Abstract
The acsD gene is involved in cellulose biosynthesis among the Acetobacter species. In the current study, we created an acsD disruption mutant in the acsABCD cellulose synthase operon of Gluconacetobacter xylinus and characterized the resulting cellulose to aid in providing insight into the function of the acsD gene. Both the wild type G. xylinus AY201 (derivative of Gluconacetobacter hansenii ATCC 23769) and the acsD disruption mutant produced crystalline cellulose I microfibrils. The cellulose produced by both appeared to be synthesized from an aggregate of pores known as a linear terminal complex; however the total cellulose synthesized was 10 % that of the wild type G. xylinus AY201. TEM observations of the acsD disruption mutant confirmed that microfibrils and bundles of microfibrils were similar in size to the G. xylinus AY201 wild type; however, the final ribbon dimensions were narrower (53.4 ± 13.1 nm wt, vs. 28.2 ± 8.2 nm). Additional TEM observations of the mutant cells incubated at 4 °C revealed an abnormal linear terminal complex orientation whereby the resulting band material could be observed in a transverse orientation as well as longitudinally to the long axis of the cell. Taken together, these data strongly suggest that acsD aids in the proper orientation of the linear terminal complexes along the longitudinal axis of the cell indicating the AcsD protein is involved in the final level of the hierarchical assembly of cellulose resulting in highly efficient cellulose synthesis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Cellulose is a crystalline biopolymer comprised of extended chains of β-1,4-linked glucose residues (Saxena and Brown 2005). It is the most abundant bio-macromolecule and is of significant economic importance. Cellulose is produced by plants, algae, tunicates, colorless protists, as well as photosynthetic and heterotrophic bacteria (Brown 1985; Ross et al. 1991; Blanton et al. 2000; Kimura et al. 2001). The gram negative bacterium, Gluconacetobacter xylinus (formerly Acetobacter xylinum), is a particularly efficient producer of pure, highly crystalline cellulose (Nishi et al. 1990; Cousins and Brown 1997; Nobles and Brown 2008). This product, termed bacterial cellulose, has distinctive properties that differentiate it from cellulose found in other organisms. Bacterial cellulose is particularly well suited for medical, industrial, and commercial applications because of its ultra-fine reticulated structure, high crystallinity, great mechanical strength, high water holding capacity, moldability during formation, and biocompatibility (Yamanaka et al. 1989; Ross et al. 1991; Yoshinaga et al. 1997; Czaja et al. 2004).
In G. xylinus, cellulose biosynthesis occurs through membrane-embedded enzyme complexes with individual catalytic subunits that are capable of polymerizing UDP-activated glucose (UDPG) into glucan chains when activated by the second messenger cyclic-di-GMP which is sensed by the bacteria through PilZ domains (Saxena et al. 1994; Weinhouse et al. 1997; Romling et al. 2005; Amikam and Galperin 2006; Morgan et al. 2013). The genes necessary for this complex process are the cellulose synthase operon, acsABCD, and flanking genes, ccpAx, cmcAx and bglAx (Saxena et al. 1994; Standal et al. 1994; Nakai et al. 2013; Deng et al. 2013).
The operon acsABCD was discovered in 1994 and codes for the proteins AcsAB, AcsC, and AcsD (Saxena et al. 1994). AcsA is an integrated inner membrane protein that contains the catalytic subunit for cellulose biosynthesis (Morgan et al. 2013). It has a PilZ domain that is responsive to c-di-GMP and, when activated, the AcsA protein changes conformation to allow for UDPG to access the catalytic site where polymerization commences (Ross et al. 1987; Romling et al. 2005; Morgan et al. 2013). The AcsB protein has a C-terminal membrane spanning domain with the major part extending into the periplasm is thought to be weakly associated with the cytoplasmic membrane and extends into the periplasm where it is suggested to be tethered to the outer membrane (Iyer et al. 2013; Morgan et al. 2013). The function of AcsB is not known, but it is suggested that the cellulose binding domains found in the protein may support the unidirectional movement of the polymerized glucose chain from the cytoplasmic to the periplasmic space. This data was gathered from Rhodobacter sphaeroides, an organism that has BcsAB (AcsAB) as two separate proteins. These proteins do not synthesize cellulose and may not reflect this exact process in G. xylinus (Morgan et al. 2013). The AcsC protein is essential for cellulose biosynthesis in vivo and is thought to be involved in the formation of an outer membrane pore complex due to its homology with other membrane and pore proteins, its predicted 18-stranded β-barrel structure, and possible localization in the outer membrane (Saxena et al. 1994; Morgan et al. 2013).
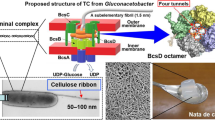
Of all the genes in the acsABCD operon, acsD is the only one not necessary for cellulose biosynthesis; however, it was shown to be necessary for the normal observed rate of synthesis of cellulose I microfibrils (Saxena et al. 1994). Previously it was suggested that the AcsD protein may play a role in the perfection of crystalline cellulose (Saxena et al. 1994). The structure and arrangement of the AcsD protein itself indicates a cylinder with the presence of a functional complex unit in the form of an octamer (Hu et al. 2010). The octamer is localized in the periplasm with an orientation parallel to the long axis of the cell where it interacts with CcpAx (Hu et al. 2010; Iyer et al. 2011; Sunagawa et al. 2013). Additionally, it has an interior space that may act as a channel through which a nascent glucan chain may pass through, thus supporting the concept that it may be a part of the pore complex; however, its true function is still unknown (Hu et al. 2010; Iyer et al. 2011).
In addition to the acsABCD operon, two genes upstream of the coding region, cmcAx, ccpAx, and one gene downstream, bglAx, are thought to play important roles in cellulose biosynthesis (Standal et al. 1994; Nakai et al. 2013; Sunagawa et al. 2013; Deng et al. 2013). The proteins CmcAx and BglAx have been suggested to aid in proper cellulose biosynthesis by cleaving tangled glucan chains when there is a failure in arrangement (Nakai et al. 2013; Deng et al. 2013) while CcpAx was determined to be essential for cellulose production and showed a significant interaction with AcsD (Standal et al. 1994; Sunagawa et al. 2013).
The highly organized enzyme assemblies coded by the acsABCD operon and flanking genes are responsible for the polymerization, crystallization, and secretion of crystalline cellulose ribbons into the external milieu (Saxena et al. 1994; Standal et al. 1994; Nakai et al. 2013; Sunagawa et al. 2013; Deng et al. 2013). Synthesis of cellulose in G. xylinus occurs as a consecutive, linked two-step process: (1) polymerization of glucose residues within the catalytic sites of the cellulose synthesizing protein complex form polymer chains (Saxena, et al. 1995; Cousins and Brown 1997) and (2) van der Waals forces which facilitate crystallization of the polymer chains into glucan mini-sheets (Cousins and Brown 1995). Following the initial crystallization of glucose polymer chains, the glucan sheets undergo hydrogen bonding to form cellulose mini-crystals that exit the pore complex (Cousins and Brown 1995, 1997). This next sequence involves the cell-directed crystallization of nascent cellulose mini-crystals into microfibrils, microfibrils into bundles, and the aggregation of these bundles into the final ribbon assembly (Haigler et al. 1982; Ross et al. 1991; Cousins and Brown 1997).
A hierarchical cell-directed self-assembly process is thought to dictate the crystallization of cellulose in G. xylinus. Terminologies used for the different hierarchical levels and assembly process were described previously (Brown 1996; Cousins and Brown 1997). For the purposes of clarity, the nomenclature used in this paper is outlined in Table 1.
Some aspects of the hierarchical crystallization process can be dissected by the use of extracellular chemical probes and energy analysis of bonding schemes. A computational energy analysis was utilized to understand cellulose assembly within and emergence from a single TC subunit (Cousins and Brown 1995). The analyses showed that van der Waals associations of the glucan chains were more favorable than hydrogen bonding for mini-sheet formation while the resulting mini-sheets favor hydrogen bonding to form the cellulose mini-crystal which exits the pore (Cousins and Brown 1995). Using this data, a model was constructed which suggested that one mini-crystal emerges from a single terminal complex (TC) subunit (Cousins and Brown 1995; Brown 1996). The fluorescent brightener Tinopal LPW (4,4′-bis[2-hydroxyethylamino-1,3,5-triazin-2-yliamino]-2,2′-stilbenedisulfonic acid, previously referred to as Calcofluor White™) was demonstrated to interrupt the in vivo assembly of crystalline cellulose I microfibrils by competing for the hydrogen bonding sites within the glucose residues of the nascent glucan mini-crystals (Haigler et al. 1980). Additionally, this disruption led to an increase in polymerization rate indicating that these two processes were separate, but coupled phases of cellulose biosynthesis (Haigler et al. 1980; Benziman et al. 1980). This suggested that one terminal complex (TC) is comprised of TC subunits which are precisely spaced into arrays that facilitate the hydrogen bonding necessary for the formation of the crystalline cellulose I microfibrils (Haigler et al. 1980; Benziman et al. 1980; Brown 1996). Carboxymethylcellulose (CMC) was used as a chemical probe to interrupt cellulose assembly by inhibiting the integration of bundles of cellulose I microfibrils into ribbons (Haigler et al. 1982). Furthermore, freeze fracture and negative staining analysis showed that TC subunits are arranged linearly along the long axis of the cell; however, the arrangement is not cohesive because the linear segments of TC subunits appear discontinuous and slightly overlapping (Haigler et al. 1982). This suggested that a bundle is produced by a single linear TC comprised of multiple TCs arranged in such a way as to allow for the fasciation of the microfibrils into bundles of microfibrils (Haigler et al. 1982; Cousins and Brown 1997).
Many organisms are capable of producing cellulose; however none are as prolific as the gram negative bacterium G. xylinus (Nobles and Brown 2008). This ability appears to be derived from a hierarchical assembly process whereby TC subunits are arranged in a precise coordination to allow for the production of microfibrils, bundles of microfibrils and the final ribbon assembly (Haigler et al. 1982); however we do not know what dictates this organizational pattern. Previously, it has been reported that the quantities of cellulose dropped precipitously when there was a disruption in the acsD gene, however cellulose biosynthesis did not cease (Saxena et al. 1994). The AcsD protein was proposed to be a component of the pore complex or to be involved in the organization of the pores contained within the linear terminal complex (TC) (Saxena et al. 1994). The purpose of this study was to re-investigate an acsD disruption mutant to determine what role this gene plays in cellulose biosynthesis. During our characterization, we observed abnormalities in the orientation of the linear TCs on the acsD mutant cells, while the components within each liner TC appeared to be aligned normally and functioned similar to that of the wild type. We used this evidence to provide additional support for the concept that there is indeed a hierarchical cellulose assembly process in G. xylinus, and it leads to highly efficient cellulose production.
Materials and methods
Bacterial strains, media, and growth conditions
The bacterial strains and plasmids used in this study are shown in Online Resource 1 and primers in Online Resource 2. Escherichia coli strains were grown at 37 °C in Luria–Bertani (LB) medium on a rotary shaker at 140 rpm or on LB plates with 2 % agar. For both G. xylinus and E. coli cultures the selection of resistance markers was performed by using the following antibiotics: ampicillin (Amp 50 μg/mL), chloramphenicol (Cm 50 μg/mL), and/or tetracycline (Tet 12.5 μg/mL). G. xylinus AY201 (derivative of Gluconacetobacter hansenii ATCC 23769, Laboratory Stock) and derivatives were grown in Schramm and Hestrin (SH) medium at 28 °C under static or rotary shaking conditions or on 2 % agar medium as previously described (Schramm and Hestrin 1954). To prepare a uniform cell suspension, 0.4 % cellulase was added to the culture medium 24 h before harvesting.
Construction of the acsD disruption mutant (acsDdm) and complementary strains (acsDCGFP and acsDNGFP)
To construct the acsDdm mutant, a 200 bp acsD fragment was PCR amplified using the primers, acsDf1-re and acsD-r1-HindIII. The taq polymerase used was ordered from NEB (Cat # M0273L) and the PCR product was purified using Qiagen QIAquick PCR purification kit. The PCR conditions were as follows: 92 °C for 40 s (denaturation), 60 °C for 40 s (annealing), and 72 °C for 60 s (elongation). The PCR product and the vector pBR325 were digested with restriction enzymes BclI and HindIII and ligated using NEB restriction enzymes (Cat# R0106S, Cat#R0104S) and NEB DNA ligase (Cat#M0202T). The ligated product was transformed to One Shot® TOP10 Chemically Competent E. coli according to the manufacturer’s instructions. The transformants were selected on LB agar medium supplemented with ampicillin and tetracycline.
The purified plasmid from E. coli was electroporated in G. xylinus to create the acsD mutant through a single cross over event on the bacterial chromosome. Wild type G. xylinus AY201 cells were grown in SH liquid medium with shaking at 140 rpm until they reached log phase (optical density of 0.7 at 600 nm). The cells were then washed in saline and suspended in 10 % glycerol. The knockout plasmid (pBR325 + ΔacsD) purified from E. coli was transformed in the wild type G. xylinus AY201 cells using Bio-Rad E. coli pulser (Cat # 165-2101) following manufacturers protocol. After electroporation the cells were grown in SH liquid medium for 3 h before they were spread on SH agar medium with tetracycline.
The two plasmids, vector with NGFP and vector with CGFP were fused, and the ligation product was transformed to E. coli strains TOP10 according to the manufacturer’s instructions. The transformants were selected by implanting on LB agar medium containing chloramphenicol. The amplified product was collected and electroporated in acsDdm strains to procure acsDNGFP and acsDCGFP strains. After electroporation, the cells were grown in SH liquid medium for 3 h before they were spread for selection on SH medium with chloramphenicol.
Screening for the disruption mutant in G. xylinus AY201
Colonies obtained after electroporation were screened using PCR. The primers F1-lig and R1-lig were used to amplify the plasmid residing within the acsD region to check for the presence of the knockout plasmid pBR325 + ΔacsD in the acsD mutant strain.
RT-PCR analysis to demonstrate the expression
Total RNA was extracted by using TRIzol max RNA isolation kit according to manufacturer’s protocol. Qiagen One-step RT-PCR kit was used to perform the reaction. The procedure was followed using the manufacturer’s instructions for 1 μg of total RNA and primers RT-F1 and RT-R1. Expression of acsC was tested using primers RT-acsC-F1 and RT-acsC-R1. The expression data for acsC was used as a control. AcsC and AcsD sequences were taken from GenBank (accession number X54676).
Construction of complement strains
The complements of the acsD disruption mutant were created by cloning a complete acsD gene. Three different PCR amplification reactions were performed simultaneously to clone the tac promoter, the acsD gene, and the GFP gene. These three amplified products were fused together with the help of a USER fusion system. Two different clones were created depending on the placement of the GFP gene in the clone at the N-terminal (acsDNGFP) or the C-terminal (acsDCGFP) region of the acsD gene. To check for the intact acsD gene fusion, the complete fusion was amplified using PCR, and the purified products were sequenced on an ABI 3730 DNA analyzer (the DNA sequencing facility, the University of Texas at Austin).
The complement strain vector (pCTP1) was prepared by Dr. D Nobles Jr. (Brown lab at UT Austin). This construct was made by using two 42 bp oligonucleotide sequences with 3′ complimentary ends of 38 bp and different 5′ ends of 5 bp. The 5′ ends have the equivalent of overhanging sticky end left after digestion by HindIII and other with XhoI. The construct was created after the oligos anneal to make double stranded vector with sticky HindIII and XhoI sticky ends on each side. The entire amplified insert is pre-digested with XhoI and HindIII to introduce into the vector. This whole construct is just a way to convert vectors to a USER-friendly form. The ratio of the amplified inserts to the vector was 4:1.
Membrane isolation, SDS-PAGE and Western Blotting of complement
Spheroplasts were obtained by adding 0.05 mg/ml of lysozyme to a 10 mM-Tris/0.75 M sucrose solution (pH 7.8) containing either acsDCGFP or acsDNGFP cells at 109 cells/ml according to the procedure in Methods in Enzymology, volume 31 (Osborn and Munson 1974). Spheroplasts were lysed by brief sonication using a 30KC Sonifer. The sonicated suspension was centrifuged at low speed for 5–7 min and the supernatant containing the membrane fraction and the cytoplasmic extract was collected and centrifuged at 40,000 rpm for 4 h at 2 °C to obtain the pellet which contained the membranes and the supernatant which contained the cytoplasmic extract.
The membranes were isolated by suspending the pellets in an ice-cold buffer solution containing 3.3 mM-Tris, pH 7.8, supplemented with 1 mM EDTA and 0.25 M sucrose. The total membrane (TM) fraction was collected through centrifugation at 40,000 rpm for 2 h. Sucrose density gradient centrifugation was employed to separate the membranes whereby the TM membrane fraction was suspended in a cold 25 % sucrose solution containing 5 mM EDTA (pH 7.5) and then layered on a 30–50 % sucrose gradient and centrifuged to equilibrium using a Beckman centrifuge with a SW41T rotor. Gradients were collected by removal of material from the top of the tube using a coarse needle. The discrete bands were recovered from the fractions by diluting the suspension with 1 mM EDTA pH 7.5, followed by centrifugation at 40,000 rpm for 2 h. The TM fractions were separated using SDS-PAGE according to the protocol described in Laemmli (1970) and then transferred from the gels to a nitrocellulose membrane (Bio-rad, Cat # 162-0114), using a Bio-Rad Semi Dry transfer cell for 1 h at 15 V. The membrane was then completely dried and the blots were performed using enhanced GFP chemiluminescent (ECL) detection kit (Roxland immunochemicals, catalogue#KCA215). The procedure was followed as per manufacturer’s protocol.
Glucose assay, quantitative cellulose measurement, cell counts and X-ray diffraction
To obtain samples for the glucose assay, quantitative cellulose measurement and X-ray diffraction; liquid cultures of wild type G. xylinus AY201 and the acsDdm mutant were grown to log phase until reaching a concentration of 106 CFU/mL. The resulting inoculum was added to 100 mL SH media and incubated at 28 °C under static conditions for 7 days. Three biological and three experimental repeats were used for each test. To determine the amount of utilized glucose, the remaining medium was collected and a glucose assay (Sigma Aldrich GAHK20) was used according to the manufacturer’s protocol for measuring the residual glucose in the spent medium. To determine quantitative cellulose growth, pellicles from the wild type, mutant and complement strains (acsDNGFP and acsDCGFP) from 3 experiments using 3 technical repeats were collected and washed with 3 % NaOH for 2 days and water for 5 days and weighed. The wet pellicle was dried on Teflon for 72 h at 40 °C and then weighed. Cell count was determined by collecting 1 g of the cellulose pellicle and 1 mL of the liquid medium from the wild type and mutant 7 day cultures. Cellulase was added to make a final solution of 1 %. The samples were incubated for 2 h on a rotary shaker set at 140 rpm. The optical density of the resulting cellulose-free cell solution was measured using a Beckman DU 640 Spectrophotometer at 600 nm. X-ray diffraction analysis using the Rigaku R-axis Spider X-ray diffraction apparatus was performed on wild type and mutant pellicles washed in 3 % Alconox detergent and autoclave sterilized. The samples were washed with water to remove the detergent and any remaining salts and then coated with mineral oil. Diffraction data was collected with CuKα radiation (A = 1.5418 Å), a beam current of 40 mA, a power of 40 kV, temperature at 25 °C, and a scan speed of 5°/min from 2θ of 0°–45° with a read out time of 30 min.
Time lapse, cell and colony light microscopy, and TEM
Plate cultures were used for time lapse, light microscopy and TEM observations. The wild type G. xylinus AY201 and the acsDdm mutant cells were grown on 1.5 % SH-agar medium and colonies were extracted between 4 and 7 days of growth. Light microscopy observations were made using a Zeiss D-7082 light microscope and a Zeiss 475057 dissecting microscope while TEM observations were made using a Philips 420 Transmission Electron Microscope available in the Brown lab. All of the light microscopy and TEM images were obtained using Image-Pro Plus software. Time lapse video was obtained by using the sequencing tool within the software which allows for multiple image capture at an interval of 1 frame every 5 s and measurements of the cellulose ribbons, bundles and microfibrils were obtained by using the measurements tool on the captured TEM images.
To obtain time lapse videos, the extracted colonies were suspended in 1 mL of SH liquid medium. The slides were prepared by pipetting a volume of 20 μL of hot 1.5 % SH-agar solution to form a base of agar medium on a microscope slide. This was followed by the addition of 15 μL of SH medium and then 15 μL of the above prepared cell sample solution. The sample was covered with a coverslip after 30 min of incubation on the slide. Time lapse videos were taken under phase contrast microscopy at 40× and 100× magnification with an optivar of 2×. The rate of movement was calculated by measuring the distance traveled by a cell divided by the time traveled (conversions and calculations are given in Online Resource 3).
For cell imaging, a 15 μL aliquot of the cell suspension was transferred to a slide and imaged under phase contrast microscopy at 40× and 100× magnification with an optivar magnification of 2×. For colony light microscopy, colonies were extracted after 7 days of growth. A thin 1 cm square of the agar containing a representative sample of the colonies was cut out using a sterile scalpel. That layer was transferred to a microscope slide and imaged without a cover slip using the Zeiss light microscope at objective magnification of 6.3× with an optivar magnification of 2× under bright field, polar extinction and first order red polarization. The same was then imaged using the Zeiss dissecting microscope at 0.8× under darkfield.
For TEM observations, the extracted colonies were suspended in 1 mL of Gluconacetobacter buffer solution comprised of 20 g/L glucose, 5.1 g/L sodium phosphate and 1.15 g/L citric acid. The samples were centrifuged for 30 s at 11 rpm. The supernatant was discarded and replaced with 1 mL dH2O. The TEM samples were prepared by adding 10 μL of the above cell solution to the Formvar coated copper grids. The cells were allowed to incubate for 30 min at 20 °C for ribbon, bundle and microfibril imaging and at 4 °C for band material imaging. A volume of 5 μL of a 0.5 % UA solution was added to the grid and extracted using filter paper. This was performed twice and the samples were allowed to dry and then observed.
Results
The G. xylinus acsD disruption mutant acsDdm
Screening for the plasmid insertion within the 470 bp acsD gene sequence (Fig. 1a) indicated the presence of a 6 Kb insert in clone acsD5 which was not present in the wild type G. xylinus AY201 (Fig. 1b). An RT-PCR analysis was performed to check gene expression (Fig. 1c). Expression of the AcsD protein was confirmed in the wild type G. xylinus AY201 but not in the acsD3 and acsD5 clones. AcsD5 was chosen for further studies.
pBR325 plasmid with a small portion of the acsD gene electroporated into G. xylinus cells and a single homologous recombination event that was promoted between the plasmid and G. xylinus cells to give rise to the acsD disruption mutant. Black arrows indicate forward and reverse primers (a); confirmation of the presence of the 6 Kb band in the acsD5 mutant in lane 1 and the absence of the band in the acsD3 clone in lane 2 and wild type G. xylinus AY201 in lane 3 by a colony PCR screen (b); RT-PCR results top row indicates the expression of the acsD gene in G. xylinus, but not acsD5 or acsD3 and bottom row the expression of acsC (c)
Glucose assays and cell counts
Cell concentrations and glucose usage were evaluated after 7 days of incubation under static conditions. Based on the glucose assay and optical density measurements, the acsDdm mutant metabolized equal amounts of glucose as the wild type and had a slightly higher concentration of cells (Table 2).
Cellulose pellicle quantification
Cellulose quantification of the resulting pellicle was determined for the wild type, mutant and complement strains after growth in 100 ml SH liquid culture medium for 7 days (Fig. 2). Dry weight measurements indicated the acsDdm mutant and acsDNGFP complement had BC-producing abilities roughly 10 % that of the wild type G. xylinus AY201 and acsDCGFP complement (Fig. 10c).
X-ray diffraction analysis
Analysis of the diffraction patterns of cellulose from the wild type G. xylinus AY201 and the acsDdm mutant indicated three peaks confirming that only cellulose I was present for both the wild type and mutant samples (Fig. 3) (Czaja et al. 2004; Liu and Hu 2008). The complement strains were not included in this analysis. The sharper peak at 22.48° is only present for cellulose I samples. The weaker reflection present for the acsDdm mutant was due to a lower sample volume.
Characterization of the acsDdm mutant using light microscopy
Time lapse videos of the cell movement were captured using phase contrast light microscopy and Image Pro Plus software (Online Resource 4 and Online Resource 5). The wild type G. xylinus AY201 cells executed typical ordered reversals while producing cellulose on an agar surface. The reversals were expressed as smooth ‘to and fro’ movements where the cell would lay down an initial ribbon in one direction and then reverse, depositing another ribbon on top in the other direction (Fig. 4a–e and Online Resource 13a).
Multiple exposure sequence under phase contrast at 35 s intervals of the wild type G. xylinus AY201. Black arrows indicate the movement of the chosen cell (a). Scale bar represents 5 μm. The diagrams b–e demonstrate the types of movements observed. Patterns of cell movement include typical linear reversal (b); sigmoid reversal (c); curved reversal (d); and circular tunnel movement (e)
On the other hand, the acsDdm mutant did not perform ordered reversals on the agar surface. Instead, the movement was erratic which was demonstrated by spinning about multiple axes, side-to-side movements, single directional non-linear advancement, advancement in one direction rotation and then advancement in the other direction, and non-ordered reversals (Fig. 5a–d and Online Resource 13b). Rate of movement measurements were calculated (Online Resource 3) using 10 cells from the wild type G. xylinus AY201 and 10 cells from the acsDdm mutant (Fig. 6). The wild type moved at an average rate of 3.61 ± 0.7 μm/min while the mutant was slower at an average rate of 1.17 ± 0.3 μm/min.
Multiple exposure sequence under phase contrast at 35 s intervals of the acsDdm mutant. Black arrows indicate the movement of the chosen cell. Scale bar represents 5 μm. The diagrams b–f demonstrate the types of movements observed. Patterns of cell movement include advancement in one direction rotation and then advancement in the other direction (b); oscillation side-to-side (c); spinning about axes (d); single directional non-linear advancement (e); and non-ordered reversals (f)
Colony morphology observations [Fig. 7 (black and white) Online Resource 6 (color)] indicated that the acsDdm mutant colonies appeared very similar to the G. xylinus AY201 wild type except that they were smaller in size. Polarization extinction and first order red polarization microscopy revealed birefringence in both the wild type and the mutant suggesting the presence of crystalline cellulose.
Characterization of the acsDdm mutant using transmission electron microscopy (TEM)
Measurements of the ribbons, bundles and microfibrils were obtained from high magnification TEM images of the cellulose. A total of 50 cells, 25 from the wild type and 25 from the mutant were compared (Fig. 8a–e; Table 3). The ribbons secreted from the wild type G. xylinus AY201 were wider as compared to the acsDdm mutant (Online Resource 7) while the bundles and microfibrils appeared to be the same size (Online Resource 8, Online Resource 9; Table 3).
Cellulose ribbon-producing G. xylinus wild type cells incubated at temperatures lower than the optimum of 28 °C produce a band-like Cellulose II assembly (Hirai et al. 1997, 2002). These unique band-like assemblies were visualized by TEM as a way to localize the cellulose secretion sites. Wild type G. xylinus AY201 and acsDdm mutant cells were incubated at 4 °C and observed (Fig. 9). The band-like assembly expressed by the wild type G. xylinus AY201 initiated from sites along the longitudinal axis and generally extended the entire length of the cell (Fig. 9a and Online Resource 10). The acsDdm mutant band-like assemblies did not have distinct orientations on the cell. The cellulose was secreted from sites on the transverse and the longitudinal axes (Fig. 9b and Online Resource 11). The orientation had no observable effect on the alignment of pores within a given linear TC as the cellulose secretions sites oriented on the transverse axis still appeared to be in a linear configuration (Online Resource 11a).
Complementation of the acsD knockout
Complementation by introduction of the replicating plasmid, pCTP1 + acsDNGFP (N term) and pCTP1 + acsDCGFP (C term), which carries acsD under control of the tac promoter (Online Resource 1) yielded transformants demonstrating cellulose yields and properties equivalent to those observed in the wild type G. xylinus AY201 (Figs. 10, 11a, b). To establish that normal cellulose production in acsDCGFP was due to complementation, SDS-PAGE and Western Blotting analyses were utilized to determine the presence of the AcsD protein tagged with GFP in acsDCGFP (Fig. 11c, d). Additionally, sequencing data verified the presence of the intact plasmid within the C-terminal complement (454 DNA Sequencer, University of Texas Austin). The sequencing data also confirmed the presence of acsD in acsDNGFP; however cellulose yield and properties of this complement more closely resembled that of the acsDdm mutant (Fig. 10) as Western blot analysis indicated that the AcsDNGFP fusion protein was not expressed (Data not shown). This may be due to a one base pair overlap between acsC and ascD, interference by the N terminal GFP tag with protein–protein interactions or inhibition of proper protein targeting and localization.
Negatively stained 4 °C incubated G. xylinus AY201 with band material extending all along the longitudinal axis of the cell (a); acsDCGFP complement with band material extrusion from the longitudinal axis of the cell (b); the SDS-Page of the acsDCGFP membrane fractions (c); Western blotting analysis of the inner membrane, periplasmic extract and total cell extract (d). Scale bar represents 0.5 μm
Discussion
Gluconacetobacter xylinus contains an operon, acsABCD, that codes for a series of proteins which make up the cellulose biosynthesizing pore complex (Saxena et al. 1994). One of the proteins coded by the operon, AcsD, has been reported to have a cylinder-shaped octamer structure localized in the periplasmic space, and it has been suggested that this structure may play a role in cellulose crystallization by functioning as a part of the cellulose synthesizing enzyme complex (Saxena et al. 1994; Hu et al. 2010; Iyer et al. 2011). The purpose of this study was to characterize the disruption mutant to aid in determining the function of AcsD. We have focused on the hierarchical levels of cellulose assembly because the properties of the cellulose product formed by the acsD disruption mutant showed significant differences from the wild type and appeared to be a result of a disruption at one of the stages of cellulose biosynthesis.
The hierarchical level of assembly has been proposed previously (Benziman et al. 1980; Haigler et al. 1980, 1982; Ross et al. 1991; Cousins and Brown 1995, 1997; Koizumi et al. 2009). Enzyme order and alignment are critical to the first stage in the development of the cellulose microfibrils. The proteins within the terminal complex (TC) subunit or pore assembly are precisely arranged to enable the nascent glucan chains to associate through van der Waals forces to form the glucan sheet and then hydrogen bond to form the glucan mini-crystal as it exits the TC subunit (Fig. 12a; Cousins and Brown 1995, 1997).
Schematic illustration of the hierarchical self-assembly process in G. xylinus emphasizing the final level differences between the wild type and the mutant. The individual enzyme complex or terminal complex (TC) subunit produces a mini-crystal of cellulose (a); three TC subunits form a TC which produces a microfibril (b); the TCs associate to form a linear TC which produces bundles of microfibrils (c); wild type linear TCs aligned together along the longitudinal axis of the cell which produces a ribbon of cellulose that propels the cell in the opposite direction to that of synthesis (d); mutant linear TCs positioned with conflicting orientations (i.e. transverse, diagonal, and/or longitudinal) to the long axis of the cell producing bundles of microfibrils, some which may associate to form a narrow ribbon, that propels the cell in an erratic manner due to the competing directionality of the cellulose synthesis (e)
Upon extrusion from the cell, the contiguous arrangement of the adjacent TC subunits into arrays of three forming a single TC; allows the mini-crystals to interact to produce the microfibril (Fig. 12b; Cousins and Brown 1995). Multiple TCs must be in close enough proximity to form a linear TC which facilitates the association of microfibrils into bundles of microfibrils (Fig. 12c; Cousins and Brown 1997). Sets of linear TCs are arranged in a precise pattern which enables the bundles of microfibrils to interact and bond to form the final ribbon assembly (Fig. 12d; Haigler et al. 1982). We suggest that AcsD may play some role at this final level of hierarchical organization of cellulose assembly although we have no exact understanding of the mechanism. We utilized the characterization of an acsD disruption mutant to aid in this determination to provide further evidence of a final stage of hierarchical cellulose assembly.
AcsD characterization
The acsDdm mutant synthesized significantly less cellulose, utilized the same amount of glucose, and produced more cells than the G. xylinus AY201 wild type. This suggests a disruption in normal cellulose biosynthesis whereby the acsDdm mutant cells re-direct glucose usage from cellulose production into cell metabolism. To determine where the disruption may be occurring, the type of cellulose synthesized by the mutant was verified using polarization microscopy and X-ray diffraction analysis. Polarization microscopy indicated the acsDdm mutant was synthesizing crystalline cellulose and X-ray diffraction analysis from statically grown pellicles confirmed it to be cellulose I (Figs. 3, 7, and Online Resource 6). These data suggest that AcsD is not a part of the cellulose synthesizing pore complex as proposed in Saxena et al. 1994. Using this initial data, it was determined that the disruption appeared not to occur at the enzyme complex level, but possibly at a higher level in the hierarchical assembly of cellulose.
We employed time lapse microscopy to characterize the movement of the acsDdm mutant cells. We hypothesized if cellulose I microfibrils are indeed being synthesized by the mutant; we should observe the same type of movement as seen in the wild type. Previous work has determined that G. xylinus cells have the ability to move in free liquid medium and on a substrate not through the use of flagella, but through a propulsive force created through cellulose biosynthesis (Brown et al. 1976). When G. xylinus wild type cells are incubated on a surface, the individual cell is directed by two forces, the inverse force of the cellulose secretion and the interaction of the deposited cellulose with other microfibrils in close proximity (Kondo et al. 2002; Hesse and Kondo 2005). This force-interaction type movement was observed in the wild type G. xylinus AY201 as the cells appeared to lay down an initial ribbon of cellulose for some distance and then at a certain point, reversed and followed that same ribbon back (Online Resource 13a). This may continue for several intervals as the cell lays down ribbon on top of ribbon. The acsDdm mutant did not show this same characteristic type of movement as the wild type; nevertheless the mutant appeared to be propelled by the cellulose it was synthesizing. Additionally, cellulose either in the form of microfibrils or narrow ribbons could be observed around the erratically moving cells (Online Resource 13b). This indicated the acsDdm mutant was synthesizing crystalline cellulose which was strong enough to propel the cell, yet too weak to perform the ordered movement seen in the wild type (Online Resource 4 and Online Resource 5). The evidence suggested a disruption in the alignment of the TCs within the linear TC or in the arrangement of the linear TCs on the cell.
Transmission electron microscopy was utilized for closer analysis of the cellulose as it exited the cell. If the disruption were at the level in which the TCs were organized within the linear TC, then microfibrils would be visualized, but not bundles or ribbons. Visual and measurement analysis indicated the acsDdm mutant synthesized microfibrils and bundles consistently similar in size to the wild type (Fig. 8e, Online Resource 8, and Online Resource 9) which can be correlated with previously reported bundle and microfibril measurement data (Haigler et al. 1982). This suggests that cellulose biosynthesis was not disrupted at this hierarchical level, as the acsDdm mutant appears to have fully functioning pores in the proper alignment allowing for the production of crystalline cellulose I in the form of microfibrils in close enough proximity to interact to form a bundle. However, ribbon measurements of the acsDdm mutant were not similar in size to the wild type (Fig. 8, Online Resource 8 and Online Resource 9). In G. xylinus, a twisting ribbon comprised of bundles of cellulose I microfibrils is synthesized and this resulting ribbon can vary in width from 60 to 40 nm as it twists down the cell and out into the external milieu (Haigler et al. 1982). The wild type ribbon variance from our tests was between 66.5 and 40.3 nm (Table 3) which corresponds to this previously determined data (Haigler et al. 1982). The mutant expressed this same kind of twisting ribbon production; however, the range variance was only 36.4–20 nm (Table 3) indicating that the largest average ribbon thickness for the acsDdm mutant was still narrower than the smallest average ribbon thickness of the wild type. The mutant cells were observed to be shorter which may account for the narrow ribbons as there may be fewer linear TCs in a shorter cell; however the cell size difference was not statistically significant and may not be a factor (Online Resource 12). The discrepancy in the ribbon data suggested that there was a possible disruption in the next level of organization; the arrangement of the linear TCs on the cell.
The 4 °C incubation techniques developed by Hirai et al. (2002) aided in localization of the linear TCs on the acsDdm mutant by visualizing the band-like assemblies on the cell surface. This enabled us to investigate the patterns of disruptions at this organization level. In G. xylinus, normal arrangement of the linear terminal complexes appears as a single row along the longitudinal axis of the cell (Kimura et al. 2001). The AcsD octamer was determined to be localized in the same linear arrangement within the cell’s periplasm (Hu et al. 2010; Iyer et al. 2011; Sunagawa et al. 2013). The linear TC orientation on the mutant cells did not have this same linear arrangement; instead, the linear TCs appeared abnormal as they were often observed to be positioned on the transverse, diagonal, or longitudinal axis (Figs. 9b, 12e) when compared to the wild type (Figs. 9a, 12d). This abnormal arrangement provided evidence that the disruption in cellulose synthesis occurs at the linear TC orientation level of organization which could be considered the highest level in the hierarchical assembly of cellulose (Fig. 12e) while the lower levels are unaffected (Fig. 12a–c).
Band material was visualized during TEM observations from both the acsDdm mutant and the wild type G. xylinus AY201 incubated at room temperature (Online Resource 15). This could be due to the washing method used to prepare the cells for grid mounting since it did include some agitation, i.e. centrifugation and/or pipette mixing. Previous studies have shown that under agitated conditions, G. xylinus AY201 as well as the acsD gene mutant will produce both cellulose I and II (Kuga et al. 1993; Saxena et al. 1994). The cellulose II band material produced under these conditions in both the mutant and wild type reaffirmed the results from the lower temperature incubation.
The competing linear TC evidence aided in clarifying three differences observed in the acsDdm mutant: (1) the erratic movement detected in the time lapse videos, (2) the reduced quantity of cellulose, and (3) the narrow ribbons. It has been determined that G. xylinus is propelled through liquid medium or on the surface of a specified medium through the propulsive force of the biosynthesis of cellulose (Brown et al. 1976; Kondo et al. 2002; Hesse and Kondo 2005). Observations (This Lab) have shown that when the G. xylinus cells encounter some obstruction (i.e. another stationary cell or a web of cellulose ribbons), the movement may cease or the cell may reverse direction. Upon observing the cells which cease moving, it appears that no more cellulose is synthesized. Perhaps the same type of phenomenon is taking place with regards to the acsDdm mutant cells. It is possible that the competing orientations of the linear TCs (i.e. transverse, diagonal, and longitudinal) (Online Resource 13b) lead to a either a slowing down or cessation of synthesis as well as inducing the erratic movement. For example (Online Resource 14), one cell may have two linear TCs oriented on the longitudinal axis (Online Resource 14a) and one on the transverse axis (Online Resource 14b). The bundles produced by the long axis linear TCs could combine into a narrow ribbon while the bundle produced by the transverse linear TC would not. The resulting direction of propulsion could be dictated by these two forces; one parallel (Online Resource 14c) to the cell and the other perpendicular (Online Resource 14d). This competition of forces combined with non-aggregation of all the bundles of microfibrils produced by the cell into a strong fully formed ribbon could cause the unusual movement (Online Resource 14e) which may hinder cellulose production. Additionally, this could provide an explanation as to why the acsDdm mutant’s ribbons were consistently narrower than the wild type. Fewer bundles of microfibrils in the final ribbon assembly due to this orientation conflict may result in narrower ribbons even if the mutant shared the same number of linear TCs as the wild type. Taken together, this evidence suggests that AcsD is involved in the proper orientation of the linear TCs along the longitudinal axis of the cell.
Based on our observations as well as previous studies, it is understood that of all the genes located in the acsABCD operon, acsD appears to be the only non-essential gene for cellulose biosynthesis. Yet, it seems necessary for the perfection of efficient cellulose biosynthesis, because without it, cellulose production is reduced by 90 %. We believe this perfection is accomplished through a complex hierarchical system in which AcsD plays an integral role at the final stage aiding in the alignment of the linear TCs along the longitudinal axis of the cell. The exact function of AcsD in this process is still unknown; however, our data suggests that this unique protein may act as an anchor that interacts with the fully formed and functional linear TC to enable localization in the cell membrane in the proper orientation and linear arrangement necessary to facilitate the formation of the final ribbon assembly making G. xylinus a unique and prolific producer of pure crystalline cellulose.
Continuing and future investigations
Standal et al. (1994) found the presence of a protein upstream of the acsABCD operon called cellulose complementing protein (CcpAx). Recently, CcpAx was discovered to be localized in the periplasm where it interacts with AcsD (Sunagawa et al. 2013). Tests using antibodies might be helpful in pinpointing a more specific location of the protein as well as its interaction with other proteins such as CcpAx and AcsC. High resolution electron microscopy methods using low concentration UA to decorate the surface of the cell may aid in determining if any pores would be visible on the surface of the CcpAx deficient cells and if their acsD mutants demonstrate the same linear TC orientation and cellulose morphology as compared to our acsDdm mutant. Furthermore, we would like to investigate whether the AcsD protein may have several bonding functions that aid in the establishment of linear TCs properly arranged to optimize efficient ribbon production. Additionally, overexpression studies of the AcsD protein may be useful in determining functionality as well as providing more insight into linear TC orientation and how they fit into the hierarchical cellulose assembly process.
References
Amikam D, Galperin MY (2006) PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6
Benziman M, Haigler CH, Brown RM Jr, White AR, Cooper KM (1980) Cellulose biogenesis: polymerization and crystallization are coupled processes in Acetobacter xylinum. Proc Natl Acad Sci USA 77:6678–6682
Blanton RL, Fuller D, Iranfar N, Grimson MJ, Loomis WF (2000) The cellulose synthase gene of Dictyostelium. Proc Natl Acad Sci USA 97:2391–2396
Brown RM Jr (1985) John Innes Symposium. Cellulose microfibril assembly and orientation: recent developments. J Cell Sci Suppl 2:13–32
Brown RM Jr (1996) The biosynthesis of cellulose. Pure Appl Chem A33:1345–1373
Brown RM Jr, Willison JHM, Richardson CL (1976) Cellulose biosynthesis in Acetobacter xylinum: visualization of the site of synthesis and direct measurement of the in vivo process. Proc Natl Acad Sci USA 73:4565–4569
Cousins SK, Brown RM Jr (1995) Cellulose I microfibril assembly: computational molecular mechanics energy analysis favours bonding by van der Waals forces as the initial step in crystallization. Polymer 36:3885–3888
Cousins SK, Brown RM Jr (1997) Photoisomerization of a dye-altered β-1,4 glucan sheet induces the crystallization of a cellulose-composite. Polymer 38:903–912
Czaja W, Romanovicz D, Brown RM Jr (2004) Structural investigations of microbial cellulose produced in stationary and agitated culture. Cellulose 11:403–411
Deng Y, Nagachar N, Xiao C, Tien M, Kao T (2013) Identification and characterization of non-cellulose-producing mutants of Gluconacetobacter hansenii generated by Tn 5 transposon mutagenesis. J Bacteriol 195:5072–5083
Haigler CH, Brown RM Jr, Benziman M (1980) Calcofluor white ST alters the in vivo assembly of cellulose microfibrils. Science 210:903–906
Haigler CH, White AR, Brown RM Jr, Cooper KM (1982) Alteration of in vivo cellulose ribbon assembly by carboxymethylcellulose and other cellulose derivatives. J Cell Biol 94:64–69
Hesse S, Kondo T (2005) Behavior of cellulose production of Acetobacter xylinum in 13C-enriched cultivation media including movements on nematic ordered cellulose templates. Carbohydr Polym 60:457–465
Hirai A, Tsuji M, Horii F (1997) Communication: culture conditions producing structure entities composed of cellulose I and II in bacterial cellulose. Cellulose 4:239–245
Hirai A, Tsuji M, Horii F (2002) TEM study of band-like cellulose assemblies produced by Acetobacter xylinum at 4 °C. Cellulose 9:105–113
Hu SQ, Gao YG, Tajima K, Sunagawa N, Zhou Y, Kawano S, Fujiwara T, Yoda T, Shimura D, Satoh Y, Munekata M, Tanaka I, Yao M (2010) Structure of bacterial cellulose synthase subunit D octamer with four inner passageways. Proc Natl Acad Sci USA 107:17957–17961
Iyer P, Catchmark J, Brown N, Tien M (2011) Biochemical localization of a protein involved in synthesis of Gluconacetobacter hansenii cellulose. Cellulose 18:739–747
Iyer P, Lui Y, Deng Y, McManus J, Kao T, Tien M (2013) Processing of cellulose synthase (AcsAB) from Gluconacetobacter hansenii 23769. Arch Biochem Biophys 529:92–98
Kimura S, Chen HP, Saxena IM, Brown RM Jr (2001) Localization of c-di-GMP-binding protein with the linear terminal complexes of Acetobacter xylinum. J Bacteriol 183:5668–5674
Koizumi S, Yue Z, Tomita Y, Kondo T, Iwase H, Yamaguchi D, Hashimoto T (2008) Bacterium organizes hierarchical amorphous structure in microbial cellulose. Eur Phys J E26:137–142
Koizumi S, Tomita Y, Kondo T, Hasimoto T (2009) What factors determine hierarchical structure of microbial cellulose: interplay among physics, chemistry and biology. Macromol Symp 279:110–118
Kondo T, Nojiri M, Hishikawa Y, Togawa E, Romanovicz D, Brown RM Jr (2002) Biodirected epitaxial nanodeposition of polymers on oriented macromolecular templates. Proc Natl Acad Sci USA 99:14009–14013
Kuga S, Takagi S, Brown RM Jr (1993) Native folded-chain cellulose II. Polymer 34:3293–3297
Laemmli U (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Liu Y, Hu H (2008) X-ray diffraction study of bamboo fibers treated with NaOH. Fiber Polym 9:735–739
Morgan JLW, Strumillo J, Zimmer J (2013) Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 493:181–187
Nakai T, Sugano Y, Shoda M, Sakaibara H, Olwa K, Tuzi S, Imai T, Sugiyama J, Takeuchi M, Yamauchi D, Mineyuki Y (2013) Formation of a highly twisted ribbons in a carboxymethylcellulase gene-disrupted strain of a cellulose-producing bacterium. J Bacteriol 195:958–964
Nishi Y, Uryu M, Yamanaka S, Watanabe K, Kitamura N, Iguchi M, Mitsuhashi S (1990) The structure and mechanical-properties of sheets prepared from bacterial cellulose. Part 2: improvement of the mechanical properties of sheets and their applicability to diaphragms of electroacoustic transducers. J Mater Sci 25:2997–3001
Nobles D, Brown RM Jr (2008) Transgenic expression of Gluconacetobacter xylinus strain ATCC 53582 cellulose synthase genes in the cyanobacterium Synechococcus leopoliensis strain UTCC 100. Cellulose 15:691–701
Osborn MJ, Munson R (1974) Separation of the inner (cytoplasmic) and outer membranes of gram-negative bacteria. In: Fleisher S (ed) Methods in enzymology. Academic Press, New York, pp 642–653
Romling U, Gomelsky M, Galperin MY (2005) c-di-GMP: the dawning of a novel bacterial signaling system. Mol Microbiol 57:629–639
Ross P, Weinhouse H, Aloni Y, Michaeli D, Wienberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M (1987) Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281
Ross P, Mayer R, Benziman M (1991) Cellulose biosynthesis and function in bacteria. Microbiol Rev 55:35–58
Saxena IM, Brown RM Jr (2005) Cellulose biosynthesis: current views and evolving concepts. Ann Bot 96:9–21
Saxena IM, Kudlicka K, Okuda K, Brown RM Jr (1994) Characterization of genes in the cellulose-synthesizing operon (acs operon) of Acetobacter xylinum: implications for cellulose crystallization. J Bacteriol 176:5735–5752
Saxena IM, Brown RM Jr, Fevre M, Geremia R, Henrissat B (1995) Multidomain architecture of glycosyl transferases: implications for mechanism of action. J Bacteriol 177:5276–5283
Schramm M, Hestrin S (1954) Factors affecting production of cellulose at the air/liquid interface of a culture of Acetobacter xylinum. J Gen Microbiol 11:123–129
Standal R, Iverson T, Coucheron DH, Fjaervik E, Blatny JM, Valla S (1994) A new gene required for cellulose production and a gene encoding cellulolytic activity in Acetobacter xylinum are colocalized with the bcs operon. J Bacteriol 176:665–672
Sunagawa N, Fujiwara T, Yoda T, Kawano S, Satoh Y, Yao M, Tajima K, Dairi T (2013) Cellulose complementing factor (Ccp) is a new member of the cellulose synthase complex (terminal complex) in Acetobacter xylinum. J Biosci Bioeng 115:607–612
Weinhouse H, Sapir S, Amikam D, Shilo Y, Volman G, Ohana P, Benziman M (1997) c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. FEBS Lett 416:207–211
Yamanaka S, Watanabe K, Kitamura N, Iguchi M, Mitsuhashi S, Nishi Y, Uryu M (1989) The structure and mechanical properties of sheets prepared from bacterial cellulose. J Mater Sci 24:3141–3145
Yoshinaga F, Tonouchi N, Watanabe K (1997) Research progress in production of bacterial cellulose by aeration and agitation culture and its application as a new industrial material. Biosci Biotechnol Biochem 61:219–224
Zaar K (1979) Visualization of pores (export sites) correlated with cellulose production in the envelope of the gram negative bacterium Acetobacter xylinum. J Cell Biol 80:773–777
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
The wild type G. xylinus AY201 (MPG 16850 kb)
Time lapse videos of the acsDdm mutant (MPG 12282 kb)
10570_2014_521_MOESM6_ESM.tif
Full color colony morphology photomicrographs of the G. xylinus AY201 wild type (a–d) and acsDdm mutant (e-h) under bright-field (a, b, e, and f), polarization extinction (c and g) and first order red polarization (d and h) with scale bar representing 100 μm (a and e) and 25 μm (b-d and f-g) (TIFF 1414 kb)
10570_2014_521_MOESM8_ESM.tif
Negatively stained cellulose synthesized from individual cells with enlargements showing examples of the ribbons (a) and bundles of microfibrils within a ribbon (b) in the G. xylinus AY201 wild type; and the ribbons (c) and bundles of microfibrils within a ribbon (d) in the acsDdm mutant. Scale bar represents 25 nm (a and c) and 10 nm (b and c) (TIFF 526 kb)
10570_2014_521_MOESM9_ESM.tif
Examples of negatively stained cellulose synthesized by the G. xylinus AY201 wild type (a-h) and acsDdm mutant (i-p) that were used to obtain measurement data. Measurement bars were added for clarification. Scale bar represents 20 nm (a, b, e, f, 1, j, m, n) and 5 nm (c, d, g, h, k, o, l, and p) (TIFF 1154 kb)
10570_2014_521_MOESM10_ESM.tif
Negatively stained band material of the wild type G. xylinus AY201 all indicating linear TC orientation along the long axis of the cell (a-e). Arrows designate longitudinal orientation (b) on an enlarged version of the cell (a). Scale bar represents 0.5 μm (TIFF 665 kb)
10570_2014_521_MOESM11_ESM.tif
Negatively stained band material of the acsDdm mutant indicating various orientations of band material on the cell. Arrows designate transverse orientation on one cell (a); transverse and longitudinal orientation on the same cell (b); transverse only (c); transverse and longitudinal (d); and diagonal orientation (e). Scale bar represents 0.5 μm (TIFF 525 kb)
10570_2014_521_MOESM12_ESM.tif
Phase contrast of the wild type G. xylinus AY201 (a and b) and the acsDdm mutant (c and d) scale bar represents 10 um. Average cell dimensions (e) (TIFF 262 kb)
10570_2014_521_MOESM13_ESM.tif
Schematic illustrations of the observed movement due to possible linear TC orientation in the wild type G. xylinus AY201 (a) and the acsDdm mutant (b) (TIFF 144 kb)
10570_2014_521_MOESM14_ESM.tif
Schematic diagram of an example of linear TC arrangement which could result in reduced cellulose biosynthesis. Liner TC orientation along the longitudinal axis of the cell (a); linear TC orientation transverse to the long axis (b); movement of the cell parallel to the long axis (c); movement of the cell perpendicular to the long axis (d); and possible circular movement of the cell as both directional forces are combined (e) (TIFF 56 kb)
10570_2014_521_MOESM15_ESM.tif
Negatively stained band material synthesized at room temperature under agitated conditions by the wild type G. xylinus (a) and the acsDdm mutant (b). Scale bar represents 0.5 μm (TIFF 561 kb)
Rights and permissions
About this article
Cite this article
Mehta, K., Pfeffer, S. & Brown, R.M. Characterization of an acsD disruption mutant provides additional evidence for the hierarchical cell-directed self-assembly of cellulose in Gluconacetobacter xylinus . Cellulose 22, 119–137 (2015). https://doi.org/10.1007/s10570-014-0521-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-014-0521-y