Sonego M, Baj G, Simonato M, Tongiorgi E (2008) BDNF mRNA splice variants display activity-dependent targeting to distinct hippocampal laminae. Mol Cell Neurosci 37:11–19
Chiaruttini C, Vicario A, Li Z, Baj G, Braiuca P, Wu Y, Lee FS, Gardossi L, Baraban JM, Tongiorgi E (2009) Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc Natl Acad Sci USA 106:16481–16486
Clark SJ, Harrison J, Paul CL, Frommer M (1994) High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22:2990–2997
Duman RS, Monteggia LM (2006) A neurotrophic model for stress-related mood disorders. Biol Psychiatry 59:1116–1127
Felsenfeld G, Groudine M (2003) Controlling the double helix. Nature 421:448–453
Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89:1827–1831
Gonzalez MM, Aston-Jones G (2008) Light deprivation damages monoamine neurons and produces a depressive behavioral phenotype in rats. Proc Natl Acad Sci USA 105:4898–4903
Hong EJ, McCord AE, Greenberg ME (2008) A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron 60:610–624
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Krishnan V, Nestler EJ (2008) The molecular neurobiology of depression. Nature 455:894–902
Kumar A, Choi KH, Renthal W, Tsankova NM, Theobald DE, Truong HT, Russo SJ, Laplant Q, Sasaki TS, Whistler KN, Neve RL, Self DW, Nestler EJ (2005) Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron 48:303–314
Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME (2008) Activity-dependent regulation of inhibitory synapse development by Npas4. Nature 455:1198–1204
Magnusson A, Partonen T (2005) The diagnosis, symptomatology, and epidemiology of seasonal affective disorder. CNS Spectr 10:625–634
Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE (2003) DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science 302:890–893
Onishchenko N, Karpova N, Sabri F, Castren E, Ceccatelli S (2008) Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem 106:1378–1387
Pollock GS, Vernon E, Forbes ME, Yan Q, Ma YT, Hsieh T, Robichon R, Frost DO, Johnson JE (2001) Effects of early visual experience and diurnal rhythms on BDNF mRNA and protein levels in the visual system, hippocampus, and cerebellum. J Neurosci 21:3923–3931
Rantamaki T, Hendolin P, Kankaanpaa A, Mijatovic J, Piepponen P, Domenici E, Chao MV, Mannisto PT, Castren E (2007) Pharmacologically diverse antidepressants rapidly activate brain-derived neurotrophic factor receptor TrkB and induce phospholipase-Cgamma signaling pathways in mouse brain. Neuropsychopharmacology 32:2152–2162
Sakata K, Woo NH, Martinowich K, Greene JS, Schloesser RJ, Shen L, Lu B (2009) Critical role of promoter IV-driven BDNF transcription in GABAergic transmission and synaptic plasticity in the prefrontal cortex. Proc Natl Acad Sci USA 106:5942–5947
Schoups AA, Elliott RC, Friedman WJ, Black IB (1995) NGF and BDNF are differentially modulated by visual experience in the developing geniculocortical pathway. Brain Res Dev Brain Res 86:326–334
Sweatt JD (2009) Experience-dependent epigenetic modifications in the central nervous system. Biol Psychiatry 65:191–197
Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME (1998) Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron 20:709–726
Timmusk T, Metsis M (1994) Regulation of BDNF promoters in the rat hippocampus. Neurochem Int 25:11–15
Tropea D, Capsoni S, Tongiorgi E, Giannotta S, Cattaneo A, Domenici L (2001) Mismatch between BDNF mRNA and protein expression in the developing visual cortex: the role of visual experience. Eur J Neurosci 13:709–721
Viegi A, Cotrufo T, Berardi N, Mascia L, Maffei L (2002) Effects of dark rearing on phosphorylation of neurotrophin Trk receptors. Eur J Neurosci 16:1925–1930
West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME (2001) Calcium regulation of neuronal gene expression. Proc Natl Acad Sci USA 98:11024–11031
Acknowledgments
The authors would like to thank Outi Nikkilä, Jesse Lindholm, and Virpi Nousiainen for excellent technical help. This study was supported by the Sigrid Juselius Foundation, and Academy of Finland Center of Excellence Program.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nina N. Karpova and Tomi Rantamäki contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Karpova, N.N., Rantamäki, T., Di Lieto, A. et al. Darkness Reduces BDNF Expression in the Visual Cortex and Induces Repressive Chromatin Remodeling at the BDNF Gene in Both Hippocampus and Visual Cortex. Cell Mol Neurobiol 30, 1117–1123 (2010). https://doi.org/10.1007/s10571-010-9544-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-010-9544-6