Abstract
Plane wave and molecular orbital density functional theory calculations with periodic models and oligomer fragments based on Iβ cellulose showed relationships among hydrogen bond (H-bond) lengths, angles, energies, and vibrational frequencies. Significantly, the 13C NMR chemical shifts (δ13C), glycosidic and hydroxymethyl torsion angles, H-bond vibrational frequencies, and H-bond geometries results all suggest the predominance of the crystallographic structure C rather than structure A of Iβ cellulose as reported by Nishiyama et al. (J Am Chem Soc 124(31):9074–9082, 2002). The results reported herein also clarified that the δ13C and δ13C′ data from Erata et al. (Cellul Commun 4:128–131, 1997) correspond to δ13C from the origin and center chains of cellulose, respectively. Moreover, this work discusses the use of cellulose oligomer fragments for their potential use in understanding cellulose assembly.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Plant cell walls (PCW) are heterogeneous structures composed of polymers such as cellulose, hemicellulose, pectins, lignins, and glycoproteins (McQueen-Mason and Cosgrove 1994; Cosgrove 2005). Ongoing investigation of the formation, molecular structure, and the potential interactions among PCW polymers could provide valuable and necessary information about PCW architecture. Improved understanding of the H-bonds occurring among PCW polymers could guide the exploration of methods to break these bonds, and could lead to the development of methods to more efficiently decompose cellulose into smaller units during biofuel production (Shen and Gnanakaran 2009). Moreover, understanding the H-bonds that occur among PCW polymers is necessary for understanding how these polymers interact during PCW assembly. The work herein used molecular orbital, and projector-augmented planewave density functional theory (DFT) (Hohenberg and Kohn 1964; Kohn and Sham 1965; Kresse and Furthmüller 1996) calculations to study the “Network A” (Net A) hydrogen-bonding (H-bonding) network that has been suggested as the dominant form in cellulose (Nishiyama et al. 2002).
The goal of the present work was to develop a method that provided precise 13C NMR chemical shifts (δ13C), then to use the method to study the relationships among O–H vibrational frequencies and molecular structure (i.e., glycosidic and hydroxymethyl torsion angles, and H-bond lengths, strengths, and angles). These calculated relationships between spectroscopic parameters and structure could then be used to interpret spectra of disordered cellulose. Although our work focused on the interactions of oligomer models of cellulose and the comparison of our results with cellulose data, the methods developed herein could be applicable for the study of cellulose surfaces, disordered cellulose, cellulose assembly, and cellulose interactions with other PCW components.
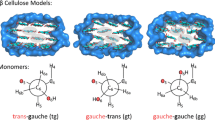
Individual cellulose molecules are linked laterally through O–H–O H-bonds (Nishiyama et al. 2002; Jarvis 2003). Moderately strong O–H–O bonds (≈10–20 kJ/mol) (Jeffrey 1997) among glycan polymers in cellulose produce H-bond networks that contribute to the recalcitrance of cellulose to defibrillation and hinder conversion of cellulose to biofuel (Himmel et al. 2007). Our work focused on the physical characteristics of the intramolecular O2–H–O6 and O3–H–O5 and intermolecular O6–H–O3 H-bonds in Net A of Iβ cellulose. Net A forms the single-layer, laterally H-bonded, sheet-like structure in cellulose. Figure 1 illustrates the Net A for two laterally H-bonded cellobiose models. The Net A structure is thought to be the predominant form (Nishiyama et al. 2008), so the current work focuses on Net A.
Nomenclature for carbon nuclei, H-bonds, and torsion angles used throughout this paper, shown for the cellobiose. Φ, Ψ, χ1, and χ2 are the O5–C1–O–C4′, C1–O–C4′–C5′, O5–C5–C6–O6, and C4–C5–C6–O6 torsion angles, respectively. Throughout, “lt” and “rt” symbolize the left and right oligomer. See the “Methods” section further information about nomenclature
Crystallographic and spectroscopic data can aid in the elucidation of cellulose structure; however, relying solely on crystallographic data could be inadequate for the elucidation. Both 13C NMR data (Blackwell 1977; Erata et al. 1997; Atalla 1999; Nishiyama et al. 2002; Sternberg et al. 2003; Zhao et al. 2007), and infrared (IR) data (Blackwell et al. 1970) are available for cellulose. However, the OH-stretching region (3,300–3,800 cm1) obtained from IR spectroscopy is difficult to interpret (Blackwell et al. 1970), because these frequencies depend on both H-bond lengths and strengths, and IR bands in the OH-stretching region are broad (Gallina et al. 2006).
Our previous work developed a method for precisely calculating NMR chemical shifts (δ13C) for cellulose-proxy models (Kubicki et al. 2013), which were compared with NMR data (Erata et al. 1997; Sternberg et al. 2003). We explored the relationships among torsion angles, H-bonding and δ13C.
The χ-torsion angles may be causing the observed upfield shift of δ13C4 for cellulose chains on the surfaces and/or disordered cellulose (Wickholm et al. 1998; Newman and Davidson 2004; Malm et al. 2010; Fernandes et al. 2011; Harris et al. 2012). We report the effect of the hydroxymethyl torsion angles χ1 (O5–C5–C6–O6) and χ2 (C4–C5–C6–O6) on the calculated C4, C5, and C6 δ13C. Moreover, this study reports the precision of the correlation between the calculated and experimental glycosidic torsion angles (Φ and Ψ), and the correlation precision between the calculated and experimental H-bond O–O lengths and O–H–O angles of the intramolecular O2-H–O6 and O3–H–O5, and intermolecular O6–H–O3 H-bonds (Nishiyama et al. 2002). In addition, we compared the calculated and ring-puckering parameter (θ) (Cremer and Pople, 1975) of our periodic Iβ cellulose model with the experimental θ values of Iβ cellulose crystal structures A and C that were reported by Nishiyama et al. (2002). Furthermore, this work compared the periodic model results with the crystallographic data (Nishiyama et al. 2002) for the intermolecular distances between the origin chain axial H atom on C1 and one of the two methylene H atoms on the hydroxymethyl group of the center chain C6 atom. The systematic study of torsion angles, bond distances, and θ, could lead to refinements in our knowledge of cellulose structure and ultimately to improved understanding of PCW chemistry.
Methods
Figure 2 shows the initial structures of the models used herein; all were built using Materials Studio 6.0 (Accelrys Inc., San Diego, CA). The 3D periodic model (P) is based on synchrotron X-ray and neutron fiber diffraction data (Nishiyama et al. 2002); all models were arranged to have the tg/Net A (Fig. 1) H-bond network. (See the Electronic Supplemental Information (ESI) for the Cartesian coordinates of the initial structures.) Hereafter, except for P, the models are referenced by the number of the glucose residues in their respective oligomeric molecules and the number of oligomers in the model. For example, the 4 × 3 model consists of three cellotetraose models arranged to give a tg/Net A H-bond network between each cellotetraose. In addition, 4 × 3 × 3 and P models (Fig. 2) were used to determine if oligomer stacking affected the calculated results. The 4 × 3 × 3 and P models were more realistic than the smaller glucan chain models with respect to cellulose, so the 4 × 3 × 3 and P models could have provided results that were more precise than the smaller models. The 4 × 3 × 3 model consists of three stacked 4 × 3 models, and the P model unit cell consists of four stacked 4 × 3 models.
Models used for the current work. The abbreviations lt, m, and rt refer to the left middle and right oligomer in the two and three oligomer models. The shaded glucose residues in each model are those analyzed for 13C NMR chemical shifts, and H-bond lengths, angles, vibrational frequencies, and energies. The periodic model contains four-4 × 3 layered models based on the Iβ tg/NetA cellulose structure (Nishiyama et al. 2002)
The shaded glucose residues in each model shown in Fig. 2 are those that were analyzed for their 13C NMR chemical shifts, glycosidic and hydroxymethyl torsion angles, and their H-bond lengths, angles, energies, and vibrational frequencies. We chose a central glucose residue unit or units within each model for analysis, because those units would best represent the bulk structure of cellulose, as compared with the glucose residues on the reducing or non-reducing ends of the glucans. The single glucose unit models (i.e., 1 × 1, 1 × 2, and 1 × 3) are obvious exceptions to this procedure. Unless otherwise stated, the notation that we used for the torsion angles and atom numbers agrees with the IUPAC notation found at a www.chem.qmul.ac.uk/iupac.
Labeling schemes from the experimental NMR data include two sets of 13C NMR chemical shifts (δ13C) labeled as either C- or C′-nuclei (Erata et al. 1997; Sternberg et al. 2003). For the crystallographic data, the C-atoms were categorized based on whether they were from the center or origin chain of cellulose (Nishiyama et al. 2002). Based on our results, the C NMR data correlated with the crystallographic data for the origin chain, whereas the C′ NMR data more precisely correlated with the crystallographic data from the center chain. Therefore, throughout the current work, the nomenclature for the periodic model results for the origin (P[o]) and center (P[c]) chains will correspond to those conventions (i.e., the P[o] results correspond to the δ13C data, and the P[c] results correspond to the (δ13C′ data).
The glucose, cellobiose, cellotriose, and cellotetraose-based models and the 4 × 3 × 3 model (Fig. 2) were energy minimized using Gaussian 09 (G09) (Frisch et al. 2009). We used the meta hybrid density functional M05-2X coupled with the 6-31G(d,p) basis set (Hohenberg and Kohn 1964; Kohn and Sham 1965; Krishnan et al. 1980; Clark et al. 1983; Zhao et al. 2006; Papajak et al. 2011). We chose the M05-2X meta-hybrid density functional based on the utility of this method to calculate the properties of biomolecules (McNamara and Hillier, 2007). In addition, M05-2X energy minimization calculations of the 3 × 3 Net A model showed that the H-bond O6–O2 and O3–O5 distances and their respective O–H–O vibrational frequencies converged for the 6-31G(d,p); additional basis functions had negligible effect on the calculated bond lengths and frequencies (ESI Figure S1).
For the DFT calculations with G09, the initial geometry of each model was subjected to one of two distinct energy minimization methods in the gas phase. One optimization was performed without symmetry or atomic constraints for a fully relaxed minimization (FR). The other method, fixed coordinate or FC, was performed using the keyword opt = modredundant in G09 (Frisch et al. 2009). The FC method froze the C1–C6 and the O5 atoms of each glucose residue, while allowing the hydroxyl groups and H-atoms to minimize unconstrained. The FC method forced the models to maintain the lateral position of the glucan chains.
Subsequent frequency calculations in G09 using M05-2X/6-31G(d,p) followed each optimization to determine the vibrational frequencies of each model, and to determine if each structure resided at a potential energy minimum. The resulting frequencies were not scaled. Note that although the 4 × 3 × 3 model underwent FR minimization, the model did not converge to an energy minimum and did not undergo a frequency calculation; however, when the calculation was terminated, the energy of this model was changing by less than 0.01 kJ/mol; therefore, the structure was likely near a stationary point on its potential energy surface (PES).
The P model was energy minimized using the Vienna Ab-Initio Simulation Package (VASP) (Kresse and Furthmüller 1996). Projector-augmented planewave pseudopotentials were used with the PBE gradient-corrected exchange correlation function for the 3D periodic DFT calculations. The choice of electron density and atomic structure optimization parameters were based on literature values (Bućko and Hafner 2005; Li et al. 2011). An energy cut-off of 77,190 kJ/mol was used with an electronic energy convergence criterion of 9.6 × 10−6 kJ/mol. Atomic structures were relaxed until the energy gradient was less than 1.93 kJ/mol/Å. 2 × 2 × 2 k-point samplings were used. Atoms were first allowed to relax with the lattice parameters constrained to the experimental values, and then the atoms and lattice parameters were allowed to relax to obtain the structures; energies and spectroscopic properties reported herein. The dispersion-correction parameters were 40 Å for the cutoff distance (Bućko and Hafner 2005), and 0.75 for the scaling factor (s6) and 20 for the exponential coefficient (d) in the damping function (Grimme 2006). Energy minimization was carried out in two stages. First, atoms were allowed to relax with the lattice parameters constrained to experimental values. Second, the atoms and lattice parameters were allowed to relax simultaneously. A subsequent frequency calculation provided the vibrational frequencies for the fully relaxed periodic model; the resulting frequencies were not scaled.
Four minimization methods were used for the VASP energy minimization calculations:
-
1.
the crystallographic structure (N) was not minimized (i.e., the structure was as determined by Nishiyama et al. 2002);
-
2.
the H-atoms were allowed to relax but the C and O atoms were constrained (FC);
-
3.
all atoms were allowed to relax during the minimization (FR). These three models were used to determine if the crystallographic assignments were accurate relative to 13C NMR data and to determine the crystallographic H-atom assignments; or
-
4.
classical MD simulation (10 ns) of a Iβ 6 × 6 cellulose microfiber followed by energy minimization both using the CHARMM force field (Guvench et al. 2009; Raman et al. 2010). This model (MM) is a cluster that was extracted from the center of a 6 × 6 cellulose model (Zhao et al. 2013). We used the MM 3 × 3 × 4 model, taken from the center of the microfibril to approximate the structure of cellulose as predicted by CHARMM.
In order to evaluate the results from these methods for reproducing details of cellulose structure, we have subjected these periodic DFT-minimized models to the same 13C NMR calculations and comparison against experimental 13C NMR data as the G09-minimized models. We also used G09 to calculate the13C NMR chemical shifts (δ13C) for the MM model that was derived from an MD simulation for comparison with the NMR data.
Single-point GIAO NMR calculations (Wolinski et al. 1990; Schreckenbach and Ziegler 1995; Cheeseman et al. 1996; Adamo et al. 1998; Buhl et al. 1999; Karadakov 2006) using the mPW1PW91/6-31G(d) method and a multistandard approach using methanol as the internal standard determined the (δ13C) of the C nuclei in each model (Sarotti and Pellegrinet 2009; Kubicki et al. 2013). The subsequent calculation of δ13C with a method that differs from the method used for the energy minimization calculation is a standard procedure (Cheeseman et al. 1996; Frisch et al. 2009; Sarotti and Pellegrinet 2009). Furthermore, our recent paper showed that energy minimization calculations with projector-augmented planewave pseudopotential with the PBE gradient-corrected exchange correlation functional in VASP followed by single-point NMR calculations using the mPW1PW91/6-31G(d) level of theory in Gaussian 09 reproduced δ13C within ±2 ppm for crystalline Iα and Iβ cellulose (Kubicki et al. 2013).
In water, the experimental NMR chemical shift of the C-atom of methanol (δ13C) is 49.5 ppm (Gottlieb et al. 1997); this value was necessary for the multistandard calculations (Sarotti and Pellegrinet 2009). To improve the precision of the NMR results, these calculations included the self-consistent reaction field (SCRF) method integral equation formalism polarized continuum model (IEFPCM) with the permittivity of water (Cancès et al. 1997; Gogonea 1998). However, the NMR calculation for the extended clusters that were extracted from the periodic models were in the gas-phase rather than a SCRF due to computational costs; moreover, because the C-nuclei of interest in the periodic models are solvent inaccessible (Fig. 2), use of the SCRF was unnecessary for this model. Comparison of the calculated and experimental δ13C shifts determined which models correlated best with the NMR data for cellulose Iβ (Erata et al. 1997; Sternberg et al. 2003).
In addition, following the energy minimization calculations with G09 or VASP, M05-2X/6-31G(d,p) NBO population analysis calculations determined the binding energies, E(2) (Panduranga et al. 2011) of the pertinent H-bonds for the oligomer models as well as extracted clusters from the periodic models (Glendening and Weinhold 1997, 1998). These analyses involved only the intra- and intermolecular H-bonded glucose residues (Fig. 1) shown in the shaded regions of Fig. 2.
The calculated NBO E(2) energies provide the energy in kJ/mol for the delocalization of electrons from an electron donor (e.g., O-atom) to an electron acceptor (e.g., hydroxyl-H atom). These calculated E(2) energies are not H-bond energies, but we related the two quantities using the following method. For example, the M05-2X/6-31G(d,p) NBO E(2) energy for the delocalization of electrons from the O-atom of one water model to a H-atom of a second H2O model in a water dimer is 79 kJ/mol. The counterpoise corrected (Boys and Bernardi, 2002) M05-2X/6-31G(d,p)//M05-2X/6-31G(d,p) binding energy for the water dimer (i.e., ΔE = Ewater dimer + EBSSE − 2Ewater monomer, where EBSSE is the energy of the counterpoise correction for basis set superposition error) is 23 kJ/mol, which is comparable to the range of reported experimental data of 20.4–12.3 kJ/mol with reported uncertainties of approximately ±3.5 kJ/mol (Fiadzomor et al. 2008). Therefore, the NBO E(2) energies are 4–7× greater than the experimentally observed H-bond energy for a water dimer, so, we divided the NBO E(2) energies by a factor of 5.5 (i.e., the average of 4 and 7) for this work.
The calculated H-bond O–O lengths, O–H–O angles, and O–H vibrational frequency results in addition to the torsion angle results for Φ (O5–C4–O–C1′), Ψ (C4–O–C1′–C5′), χ1 (O5–C5–C6–O6), and χ2 (C4–C5–C6–O6) were obtained with GaussView version 5.0.9 (Frisch et al. 2009) from the energy minimized models. The ring-puckering parameter (θ) (Cremer and Pople 1975) for the periodic model was calculated using the online θ calculator of Shinya Fushinobu (http://www.ric.hi-ho.ne.jp/asfushi/) and was compared with the experimentally derived θ values of Nishiyama et al. (2002).
To evaluate the precision of the relationship between the δ13C results and data, we used the mean unsigned error (MUE) and root mean-squared error (RMSE), as used previously (Watts et al. 2011).
Synthetic infrared spectra were generated using the results from G09 and the program Molden (Schaftenaar and Noordik 2000).
Results and discussion
NMR results
NMR benchmarking
This section focuses on the structures of the models and the correlations between the calculated 13C chemical shifts (δ13C) with the cellulose NMR data. Developing a method that accurately reproduces experimental δ13C and crystallographic data is important, because if the method can precisely reproduce those data, then we can have greater certainty in the accuracy of our study of cellulose H-bond chemistry. Furthermore, if the method can reproduce the spectroscopic and structural data precisely, then the method could be used for modeling other systems of interest that are not as well constrained by experimental data.
Previous work (Kubicki et al. 2013) showed that periodic DFT calculations resulted in structures that replicate experimental δ13C for both cellulose Iα and Iβ when subjected to NMR calculations in Gaussian 09 (Frisch et al. 2009). However, Table 1 shows that the precision of the calculated δ13C with respect to the NMR data (Erata et al. 1997; Sternberg et al. 2003) depends on the geometry of the Iβ cellulose model. In comparison with observed interatomic distances in Iβ cellulose structure A (Nishiyama et al. 2002), we have determined the glycosidic O, the C1–C1 inter-chain, and C1–C1 inter-layer repeat distances for our models. The experimental values are 10.38, 8.20 and 7.78 Å, respectively. For the P FR model, the measured results are 10.40, 8.14, and 7.55 Å, respectively, which agree well with the results for structure A (Nishiyama et al. 2002). In general, the molecular cluster models have similar values, where the three distances for the 3 × 1 through 4 × 3 FR, and 4 × 3 × 3 models are 10.44(±0.07), 8.08(±0.19), and 7.71 (for the 4 × 3 × 3 model) Å, respectively.
Significantly, the δ13C results from the N model, the non-minimized model that Nishiyama et al. (2002) reported as the likely structure of cellulose Iβ, do not correlate precisely with the NMR data (Table 1). The results in Table 1 show that it was necessary to fully relax (model FR) model N during minimization in order to reproduce the experimental δ13C. The δ13C RMSEs for the FR, FC, and N periodic models were, 1.1, 1.4, and 10.3 ppm, respectively; therefore, N shows the poorest correlation of the three models with the NMR data of cellulose Iβ. In addition, model FC, which was minimized with relaxed H-atoms did not replicate the NMR data as precisely as the FR model did (Table 1 and Fig. 3). The results from model FC suggest that there was likely uncertainty in the assignment of the H-atom positions in model N by Nishiyama et al. (2002), and the results from model FR show that there was also uncertainty in the C- and O-atom positions in model N.
δ13C NMR results from models obtained from the data of Nishiyama et al. (2002). The CHARMM model results were obtained from a low energy result of a MD simulation. The N, FC, and FR periodic structures underwent energy minimization calculations using VASP with fully constrained atoms (N), constrained backbones and relaxed H-atoms (FC), and with full relaxation of all atoms (FR). The 3 × 1 FR model underwent unconstrained minimization in Gaussian 09. a shows the origin chain results and b shows the center chain results, based on the δ13C and δ13C′ of Erata et al. (1997) and Sternberg et al. (2003). The error bars show the RMSEs of the models; the RMSE for the FC, FR, and 3 × 1 FR are significantly less than those for CHARMM and N. See Table 2 for the respective δ13C NMR and errors
Notably, the model extracted from the MD simulation (MM) exhibited poorer precision with regard to the δ13C data than the N, FC, or FR models (Table 1 and Fig. 3). Therefore, although the computational cost of obtaining the MM model was low, the δ13C results obtained from that models suggest that it is inaccurately reproducing the structure of cellulose.
In addition, we found that the δ13C NMR results from the origin chain of the periodic structure (P[o]) (Fig. 2) correlated most precisely with the C-atom data (Erata et al. 1997; Sternberg et al. 2003), and the results from the center chain of the periodic structure (P[c]) correlated most precisely with the C′-atom data (Table 1 and Fig. 3). These assignments for the origin and center chains were ambiguous in Erata et al. (1997) and Sternberg et al. (2003); that is, it was uncertain whether the C-atom assignments were for particular glucose residues within a single glucan chain in cellulose, or whether the δ13C represented C-atoms from different glucan chains within cellulose. Our results provided a clarified interpretation of the NMR data.
Although the oligomer fragment models (Fig. 2) used for this work do not represent crystalline cellulose, they were used to model structures that could occur during cellulose assembly or degradation. When compared with the NMR data, the results from models that were energy minimized with G09 using the FC models did not correlate as precisely as the fully relaxed (FR) model results did (Table ESI 1).
Among the G09-minimized models, the δ13C results for FR 3 × 1 exhibited the most precise correlation (i.e., linearity and low errors) with the NMR data for cellulose Iβ (Table 1 and Fig. 3). This result suggests that once a glucose unit in a cellulose chain is H-bonded to other cellulose chains, it could exhibit δ13C that are similar to those of the bulk crystalline cellulose. Although the periodic models (FC and FR) provided the most precise correlation with the NMR data, the 3 × 1 provided remarkable precision, considering its structural simplicity relative to cellulose. We conclude that the computational methodology used here to predict δ13C values for cellulose is precise to within ± 2 ppm, based on the maximum errors (Table 1), provided that the model structure is realistic.
Comparison to crystalline cellulose 13C NMR
Table 2 shows the χ1 and χ2 torsion angles for the 3 × 1, P, and MM models, and the data for the crystal structures A and C (Nishiyama et al. 2002). Table 2 shows that the χ1 and χ2 results from model FR P agree more precisely with experimental origin chain data from structure C but not structure A (Nishiyama et al. 2002), than do the results from model FR 3 × 1 and MM. These results for the origin chain further corroborate with the NMR results. Although neither of the χ1 or χ2 results agree precisely with the corresponding center chain data from structure A or C, the δ13C values of the center chain reproduce observed δ13C values as precisely as the origin chain. Thus, the discrepancy between the structure A and C χ1 and χ2 with the calculated P values could be due to error in the calculated structure, error in the experimentally-derived χ1 and χ2 values, or insensitivity of the δ13C to χ1 and χ2.
Nishiyama et al. (2002) suggested that their structure A fit the observed X-ray data for cellulose Iβ better than structure C did; however, the calculated torsion angles χ1 and χ2 for periodic model FR fit the data from structure C better than they do for structure A. The initial structure of model P was that of the experimentally-based structure of structure A obtained from Nishiyama. Our results indicate that during the energy minimization calculation, structure A became more like structure C. Nishiyama et al. (2002) chose structure A over structure C because structure C had a close contact (2.138 Å) between the axial H on C1 of the origin chain (C1Ho) and the one of the methylene H atoms of the center chain (C6Hc) of cellulose Iβ. However, the DFT-D2 energy-minimized structure A of Nishiyama et al. (2002) with full atomic and lattice parameter relaxation (i.e., model P FR), the interaction distance between the H-atoms of C1Ho and C6Hc was 2.139 Å, which differs from the structure C distance by only 0.001 Å (Fig. 4). Thus, the DFT-D2 calculations are predicting that structure C is lower in energy than structure A and that the 2.138 Å C1Ho–C6Hc H-atom distance does not cause prohibitive steric interactions. This result suggests that structure C from Nishiyama et al. (2002) more likely describes the structure of cellulose Iβ than structure A does.
The dashed line represents the interaction distance between the axial H atom of C1 on the origin chain (C1o) and one of the two methylene-H atoms on C6 of the center chain (C6c). For cyrstallographic structure C of Nishiyama et al. (2002) the C1oH–C6cH interaction distance was 2.138 Å, which agrees precisely with the results form model P FR from this work (2.139 Å). For model N, which was based on structure A of Nishiyama et al. (2002), the calculated C1oH–C6cH interaction distance was 2.358 Å. This model in this figure comes from the shaded region of the periodic model (Fig. 2)
Significantly, further evidence that supports structure C as being a better approximation of cellulose Iβ structure than structure A from Nishiyama et al. (2002) is the ring puckering parameter (θ). Nishiyama et al. (2002) reported θ for the origin chain and center chains of 10.2 and 6.7 for structure A (Table 2), respectively, and θ of 3.2 for both the origin and center chains of structure C. We reported periodic DFT-calculated θ values (model P FR) of 1.2 and 2.8 for the origin and center chains, respectively (Kubicki et al. 2013). Therefore, the calculated θ results from P FR correlate better with those from structure C than they do with those from structure A of Nishiyama et al. (2002).
The effect of hydroxymethyl torsion angles (χ) on δ13C4, δ13C5, and δ13C6
Prior NMR data has shown that surface δ13C4 (Wickholm et al. 1998; Newman and Davidson, 2004; Malm et al. 2010; Fernandes et al. 2011; Harris et al. 2012) values are upfield shifted from the bulk cellulose by 4 to 5 ppm. This observation has been interpreted as rotations of χ1 and χ2. Previous calculations have shown that δ13C5 and δ13C6 are relatively insensitive to changes in χ1 and χ2 (Kirschner and Woods 2001; Gonzalez-Outeiriño et al. 2006). Therefore, using the energy minimization and NMR calculation methodologies that produced precise agreement between calculated and observed δ13C chemical shifts, we explored the relationship between those δ13C and the hydroxymethyl torsion angles (Fig. 5). For these comparisons, we used all of the models (i.e., FR, 3 × 1 and larger), rather than just those that correlated favorably with the NMR data, because the other models used for this work could be useful for elucidating the structure of other cellulose allomorphs, surface or internal glucans, and disordered cellulose. See ESI Table S2 for the numerical results plotted in Fig. 5.
Relationship between the torsion angles χ1 and χ2 and the calculated δ13C of the nuclei C4, C5, and C6 for the 3 × 1 through 4 × 3 × 3 G09 minimized and the periodic (fully relaxed) VASP cellulose models. The results show that the chemical shifts for C4, C5, and C6 do not vary significantly over the range of the torsion angles χ1 and χ2
Figure 5 shows the relationship between the torsion angles χ1 and χ2 with the calculated δ13C4, δ13C5, and δ13C6 for the DFT minimized models. These results show that χ1 and χ2 range from 153° to 170° and -90° to -72°, respectively for the models ranging in size from 3 × 1 to 4 × 3, the 4 × 3 × 3 model, and the fully relaxed P model. However, δ13C4, δ13C5, and δ13C6 remain relatively constant over the ranges of χ1 and χ2: 86.2(±1.4), 71.0(±0.9), and 63.8(±1.3) ppm, respectively. These standard deviations approach the RMSE of the fully relaxed periodic model (Fig. 3), so ascertaining these shifts using DFT could be problematic. These results reaffirm that δ13C5, and δ13C6 are insensitive to χ1 and χ2 rotations for the tg conformation (Kirschner and Woods 2001; Gonzalez-Outeiriño et al. 2006) and do not support the hypothesis that χ1 and χ2 change are necessarily the cause the observed upfield shift for δ13C4 (Wickholm et al. 1998; Davidson et al. 2004; Malm et al. 2010; Fernandes et al. 2011; Harris et al. 2012).
Prior NMR and molecular dynamics studies suggested that polar solvents such as water can disrupt H-bonding networks in oligosaccharides and cause changes in hydroxymethyl torsion angles (Kirschner and Woods 2001; Gonzalez-Outeiriño et al. 2006). The current work did not incorporate explicit solvent molecules and the initial structures were based on crystallographic data. Calculations on the effects of explicit solvation on χ1 and χ2 angles in periodic and oligomer proxies of cellulose would be worthwhile, but are outside the scope of this study.
Glycosidic Torsion Angles (Φ,Ψ) effect on δ13C1 and δ13C4
The Φ and Ψ results from FR P origin and center chains agree more precisely with the data from structure C than with the data for structure A (Table 2) from Nishiyama et al. (2002). The Φ and Ψ results further suggest the full relaxation of the periodic model during its energy minimization produced torsion angles that agree better with the experimental data of structure C than with the data from structure A. The P model Φ and Ψ, χ1 and χ2, and δ13C results all suggest that structure C, rather than structure A, is the observed structure of cellulose Iβ. The FR 3 × 1 model results for the torsion angles did not agree with the data as well as the FR P model results did; this discrepancy could be due to the lack of intermolecular interactions between glucan chains in the FR 3 × 1 model. The MD simulation Φ and Ψ results from model MM also fit the data for structure C with good precision, except that Φ (−98°) for the center chain underestimates Φ (−92°) for structure C by 6°.
The differences between the non-minimized structure A of Nishiyama et al. (2002) and the DFT-minimized structure A that better correlated with structure C of Nishiyama et al. (2002) are subtle. When superimposed, the shaded regions of model P (Fig. 2) from structure A and the P FR model that better correlates with structure C showed minor differences in ring positions, hydroxymethyl torsion angles, and hydroxyl group torsion angles. However, based on the NMR results, torsion angles, θ, and the C1Ho–C6Hc H-atom distance, as discussed in the previous paragraphs, these minor structural differences culminated in structure C correlating more precisely to the structure of cellulose Iβ than structure A does.
The glycosidic torsion angles from model FR P agree with both the crystallographic data for the glycosidic torsion angles and NMR data for cellulose Iβ; however, although the results from FR 3 × 1 agree precisely with the NMR data, they do not agree as precisely with the crystallographic data for the glycosidic torsion angles, likely because the FR 3 × 1 model did not capture the chemistry of cellulose. However, the small glucan chain models that we used for this work could provide data that is useful for interpreting data obtained from fragments of cellulose that occur during cellulose degradation or to better understand cellulose assembly, because cellulose would be unlikely to have the properties of crystalline cellulose during formation and degradation.
Therefore, we explored (Fig. 6) the relationship between the glycosidic torsion angles (Φ, Ψ) and NMR data for δ13C1 and δ13C4 for the DFT minimized models. The calculated glycosidic torsion angles for all of the models used for our work range from −101º to −88º for Φcalc, and from −150º to −133º for Ψcalc, and the calculated δ13C1 and δ13C4 range from 83–87 to 101–106 ppm. (ESI Table S3 provides the chemical shift and glycosidic torsion angle results shown in Fig. 6.) Although δ13C1 and δ13C4 both vary with Φ and Ψ, the variation is scattered. These results suggest that it could be problematic to correlate 13C NMR spectra with glycosidic torsion angles. Consequently, we did not pursue using them as a parameters for linking 13C NMR and structure in disordered cellulose.
Relationship between the glycosidic torsion angles (Φ, Ψ), and the calculated δ13C of the nuclei C1 and C4 for the 3 × 1 through 4 × 3 × 3 G09 minimized and the periodic (fully relaxed) VASP cellulose models. The NMR chemical shifts for the two nuclei with range from 83 to 87 ppm and 101 to 106 ppm for C1 and C4, respectively, could aid in the interpretation of the glycosidic torsion angles observed for cellulose
H-bond lengths, angles, energies, and vibrational frequencies
H-bond geometries of the DFT minimized models (Fig. 2) could represent stages in cellulose crystallization or amorphization. ESI Table S4 shows the numerical results for the H-bond lengths, angles, energies, and vibrational frequencies discussed in this section.
H-bond O–H–O angles and O–O lengths obtained with neutron fiber diffraction crystallography could exhibit uncertainties for H-atom positions because the reported H-bond geometry data is based on SHELX-97 fitted data (Nishiyama et al. 2002), which makes correlating the data and results a challenge. Table 3 shows the comparisons of the O–O bond H-bond lengths and O–H–O H-bond angles for the intramolecular (O2–H–O6 and O3–H–O5) and intermolecular (O6–H–O3) H-bonds from the FR P, FR 3 × 1, MM, and model N (structure A) from Nishiyama et al. (2002). Unlike the torsion angle data, the crystallographers did not report experimental uncertainties for the bond lengths and angles, and they did not report H-bond data for their structure C (Nishiyama et al. 2002).
For the intramolecular H-bond O3–O5 distance and H-bond O3–H–O5 angle, Table 3 shows that there is precise agreement between the results (FR P) and data (N) structure A (A) for both the origin [o] and center [o] models. However, the corresponding results for the intramolecular H-bond O2–O6 and O6–O3 distances, and the intermolecular H-bond O2–H–O6 and O6–H–O3 angles do not agree as precisely as the H-bond O3–O5 distances and H-bond O3–H–O5 angles did. For instance, the H-bond O2–O6 distance (2.75 Å) result for the FR P[o] model correlates precisely with the datum from N A[o] (2.77 Å), but the H-bond O2–H–O6 angles from the FR P[o] model (172º) and N A[o] (159º) do not agree as precisely (Table 3). The MM and 3 × 1 results show similar discrepancies with the N data for the three H-bonds. If model FR P is reproducing structure C of Nishiyama et al. (2002), rather than structure A, then it could be that the H-bond O3–O5 distance and H-bond O3–H–O angle are similar for both structures A and C. The differences between the H-bond O3–O5 lengths and O3–H–O5 angles for structures A and C could be subtle, and could exhibit a degree of uncertainty, because H-bond positions are generated through modeling rather than measurements when using neutron fiber diffraction crystallography (Nishiyama et al. 2002).
Comparison of the calculated bond lengths and bond angles from the DFT-minimized models with the vibrational frequencies and the calculated energies of the H-bonds could be useful for studying cellulose assembly during biosynthesis and disordering. The calculated vibrational frequencies could provide insight into the interactions of cellulose polymers during aggregation or degradation and the energies of the H-bonds. Furthermore, because IR spectra are difficult to interpret because of broad peaks in the O–H stretching region for cellulose (Blackwell et al. 1970; Gallina et al. 2006), these calculated results could be applied to the interpretation of cellulose IR spectra. Significantly, the methods developed here could also be applied to determine the interactions that occur between other PCW polymers.
Figures 7, 8 and 9 show examples of only those origin (P[o]) and center (P[c]) chain frequencies that were not coupled to other frequencies. (Note that the vibrational frequencies for the 4 × 3 × 3 model are not included in this discussion because it was not possible to complete the frequency calculations for this structure.) The intermolecular O6–H–O3 H-bond vibration was coupled in all instances, whereas there were eight instances where the O3–H–O5 and O2-H–O6 O–H vibrations were not coupled to another H-bond or were not degenerate with respect to the P[o] and P[c] chains. The FR P model exhibited a large number of coupled vibrational frequencies shown in ESI Table S5. The coupled frequencies seen in the periodic models could explain the complexity of observed IR spectra, because the coupling of these H-bonds likely occurs under experimental conditions.
Figures 7–9 show the relationship between H-bond lengths, angles, NBO E(2) energies, and vibrational frequencies for the intramolecular O2–H–O6 (Fig. 7) and O3–H–O5 (Fig. 8) H-bonds, and the intermolecular O6–H–O3 (Fig. 9) H-bonds.
The NBO E(2) energies range from −57 to −102, −33 to −141, and −39 to −127 kJ/mol for the O2–H–O6, O3–H–O5, and O6–H–O3 H-bonds, respectively (Figs. 7a, 8a, 9a). As discussed in the Methods section, we divided the E(2) energies by 5.5 to obtain H-bond energies that compare with the available data for the energy of a water dimer H-bond. This division gives estimated O2–H–O6, O3–H–O5, and O6–H–O3 H-bond energies of −10 to −19, −6 to −26, and −7 to −23 kJ/mol, respectively. Therefore, each of these H-bonds is moderately strong, as expected for organic molecules (Jeffrey 1997). Because these E(2) energies were for a single, central glucose residue of each respective model, they provide approximate H-bond energies per glucose residue. Correlation of these H-bond energy results with the precise NMR data obtained for these models could provide information about the energy gained from cellulose assembly and the energy required to disassemble cellulose microfibrils (Zhao et al. 2013).
Figures 7a, c and 8a, c also show that intramolecular H-bond NBO E(2) energies for the 3 × 2, 3 × 3, 4 × 2, 4 × 3, and periodic structure are stronger than those for the 3 × 1 and 4 × 1 models. These results suggest that the intermolecular H-bonds in the former structures could be adding stability to the intramolecular H-bond of those models. Therefore, the intermolecular H-bonds present in cellulose microfibrils could add thermodynamic stability of the intramolecular H-bonds in the microfibril, if these trends are correct. If experimentalists are able to discern greater intramolecular H-bond energies in cellulose fragments due to intermolecular H-bonds, then it could be possible to determine if cellulose assembles (disassembles) through the addition (loss) of single chains of cellulose (e.g., 3 × 1, 4 × 1, etc.), or through the addition (loss) of fragments (e.g., 3 × 2, 3 × 3, etc.) that exhibit intermolecular H-bonds.
Generally, as H-bond O–O lengths shorten (Figs. 7a, 8a, 9a) and O–H–O bond angles become more linear (Figs. 7b, 8b, 9b), the NBO E(2) energies increase for the intramolecular O2–H–O6 and O3–H–O5 H-bonds, which agrees with general chemistry principles. However, there is no discernible relationship among the model sizes, and their respective H-bond O–O lengths (or O–H–O bond angles) and NBO E(2) energies. Therefore, the relationship between H-bond lengths, angles, and energies might be ambiguous when studying cellulose assembly and disassembly.
Figures 7b, 8b, and 9b show the relationship between the H-bond O–O lengths and the unscaled vibrational frequencies, and Figs. 7d, 8d, and 9d show analogous trends between the H-bond O–H–O angles and unscaled vibrational frequencies. The frequencies for the FR P model were calculated using VASP, while those for the 3 × 1 through 4 × 3 models were calculated with G09. In general, the 3 × 1 and 4 × 1 models, which exhibit only intramolecular H-bonds, showed higher vibrational frequencies coupled with longer H-bond O–O distances and less linear O–H–O bond angles than did the 3 × 2, 3 × 3, 4 × 2, and 4 × 3 models, which contain intermolecular H-bonds. The trends follow previously reported trends between H-bond lengths and angles with vibrational frequencies. These results suggest that it could be possible to determine if cellulose assembles (degrades) through the addition (loss) of single chains, or multiple chains that exhibit intermolecular H-bonds; higher frequency/longer H-bonds on the fragments would suggest the presence of single chains, and lower frequency/shorter H-bonds would suggest the presence of multiple chains.
The O–H–O H-bond angle results could be useful for interpreting experiment IR data, because the results from the cellulose fragment models show that the intermolecular O–H–O H-bond angles are less linear than the intramolecular H-bond angles are for the 3 × 2, 3 × 3, 4 × 2, and 4 × 3 models. A similar but less distinct trend is present for the H-bond types and their respective H-bond O–O lengths. Therefore, combining crystallographic data, IR spectroscopic data, and NMR results and data could aid in distinguishing the types of H-bonds observed with IR spectroscopy.
Although the interpretation of hydroxyl-H bonds with vibrational spectroscopy can be difficult due to broad spectral bands, these results could be useful for differentiating cellulose assembly and degradation products that exhibit intermolecular H-bonds from those that do not. Figure 10 shows the molecular DFT-calculated plot of the IR intensities versus the unscaled vibrational frequencies for the 3 × 1, 3 × 2, and 3 × 3 models in Fig. 10a, and the analogous plot for the 4 × 1, 4 × 2 and 4 × 3 models in Fig. 10b. As seen in Fig. 10, when intermolecular H-bonds are present (models 3 × 2, 3 × 3, 4 × 2, and 4 × 3), the frequencies in the 3,400–3,650 cm−1 range appear, and the frequencies at approximately 3,700 and 3,800 cm−1 become more intense. In general, each IR frequency is attributable to multiple stretching modes, rather than a single mode. For example, the calculated frequencies for the 3 × 2 model (Fig. 10a) from approximately 3,500–3,650 cm−1 all correspond to O2–H, O3–H, and O6–H stretches. However, the trends seen in Fig. 10 could be useful, if they are experimentally observable, and this method could be useful for studying the formation of intermolecular interactions among other PCW polymers.
Synthetic infrared spectra for the a 3 × 1, 3 × 2, and 3 × 3 models and the b 4 × 1, 4 × 2, and 4 × 3 models. Labeled O–H stretches (e.g., O2–H) indicate which hydroxyl groups modes are active at a particular frequency. Bracketed frequencies indicate that multiple modes are vibrating in a particular frequency range
Conclusion
The results with the Iβ cellulose tg/Net A H-bond network suggest that further DFT studies using periodic structures and small models for cellulose proxies could be useful for distinguishing other H-bond networks that could be present in cellulose during assembly and/or degradation. Those studies could lead to the differentiation of ordered (crystalline), disordered (amorphous), and surface cellulose samples. In addition, the calculated and correlated relationships among H-bond lengths, strengths, angles, and vibrational frequencies, coupled with precise NMR results could aid in the differentiation of cellulose Iβ from other allomorphs of cellulose. These results also provided information about relative H-bond strengths in Iβ cellulose that could be useful for determining the mechanisms of cellulose formation and degradation. Finally, the method developed by this work could also be useful for interpreting the interactions among other plant cell wall polymers.
Based on the results of this work, we argue that Nishiyama et al. (2002) should have reported their structure C as the structure of cellulose Iβ rather than their structure A because the NMR results for FR fit the NMR data better than those data from N. In addition, the χ1 and χ2 torsion angle results for FR fit the data for structure C better than those for structure A. Moreover, the Φ and Ψ torsion angle results for FR fit the data for structure C better than those for structure A. Significantly, the presence of the 2.139 Å bond distance between the H-atoms of C1Ho and C6Hc for periodic model FR, which Nishiyama et al. (2002) reported for their structure C but not for their structure A. Also, the precise correlation between the calculated ring-puckering parameters (θ) with structure C but not structure A. Finally, the H-bond lengths and angles for N structure A do not agree precisely with the results from the periodic model.
Additional results from this work showed that the effect of hydroxymethyl torsion angles (χ1, χ2) rotation by as much as 15º had minimal effect on the δ13C4, δ13C5 and δ13C6. However, the effect of hydroxymethyl torsion angle (Φ, Ψ) rotation had discernable effects on δ13C1 and δ13C4, which could be useful for data interpretation for crystalline and non-crystalline cellulose, cellulose assembly constituents, and cellulose degradation products.
References
Adamo C, Barone V, Introduction I (1998) Exchange functionals with improved long-range behavior and adiabatic connection methods without adjustable parameters: the mPW and mPW1PW models. J Chem Phys 108:664–675
Atalla RH (1999) Celluloses. In: Meth-Cohn O, Barton D, Nakanishi K (eds) Comprehensive natural products chemistry. Elsevier, New York, pp 529–598
Blackwell J (1977) Infrared and Raman spectroscopy of cellulose. In: Arthur J (ed) Cellulose chemistry and technology: ACS symposium series. ACS Publications, Washington, DC, pp 206–218
Blackwell J, Vasko PD, Koenig JL (1970) Infrared and Raman spectra of the cellulose from the cell wall of Valonia ventricosa. J Appl Phys 41:4375–4379
Boys SF, Bernardi F (2002) The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys 100:65–73
Bućko T, Hafner J (2005) No geometry optimization of periodic systems using internal coordinates. J Chem Phys 122:124508–124517
Buhl M, Kaupp M, Malkina OL, Malkin VG (1999) The DFT route to NMR chemical shifts. J Comput Chem 20:91–105
Cancès E, Mennucci B, Tomasi J (1997) A new integral equation formalism for the polarizable continuum model: theoretical background and applications to isotropic and anisotropic dielectrics. J Chem Phys 107:3032–3041
Cheeseman JR, Trucks GW, Keith TA, Frisch MJ (1996) A comparison of models for calculating nuclear magnetic resonance shielding tensors. J Chem Phys 104:5497–5509
Clark T, Chandrasekhar J, Spitznagel GW, Schleyer PVR (1983) Efficient diffuse function-augmented basis sets for anion calculations. III. The 3-21 + G basis set for first-row elements, Li–F. J Comput Chem 4:294–301
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6:850–861
Cremer D, Pople J (1975) A general definition of ring puckering coordinates. J Am Chem Soc 97:1354–1358
Davidson TC, Newman RH, Ryan MJ (2004) Variations in the fibre repeat between samples of cellulose I from different sources. Carbohydr Res 339:2889–2893
Erata T, Shikano T, Yunoki S, Takai M (1997) The complete assignment of the 13C CP/MAS NMR spectrum of native cellulose by using 13C labeled glucose. Cellul Commun 4:128–131
Fernandes AN, Thomas LH, Altaner CM, Callow P, Forsyth VT, Apperely DC, Kennedy CJ, Jarvis MC (2011) Nanostructure of cellulose microfibrils in spruce wood. Proc Natl Acad Sci USA 108:E1195–E1203
Fiadzomor PAY, Keen AM, Grant RB, Orr-Ewing AJ (2008) Interaction energy of water dimers from pressure broadening of near-IR absorption lines. Chem Phys Lett 462:188–191
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery Jr JA, Vreven T Kudin KN, Burant JC et al (2009) Gaussian 09 Revision B.01. Wallingford, CT
Gallina ME, Sassi P, Paolantoni M, Morresi A, Cataliotti RS (2006) Vibrational analysis of molecular interactions in aqueous glucose solutions. Temperature and concentration effects. J Phys Chem B 110:8856–8864
Glendening ED, Weinhold F (1997) Natural resonance theory: II. Natural bond order and valency. J Comput Chem 19:610–627
Glendening ED, Weinhold F (1998) Natural resonance theory: II. Natural bond order and valency. J Comput Chem 19:610–627
Gogonea V (1998) Self-consistent reaction field methods: cavities. In: von Rague Schleyer P, Allinger NL, Clark T, Gastiger J, Kollman PA, Schaefer HF, Schreiner PR (eds) Encyclopedia of computational chemistry. Wiley, New York, NY, pp 2560–2574
Gonzalez-Outeiriño J, Kirschner KN, Thobhani S, Woods RJ (2006) Reconciling solvent effects on rotamer populations in carbohydrates: a joint MD and NMR analysis. Can J Chem 84:569–579
Gottlieb HE, Kotlyar V, Nudelman A (1997) NMR chemical shifts of common laboratory solvents as trace impurities. J Org Chem 62:7512–7515
Grimme S (2006) Semiempirical GGA-type density functional constructed with a long-range dispersion correction. J Comput Chem 27:1787–1799
Guvench O, Hatcher ER, Venable RM, Pastor RW, MacKerell AD Jr (2009) CHARMM additive all-atom force field for glycosidic linkages. J Chem Theory Comput 5:2353–2370
Harris DM, Corbin K, Wang T, Gutierrez R, Bertolo AL, Petti C, Smilgies D-M, Estevez JM, Bonetta D, Urbanowicz BR et al (2012) Cellulose microfibril crystallinity is reduced by mutating C-terminal transmembrane region residues CESA1A903 V and CESA3T942I of cellulose synthase. Proc Natl Acad Sci USA 109:4098–4103
Himmel ME, Di S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD (2007) Biomass recalcitrance: engineering plants and enzymes for biofuels. Science 315(80):804–807
Hohenberg P, Kohn W (1964) Inhomgeneous electron gas. Phys Rev 136:B864–B871
Jarvis M (2003) Cellulose stacks up. Nature 426:1–2
Jeffrey GA (1997) An introduction to hydrogen bonding. Oxford University Press, New York
Karadakov PB (2006) Ab initio calculation of NMR shielding constants. In: Webb GA (ed) Modern magnetic resonance. Springer, Netherlands, pp 63–70
Kirschner KN, Woods RJ (2001) Solvent interactions determine carbohydrate conformation. Proc Natl Acad Sci USA 98:10541–10545
Kohn W, Sham LJ (1965) Self-consistent equations including exchange and correlation effects. Phys Rev 140:A1133–A1138
Kresse G, Furthmüller J (1996) Efficient iterative schemes for ab initio total-energy calculations using a plane-wave basis set. Phys Rev B 54:11169–11186
Krishnan R, Brinkley JS, Seeger R, Pople JA (1980) Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J Chem Phys 72:650–654
Kubicki JD, Mohamed MN-A, Watts HD (2013) Quantum mechanical modeling of the structures, energetics and spectral properties of Iα and Iβ cellulose. Cellulose 20:9–23
Li Y, Lin M, Davenport JW (2011) Ab initio studies of cellulose I: crystal structure, intermolecular forces, and interactions with water. J Phys Chem C 115:11533–11539
Malm, E., Bulone, V., Wickholm, K., Larsson, P., and Iversen, T. (2010). The surface structure of well-ordered native cellulose fibrils in contact with water. Carbohydr. Res. 345
McNamara JP, Hillier IH (2007) Semi-empirical molecular orbital methods including dispersion corrections for the accurate prediction of the full range of intermolecular interactions in biomolecules. Phys Chem Chem Phys 9:2362–2370
McQueen-Mason S, Cosgrove DJ (1994) Disruption of hydrogen bonding between plant cell wall polymers by proteins that induce wall extension. Proc Natl Acad Sci USA 91:6574–6578
Newman RH, Davidson TC (2004) Molecular conformations at the cellulose:water interface. Cellulose 11:23–32
Nishiyama Y, Langan P, Chanzy H (2002) Crystal structure and hydrogen-bonding system in cellulose Iβ from synchrotron X-ray and neutron fiber diffraction. J Am Chem Soc 124:9074–9082
Nishiyama Y, Johnson GP, French AD, Forsyth VT, Langan P (2008) Neutron crystallography, molecular dynamics, and quantum mechanics studies of the nature of hydrogen bonding in cellulose Iβ. Biomacromolecules 9:3133–3140
Panduranga S, Chamundeeswari V, Rajan E, Jebaseelan J, Sundaraganesan N (2011) Theoretical and experimental studies on 2-(2-methyl-5-nitro-1-imidazoyl)ethanol. Eur J Chem 2:136–145
Papajak E, Zheng J, Xu X, Leverentz HR, Truhlar DG (2011) Perspectives on basis sets beautiful: seasonal plantings of diffuse basis functions. J Chem Theory Comput 7:3027–3034
Raman EP, Guvench O, MacKerell AD (2010) CHARMM additive all-atom force field for glycosidic linkages in carbohydrates involving furanoses. J Phys Chem B 114:12981–12994
Sarotti AM, Pellegrinet SC (2009) A multi-standard approach for GIAO (13)C NMR calculations. J Org Chem 74:7254–7260
Schaftenaar G, Noordik JH (2000) Molden: a pre- and post-processing program for molecular and electronic structures. J Comput Aided Mol Des 14:123–134
Schreckenbach G, Ziegler T (1995) Calculation of NMR shielding tensors using gauge-including atomic orbitals and modern density functional theory. J Phys Chem 99:606–611
Shen T, Gnanakaran S (2009) The stability of cellulose: a statistical perspective from a coarse-grained model of hydrogen-bond networks. Biophys J 96:3032–3040
Sternberg U, Koch F, Prieß W, Witter R (2003) Crystal structure refinements of cellulose polymorphs using solid state 13C chemical shifts. Cellulose 10:189–199
Watts HD, Mohamed MNA, Kubicki JD (2011) Comparison of multistandard and TMS-standard calculated NMR shifts for coniferyl alcohol and application of the multistandard method to lignin dimers. J Phys Chem B 115:1958–1970
Wickholm K, Larsson PT, Iversen T (1998) Assignment of non-crystalline forms in cellulose I by CP/MAS 13C NMR spectroscopy. Carbohydr Res 312:123–129
Wolinski K, Hinton JF, Pulay P (1990) Efficient implementation of the gauge-independent atomic orbital method for NMR chemical shift calculations. J Am Chem Soc 112:8251–8260
Zhao Y, Schultz NE, Truhlar DG (2006) Design of density functionals by combining the method of constraint satisfaction with parametrization for thermochemistry, thermochemical kinetics, and noncovalent interactions. J Chem Theory Comput 2:364–382
Zhao H, Kwak JH, Zhang ZC, Brown HM, Arey BW, Holladay JE (2007) Studying cellulose fiber structure by SEM, XRD, NMR and acid hydrolysis. Carbohydr Polym 68:235–241
Zhao Z, Shklyaev OE, Nili A, Mohamed MNA, Kubicki JD, Crespi VH, Zhong L (2013) Cellulose microfibril twist, mechanics, and implication for cellulose biosynthesis. J Phys Chem A 117:2580–2589
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Watts, H.D., Mohamed, M.N.A. & Kubicki, J.D. A DFT study of vibrational frequencies and 13C NMR chemical shifts of model cellulosic fragments as a function of size. Cellulose 21, 53–70 (2014). https://doi.org/10.1007/s10570-013-0128-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10570-013-0128-8