Abstract
Alternative hydrogen-bond structures were found for cellulose II and IIII based on molecular dynamics simulations using four force fields and energy optimization based on density functional theory. All the modeling results were in support to the new hydrogen-bonding network. The revised structures of cellulose II and IIII differ with the fiber diffraction models mainly in the orientation of two hydroxyl groups, namely, OH2 and OH6 forming hydrogen-bond chains perpendicular to the cellulose molecule. In the alternative structures, the sense of hydrogen bond is inversed but little difference can be seen in hydrogen bond geometries. The preference of these alternative hydrogen bond structures comes from the local stabilization of hydroxyl groups with respect to the β carbon. On the other hand when simulated fiber diffraction patterns were compared with experimental ones, the current structure of cellulose II with higher energy and the alternative structure of cellulose IIII with lower energy were in better agreement.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Hydrogen bonding (HB) is considered to be important for the structural stability of cellulose and other polysaccharides. However, due to the small X-ray scattering power of hydrogen, the hydrogen positions in a crystalline structure are difficult to determine. Furthermore, hydrogen can be disordered in the structure even when heavier atoms are ordered as can be seen in hexagonal water (Peterson and Levy 1957) or cyclodextrins (Saenger et al. 1982). Neutron scattering is adapted to probe hydrogen positions due to the strong scattering amplitudes of hydrogen and deuterium atoms as well as the high scattering length contrast between hydrogen and deuterium but suffers from relatively small flux and thus poor counting statistics.
During the last decade, the atomic coordinates of a series of cellulose allomorphs [Iα (Nishiyama et al. 2003), Iβ (Nishiyama et al. 2002), II (Langan et al. 1999, 2001), IIII (Wada et al. 2004)] including the hydroxyl groups’ hydrogen positions have been proposed based on X-ray and neutron fiber diffraction data of highly crystalline samples. So far, the proposed structural details have not encountered any contradiction with experimental data, and are used as reference structures. Modeling studies are often based on these X-ray/neutron structures that are currently the most reliable models. Nevertheless, some structures contain apparent hydrogen bond disorder and the exact positions of hydrogen atoms remains less established compared to those of the carbon and oxygen atoms that are less mobile and for which X-ray data can give reliable positions.
While studying the thermal behavior of cellulose II by molecular dynamics (MD) simulation, we observed an irreversible flipping of the donor–acceptor hydroxyl groups in the hydrogen bond pattern without any significant change of the carbon and oxygen positions in the structure. We further confirmed this behavior with a series of MD simulations using different force fields and by energy minimization using a more universal density functional theory with dispersion corrected potentials. We also examined the agreement with reported neutron diffraction intensity and propose a new structure as a viable alternative to the current cellulose II model. Similar approach was applied on cellulose IIII for which a similar alternative could be found.
Computational details
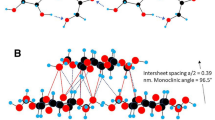
The X-ray/neutron structure coordinates of cellulose II by Langan et al. (1999, 2001) and IIII by Wada et al. (2004) were used to as starting models. P1 super cells with 40-chains (5 × 8) for cellulose II and 24-chains (4 × 6) for cellulose IIII and 8 glucose-per-chain (Fig. 1), with periodic boundary parallel to 0 1 0, 1 −1 0, 0 0 1 plane of II and 1 0 0, 0 1 0, 0 0 1 plane of IIII were constructed by symmetry operations. The end of each chain was covalently bonded with the other end of the periodic image of the same molecule.
All MD simulations were carried out using the GROMACS 4.5.5 package (Hess et al. 2008). Four carbohydrate force fields, namely GROMOS 56Acarbo (Hansen and Hünenberger 2011) with modifications in Lennard-Jones parameters (GROMOS r56Acarbo (Chen et al. 2014a), GLYCAM 06 (Kirschner et al. 2008), OPLS-AA (Damm et al. 1997) and CHARMM C36 (Guvench et al. 2009) were employed using our standard computational conditions (Chen et al. 2012, 2014b). The cutoff for coulomb and Lennard-Jones interactions was set to 0.9 nm. The MD simulations were carried out after optimization by steepest-descent followed by conjugate gradient method. System was heated up from 0 to 300 K in 6 ns in a stepwise manner with a heating rate of 10 K/100 ps and 100 ps equilibration every 10 ps. Between 300 and 500 K the heating/cooling rate and equilibration time was 10 K/200 ps and 200 ps respectively. Production runs of 10–24 ns were carried out at 300 and 500 K.
Periodic DFT calculations were performed by using the Quantum ESPRESSO 5.0 package (Giannozzi et al. 2009). We used the ultrasoft pseudopotentials and plane wave basis sets with a kinetic energy cutoff of Ecut = 70 Ry. A Monkhorst–Pack k-point grid with 2 × 2 × 2 points was used for sampling the Brillouin zone. The exchange correlation functional was described by the PBE generalized gradient approximation (Perdew et al. 1996; Grimme 2006). To take into account of the London dispersion correction in the calculation the DFT-D2 approach was used (Grimme2006). Two structures, the experimental coordinates and the coordinates after annealing using GROMOS r56Acarbo force field were taken as starting models. The structure optimization was carried out with the cell parameters relaxed.
A fiber diffraction diagram was simulated from calculated diffraction intensities for each model. The thermal parameters were set to be uniform and isotropic with U = 0.05 Å2 for all non-hydrogen (deuterium) atoms and U = 0.07 Å2 for hydrogen and deuterium. The diffraction patterns were convoluted with Gaussian functions in the directions perpendicular and along the chain axis to account for the limited crystal sizes and instrument broadening. The c-axis of the crystals was supposed to follow Gaussian distribution centered at fiber axis while a- and b-axis were randomly distributed around the c-axis using a procedure described previously (Nishiyama et al. 2012). Under such conditions, the simulated patterns are directly comparable to experimental data.
Entropy differences between structures with HB pattern A and B of cellulose II and IIII were estimated using a quantum harmonic oscillator approximation of their vibrational density of state (Berens et al. 1983). MD simulations for entropy calculations were performed for 100 ps in double precision for each force field starting from the equilibrated structures of cellulose II and IIII with HB pattern A and B. The time step was 0.5 fs and velocities were recorded every 1.5 fs. The vibrational density of state was calculated with the g_dos (Caleman et al. 2012) program of GROMACS.
Results and discussion
Cellulose II
Using GROMOS r56Acarbo force field, the root of mean square deviation with respect to the experimental structure of each residue was <0.3 Å at 300 K, and the basic conformational parameters remained unchanged.
Cellulose II has two non-equivalent chains in the unit cell. The one with its reducing end running in the direction of the c-axis (origin chain in Langan et al. 1999, 2001) is characterized by a smaller ring puckering polar angle. This parameter was found to be in the range of Θ = 4°–8° in the simulations whereas the other one, running in the opposite direction (center chain), was measured to be Θ = 12°–16°. In the latest X-ray structure (Langan et al. 2001), these angles are 5.0° and 10.2° respectively.
When heated up to 500 K, a remarkable discontinuity was seen between 350 and 400 K on different monitored structural parameters, such as unit cell parameters (Fig. 2) or torsion angle distributions of hydroxyl groups τ2 and τ6 corresponding to C1–C2–O2–H and C5–C6–O6–H (Fig. 3). The hydroxymethyl group torsion angle ω was 100 % in gt conformation (dihedral angle of around 60°) at 300 K, although alternative conformations appeared early in the heating stage with a non-monotonical variation upon heating.
When cooled down to 300 K, the unit cell parameters did not show any transition and reached values only slightly different from the starting structure (Fig. 2). The values of τ2 and τ6 also remained essentially the same as at 500 K (Fig. 3). On the other hand, the hydroxymethyl groups went back to 100 % gt conformation when cooled to 300 K (Fig. 3). The torsion angle τ3 showed little variation during both heating and cooling.
A glimpse at simulation snapshots of the two structures before and after annealing (Fig. 4) clarifies that they are similar although not identical. The infinite hydrogen-bonding chains in the direction perpendicular to the pyranose plane, engaging hydroxyl groups in 2 and 6 positions in the sequence of …(O2o…O2c…O6c…O6o…) were present before and after the change. Here, the suffix ‘o’ and ‘c’ represents the origin and center chain respectively. The difference between the two structures before and after annealing is primarily characterized by the hydrogen bonds: (HO2o–O2o…HO2c–O2c…HO6c–O6c…HO6o–O6o…) in the structure before and (O2o–HO2o…O2c–HO2c…O6cHO6c…O6oHO6o…) after annealing. We name the former hydrogen bond pattern A and the latter pattern B (Fig. 4). In terms of conformation, the HO2 atom (hydroxyl hydrogen at position 2) is in the trans position with respect to the aliphatic hydrogen H2 in pattern A whereas during the annealing it rotated to nearly eclipse position (pattern B). This hydrogen pattern was maintained during the cooling process. The total potential energy of the annealed structure at 300 K was about 5.7 kJ/mol per glucosyl unit (Table 1) lower than the total potential energy of the structure in the heating process at the same temperature.
Interestingly, simulations with the force-fields GLYCAM 06 and CHARMM C36 resulted in a similar flip transition of HO2 and HO6 although at a lower temperature around 300 K. The OPLS-AA force-field also gave a similar transition between 300 and 430 K (figure S1). The total potential energies (ΔEpot) at either 100 K or 300 K before and after annealing are listed in Table 1. The energy difference between pattern A and B was even larger with these three force fields. The entropy contribution for the stabilization of structures was almost negligible (supplementary files Table S1). Thus the enthalpy difference, which equals to the energy difference here, can be regarded as the scale of free energy and is corroborated with the transition temperature.
The energy minimization process using density functional theory conserved the hydrogen bond patterns. The energy after optimization of pattern A was 8.14 kJ/mol per glucosyl unit higher than that of pattern B. The structures optimized by DFT followed the P21 symmetry within 0.01 Å accuracy (Table S4) and thus the fractional coordinates of the asymmetric unit are given as cif files in supplementary files.
Cellulose IIII
Simulations of the allomorph cellulose IIII showed similar behavior. In contrast to cellulose II, the unit cell of cellulose III I contains only one chain, and the infinite hydrogen bond chain (O2…O6…), perpendicular to the chain direction, is present in the crystallographic model. In the current crystallographic model, the HO2 is in trans configuration with respect to H2, while the alternative hydrogen bonding scheme, corresponding to pattern B, was also considered during the refinement.
Cellulose IIII is characterized by having its hydroxymethyl groups in gt and a non-staggered chain arrangement. At 300 K, these features were unchanged with GROMOS, OPLS and CHARMM. On the other hand, the GLYCAM06 force-field caused chain sliding that led to a staggered arrangement similar to cellulose I, while simultaneously maintaining the gt conformation of the hydroxymethyl group. When heated to 500 K, similar chain sliding was observed between 400 and 450 K even with the GROMOS and CHARMM force-fields. One could relate this phenomenon to the experimental observation that cellulose IIII converts to cellulose I by hydrothermal treatment (Chanzy et al. 1987). However, spontaneous flip-flop from pattern A to pattern B could not be observed. With OPLS the basic structure remained stable at 600 K and a flip from pattern A to pattern B occurred at around 570 K. When pattern B was used as initial structure, only CHARMM and OPLS gave basic features of cellulose IIII at 300 K described above.
Since the structures with pattern A or pattern B departed too far from the experimental structure during MD simulation, the energy components using GLYCAM and GROMOS force fields were compared only from energy minimization (EM) (Table 2). In cellulose IIII again, the energy of pattern B was lower than the energy of pattern A. However, the energy difference was not as significant as the difference for cellulose II. From DFT, the structure with pattern B was 2.95 kJ/mol per residue lower than that with pattern A, close to k B T at 300 K.
Effect of local geometry
To identify the factors favoring pattern B over A the different potential energy contributions were compared. The four force fields employed here agree that the 1–4, 1–5 Coulomb interaction favors pattern B (Table 1). The smaller difference of Coulomb 1–4 energy with CHARMM and OPLS force field can be partially explained by the positive charge (0.09 e in CHARMM and 0.06 e in OPLS) attributed to H2 causing Coulomb repulsion with HO2 in cis configuration. Since aliphatic hydrogen atoms have zero charge in GLYCAM, and GROMOS does not even have explicit aliphatic hydrogen, the difference in 1–4, 1–5 Coulomb interaction in those force-fields probably originates from the repulsion between carbon and hydroxyl hydrogen. The contribution of short-range Coulomb interaction that involves hydrogen bond energy was either positive or negative depending on the force field and the hydrogen bonding geometry within the two structures were not very different (supplementary Table S2 and S3).
The effect of local geometry can be further found in analogue structures found in a crystallographic database (Fig. 5). The hydroxyl hydrogen is found in three staggered positions but has a strong tendency to avoid the side of the aliphatic carbon. Especially the secondary hydroxyl groups have small narrow distribution when the hydroxyl atom is on the aliphatic carbons side. The pattern A corresponds to the situation where the secondary hydroxyl groups are on the aliphatic carbon while they are in the opposite side in pattern B.
Polar histograms of H–C–O–H dihedral angles of secondary alcohol (left) and C–C–O–H dihedral angles of primary alcohol (right) found in cambridge structural database (Allen 2002; Bruno et al. 2004). The solid lines are the dihedral angles in cellulose II structures and dashed lines in cellulose IIII structures after DFT optimization
The flipping of hydrogen bond from pattern A to B implies a movement of nearly 1.7 Å, but the distance to the tautomer hydrogen falls between 0.7 and 0.77 Å for cellulose II and 0.51–0.57 Å for cellulose IIII.
Given the limited resolution and uncertainty of the carbon and oxygen atomic positions, and given the fact that neutrons themselves do not distinguish the chemical bonds, it is difficult to conclude between the two competing models based on the current experimental data only. Indeed, close inspection of the structural models of β-D-cellotetraose hemihydrate based on single crystal X-ray diffraction studies that were independently published by two groups almost simultaneously, we find discrepancy corresponding to the difference between pattern A and B. Gessler and coworkers could locate 12 of the hydroxyl hydrogen atoms directly on the Fourier map (Gessler et al. 1995), the orientations of HO2 and HO6 are similar to our pattern B. On the other hand, Raymond and his co-workers placed the hydrogen atoms at the position of maximum electron density on a circle of standard bond angle and bond length (Raymond et al. 1995), resulting in the orientation of HO2 and HO6 similar to the pattern A here.
The simulated neutron fiber diffraction data of the DFT-optimized structure of cellulose II with pattern A and pattern B is shown in Fig. 6 together with the experimental data. In the simulated diffraction patterns, remarkable differences can be found close to the meridian where the relative intensities of 0 0 4 and 0 0 6 were really strong in pattern A compared to pattern B, both in the hydrogenated and the deuterated version. In the experimental data 0 0 4 and 0 0 6 are stronger than other layer line spots which supports pattern A, contrary to the energetic considerations based on force fields or DFT optimization. The cellulose II samples used to record fiber diffraction have limited crystallite size and crystallinity as is obvious from the line broadening and the limited resolution of the diffraction pattern. The structure is thus probably not attaining the organization of lowest energy due to the kinetics of regeneration process. The strong meridian in the experimental data can also arise from domains with poor lateral order but with a rather strict P21 symmetry within each individual chain as indicated by the absence of odd-order meridian reflections. A quarter stagger between two adjacent chains would also give relatively strong 0 0 4.
Simulated neutron fiber diffraction pattern of DFT optimized cellulose II structures with hydrogen bond pattern A (left) and pattern B (middle) compared with experimental data (Langan et al. 1999)
The statistical test on neutron data had not ruled out the co-existence or existence of the pattern B, and our DFT result suggests that it is highly probable that pattern B at least should co-exist in the structure.
The simulated diffraction pattern A and B of cellulose IIII were similar. However, some differences can be visually pointed as in the circled region of Fig. 7. In the neutron fiber diffraction data, due to the relatively low signal-to-noise ratio, imperfect absorption corrections and the change of sample volumes in the beam, it is difficult to evaluate with precision the relative intensities of distant spots. However, if the intensity ratio of neighboring spots were inversed as indicated in the circle, a good counting statistics would discriminate the two situations. For example, in the hydrogenated version of cellulose IIII, different peak appear in the circle between pattern A and pattern B. Visual inspection of the experimental data suggests that the pattern B is more likely.
Simulated neutron fiber diffraction data of DFT optimized cellulose IIII with hydrogen bond pattern A (left), pattern B (middle) and experimental data (Wada et al. 2004). Differences can be visually noticed in the circled area
Conclusion
A new plausible hydrogen bonding structure that was earlier omitted during the refinement process of cellulose II and IIII is here suggested from MD simulations using various force fields. The density functional theory with dispersion correction further supported this optional model. The preference for the alternative hydrogen-bonding scheme was found to be due to the local geometry dictated by second neighboring atoms. The simulated neutron fiber diffraction pattern of cellulose IIII supported the new structure while that of cellulose II showed discrepancies with the reported experimental diffraction pattern.
References
Allen FH (2002) The chambridge structural database: a quarter of a million crystal structure and rising. Acta Crystallogr Sect B Struct Sci 58:380–388
Berens PH, Mackay DHJ, White GM, Wilson KR (1983) Thermodynamics and quantum corrections from molecular dynamics for liquid water. J Chem Phys 79:2375–2389
Bruno IJ, Cole JC, Kessler M, Luo J, Motherwell WDS, Purkis LH, Smith BR, Taylor R (2004) Retrieval of crystallographically-derived molecular geometry information. J Chem Inf Comput Sci 44:2133–2144
Caleman C, van Maaren PJ, Hong M, Hub JS, Costa LT, van der Spoel D (2012) Force field benchmark of organic liquids: density, enthalpy of vaporization, heat capacities, surface tension, isothermal compressibility, volumetric expansion coefficient, and dielectric constant. J Chem Theory Comput 8:61–74
Chanzy H, Henrissat B, Vincendon M, Tanner SF, Belton PSB (1987) Solid state 13C NMR an electron microscopy study on the reversible cellulose I->cellulose III transformation in Valonia. Carbohydr Res 160:1–11
Chen P, Nishiyama Y, Mazeau K (2012) Torsional entropy at the origin of the reversible temperature-induced phase transition of cellulose. Macromolecules 45:362–368
Chen P, Nishiyama Y, Mazeau K (2014a) Atomic partial charges and one Lennard-Jones parameter crucial to model cellulose allomorphs. Cellulose 21:2207–2217
Chen P, Nishiyama Y, Putaux JL, Mazeau K (2014b) Diversity of potential hydrogen bonds in cellulose I revealed by molecular dynamics simulation. Cellulose 21:897–908
Damm W, Frontera A, Tirado-Rives J, Jorgensen WL (1997) OPLS all-atom force field for carbohydrates. J Comput Chem 18:1955–1970
Gessler K, Krauss N, Steiner T, Betzel C, Sarko A, Saenger W (1995) β-D-cellotetraose hemihydrate as a structural model for cellulose II. An X-ray diffraction study. J Am Chem Soc 117:11397–11406
Giannozzi P, Baroni S, Bonini N, Calandra M, Car R, Cavazzoni C, Ceresoli D, Chiarotti GL, Cococcion M, Dal Dabo I, Corso A, de Gironcoli S, Fabris S, Fratesi G, Gebauer R, Gerstmann U, Gougoussis C, Kokalj A, Lazzeri M, Martin-Samos L, Marzari N, Mauri F, Mazzarello R, Paolini S, Pasquarello A, Paulatto L, Sbraccia C, Scandolo S, Sclauzero G, Seitsonen AP, Smogunov A, Umari P, Wentzcovitch RM (2009) QUANTUM ESPRESSO: a modular and open-source software project for quantum simulations of materials. J Phys Condens Matter 21:395502
Grimme S (2006) Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J Comput Chem 27:1787–1799
Guvench O, Hatcher E, Venable RM, Pastor RW, MacKerell J, Alexander D (2009) CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J Chem Theory Comput 5:2353–2370
Hansen HS, Hünenberger PH (2011) A reoptimized GROMOS force field for hexopyranose-based carbohydrates accounting for the relative free energies of ring conformers, anomers, epimers, hydroxymethyl rotamers, and glycosidic linkage conformers. J Comput Chem 32:998–1032
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Kirschner KN, Yongye AB, Tschampel SM, Gonzalez-Outeirino J, Daniels CR, Foley BL, Woods RJ (2008) GLYCAM06: a generalizable biomolecular force field carbohydrates. J Comput Chem 29:622–655
Langan P, Nishiyama Y, Chanzy H (1999) A revised structure and hydrogen-bonding system in cellulose II from a neutron fiber diffraction analysis. J Am Chem Soc 121:9940–9946
Langan P, Nishiyama Y, Chanzy H (2001) X-ray structure of mercerized cellulose II at 1 Å resolution. Biomacromolecules 2:410–416
Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124:9074–9082
Nishiyama Y, Sugiyama J, Chanzy H, Langan P (2003) Crystal structure and hydrogen bonding system in cellulose Iα from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 125:14300–14306
Nishiyama Y, Johnson GP, French AD (2012) Diffraction from nonperiodic models of cellulose crystals. Cellulose 19:319–336
Perdew JP, Burke K, Ernzerhof M (1996) Generalized gradient approximation made simple. Phys Rev Lett 77:3865–3868
Peterson SW, Levy HA (1957) A single-crystal neutron diffraction study of heavy ice. Acta Cryst 10:70–76
Raymond S, Heyraud A, Qui DT, Kvick Å, Chanzy H (1995) Crystal and molecular stucture of β-D-cellotetraose hemihydrate as a model of cellulose II. Macromolecules 28:2096–2100
Saenger W, Betzel C, Hingerty B, Brown GM (1982) Flip-flop hydrogen bonding in a partially disordered system. Nature 296:581–583
Wada M, Chanzy H, Nishiyama Y, Langan P (2004) Cellulose IIII crystal structure and hydrogen bonding by synchrotron X-ray and neutron fiber diffraction. Macromolecules 37:8548–8555
Acknowledgments
We thank Dr. Jakob Wohlert for discussion and providing GLYCAM 06 and CHARMM C36 force field parameters in GROMACS format. P. C. thanks funding support from Chinese government to study in France. Y. O. was supported by a Grant-in-aid for JSPS research fellow (23-2362). M. B. W. Thanks the Swedish Foundation for Strategic Research (SSF) for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Supplementary figures and tables. The cif files containing fractional coordinates of DFT optimized structures of cellulose II and IIII with pattern A and pattern B.
Rights and permissions
About this article
Cite this article
Chen, P., Ogawa, Y., Nishiyama, Y. et al. Alternative hydrogen bond models of cellulose II and IIII based on molecular force-fields and density functional theory. Cellulose 22, 1485–1493 (2015). https://doi.org/10.1007/s10570-015-0589-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-015-0589-z