Abstract
Synovial mesenchymal stem cells (SMSCs) have the potential to attenuate osteoarthritis (OA)-induced injury. The role and mechanism of SMSC-derived exosomes (SMSC-Exos), pivotal paracrine factors of stem cells, in OA-associated injury remain unclear. We aimed to confirm the effect of SMSC-Exos with specific modifications on OA-induced damage and to investigate the potential molecular mechanisms. Exosomes derived from miR-155-5p–overexpressing SMSCs (SMSC-155-5p-Exos) and SMSCs (SMSC-Exos) were isolated and characterized. CCK-8, Transwell, and Western blot analyses were used to detect proliferation, migration, extracellular matrix (ECM) secretion, and apoptosis of osteoarthritic chondrocytes. The therapeutic effect of exosomes in a mouse model of OA was examined using immunohistochemical staining and OARSI scores. SPSS 17.0 and GraphPad software were used for all statistical analyses in this study. The SMSC-Exos enhanced the proliferation and migration and inhibited the apoptosis of osteoarthritic chondrocytes but had no effect on ECM secretion. The miR-155-5p–overexpressing exosomes showed common characteristics of exosomes in vitro and further promoted ECM secretion by targeting Runx2. Thus, the SMSC-155-5p-Exos promoted proliferation and migration, suppressed apoptosis and enhanced ECM secretion of osteoarthritic chondrocytes, and effectively prevented OA in a mouse model. In addition, overexpression of Runx2 partially reversed the effect of the SMSC-155-5p-Exos on osteoarthritic chondrocytes. Given the insufficient effect of the SMSC-Exos on the ECM secretion of osteoarthritic chondrocytes, we modified the SMSM-Exos and demonstrated that the SMSC-155-5p-Exos could prevent OA. Exosomes derived from modified SMSCs may be a new treatment strategy to prevent OA.

Graphical abstract
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoarthritis (OA) is the main cause of disability in the elderly population, leading to serious joint disorders (Owens and Conaghan 2016). Studies have found that many factors, such as trauma, abnormal mechanical load, lack of nutrient supply, and genetic tendency, can lead to OA (Solmi et al. 2019). At present, there are still many difficulties to overcome in the treatment of OA. Most of the drugs used to treat OA only relieve joint pain, and therapies that improve joint damage are not available (Solmi et al. 2019; Clarke et al. 2017).
The development of OA mainly involves a decrease in chondrocyte number and an increase in apoptosis and metabolic disorder of the extracellular matrix (ECM) in articular tissue (Li et al. 2019a; Wang et al. 2019; Sun et al. 2019). Based on the mechanism of OA, researchers have explored new treatment and prevention methods for this condition. Current studies on the mechanisms of multiple pathways may result in new strategies to treat OA. During the progression of the disease, areas with severe matrix degradation and destruction often show excessive apoptosis of chondrocytes (Wang et al. 2019). In addition, chondrocytes continuously produce and secrete ECM, such as collagen II (CoII) and SOX9, which are the common structural constituents of cartilage ECM and key indicators to assess the normal physiological function of chondrocytes (Neybecker et al. 2018; Hausburg et al. 2018). During the pathogenesis of OA, the synthesis and secretion of collagen are highly abnormal, which causes irreversible destruction of the cartilage ECM and prevents timely repair. Thus, recent studies have examined therapeutic targets of OA from the perspective of promoting cartilage regeneration, retarding apoptosis and maintaining ECM secretion.
In recent years, the potential of cell therapy in the field of tissue repair has been gradually revealed. Studies extracting mesenchymal stem cells (MSCs) from various tissues and applying them in tissue repair have significantly increased (Bornes et al. 2014). Synovially derived mesenchymal stem cells (SMSCs) have strong vitality and can maintain their unique cell morphology and differentiation potential (cartilage, bone, muscle, fat differentiation, etc.) after more than ten generations (Reisbig et al. 2019). Research has shown that transplanted cells for tissue repair in cell therapy mainly stimulate the activity of recipient cells around the injured site through paracrine mechanisms rather than directly converting to specific parenchymal cells and replacing damaged tissues (Qi et al. 2019; Liu et al. 2018). Subsequent studies have found that exosomes (Exos) play an important role in these paracrine mechanisms (Liu et al. 2018). Exos are membranous vesicles (diameter: 30–100 nm) that move into the extracellular matrix after the integration of intracellular polyvesicles and cell membranes (Liu et al. 2018). These vesicles are widely distributed and can be secreted by many kinds of cells (Rai et al. 2018). An increasing number of basic and clinical studies have shown that Exos secreted by SMSCs are closely related to OA and can promote chondrocyte regeneration, inhibit apoptosis, and improve ECM balance, thus affecting the fate of cells or tissues (Reisbig et al. 2019; Tao et al. 2017). Moreover, Exos contain many functional microRNAs (miRNAs), and multiple studies have confirmed that miRNAs are critical in the pathological process of OA (Tao et al. 2017; Mao et al. 2018).
In this study, through the exploration of Exos in SMSCs and functional miRNAs in Exos, we aimed to provide a new perspective for a better understanding of OA. An in-depth study of Exos will help elucidate the molecular mechanism underlying the formation and development of OA and provide new strategies for the clinical diagnosis and treatment of OA.
Methods
Isolation and incubation of SMSCs
Human cartilage was obtained from knee articular cartilage of OA patients during total knee arthroplasty (THA), and this protocol was approved by the Research Ethics Committee in our hospital (No. ZH[2018]012). The OA patients also signed informed consent, and no other systemic diseases were found. Primary chondrocytes were extracted from fragmented cartilage tissue using collagenase type II (Gibco). The cells were incubated in DMEM which contains 10% FBS, 25 μg/mL ascorbic acid 2-phosphate and added 1% penicillin–streptomycin at 37 °C with 5% CO2 for the follow-up study. Morphological features of the cell were observed by inverted light microscopy (Olympus, Shinjuku-ku, Tokyo). For determination of whether SMSC-Exos could be taken up by chondrocytes, SMSCs were cultured with green fluorescent lipophilic dye in advance, and chondrocytes were cultivated for 12 h with Exos collected from the labeled SMSCs. The nuclei of the chondrocytes were stained with DAPI.
Preparation and identification of SMSC Exos
Human synovial-derived MSCs were cultured in DMEM which contains 10% FBS. The cells were cultivated for 3 days at 37 °C with 5% CO2, and then, Exos were isolated from SMSCs with different conditioned DMEM. The concentration, size, and density of Exos were measured using a nanoparticle tracking analyzer (PMX, Germany). The morphology was assessed by transmission electron microscopy (Hitachi, Japan). Exo surface markers (CD81, CD63) and the riboprotein marker TFIIB were detected by Western blot analysis.
Plasmid or adenovirus construction and transfection
Human osteoarthritic chondrocytes were cultured and transfected with miR-7a, miR-206, miR-320a, miR-155-5p mimic (GenScript; Nanjing, China), and miR-155-5p inhibitor (GenScript; Nanjing, China) at a concentration of 100 nM by Lipofectamine® 2000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). For control, chondrocytes were transfected with a scrambled control miRNA using Lipofectamine® RNAiMAX (Invitrogen). The adenovirus-overexpressing Runx2 was from GeneChem Biological Co., Ltd. (GeneChem, Shanghai, China); this vector was transfected at a multiplicity of infection (MOI) of 50 in FBS-free DMEM for 6 h and then added to plates with 5% FBS of DMEM for 36 h.
Animal studies
Twenty specific pathogen-free (SPF) BALB/C mouse were randomly allocated to four groups after 5 days of adaptive feeding. The detailed method was described previously (Tao et al. 2017). The groups were as follows: (1) Normal group (without cold water stimulation; 5 mice, 10 knee joints, n = 10); (2) OA group (mice were given 4 °C cold water stimulation for 2 h twice a day to construct the OA models and injected with normal saline into the articular cavity 20 days later; continuous injection for 2 weeks, once a day; 5 mice, 10 knee joints, n = 10); (3) OA + SMSC-Exo group (osteoarthritic mice were administered SMSC-Exos [30 μL; 1011 Exo particles/mL] and saline by articular cavity injection, 5 mice, 10 knee joints, n = 10); and (4) OA + SMSC-155-5p-Exo group (mice were administered SMSC-155-5p-Exos [30 μL; 1011 Exo particles/mL] and saline by articular cavity injections, 5 mice, 10 knee joints, n = 10). Two weeks later, the articular tissues were sampled, and the related indicators were detected.
Reverse transcription-quantitative polymerase chain reaction
After harvesting the chondrocytes and separating the total RNA by TRIzol Reagent (Invitrogen), we performed reverse transcription of cDNA of miRNAs, and reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was conducted with a detection kit for miR-7a, miR-206, miR-320a, miR-155-5p, and so on (GenScript; Nanjing, China). Real-time RT-qPCR was conducted with TransStart® Top Green qPCR SuperMix (Transgen Biotech), and GAPDH was then assessed. The primer sequences of these miRNAs are provided in Table 1. After RNA isolation, miRNA microarray analysis and small RNA sequencing were conducted.
Western blot analysis and antibodies
According to a previously reported paper (Li et al. 2019b), protein was extracted from chondrocytes using lysis buffer containing 1% phosphatase inhibitors, 0.5% PMSF, and 0.1% protease inhibitors. Boiled protein (30 μg) was added to SDS-PAGE gels and transferred to PVDF membranes (Millipore, CA, USA). Antibodies against Runx2 (cat. no. sc-390351), Bcl-2 (cat. no. sc-166943), caspase-3 (cat. no. sc-5273), Bax (cat. no. sc-7480), CoII (cat. no. sc-514489), SOX9 (cat. no. sc-166505), and GAPDH (cat. no. sc-137179) at a dilution of 1:1000 (Santa Cruz Biotechnology, CA, USA) were used in this study. The membranes were cultured with primary antibody (1:1000) overnight at 4 °C following the manufacturers’ instructions. Then, the membranes were cultured with secondary antibodies for 2 h at room temperature, and proteins were examined by SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific).
Flow cytometry
To explore the effect of different treatments on cell apoptosis of MSCs, we analyzed the MSCs in different groups by flow cytometry. Cells were digested by 0.25% trypsin-EDTA, labeled using Annexin V-FITC (BD Biosciences, 1:200) for 20 min at 4 °C and detected by flow cytometry and BD FACSuite software (BD Biosciences, San Jose, CA, USA).
Migration assay
Migration in conditioned chondrocytes was detected using a Transwell system according to a previous study (Qi et al. 2019). Briefly, 5 × 104 osteoarthritic chondrocytes were placed in a 24-well transwell plate with 3 μm pore size inserts (Corning, NY, USA, order no. 07-200-166). Then, 500 μL of DMEM containing 0.5% FBS and 1% PBS with 5 μg Exos (10 × 1011 particles/mL) was added to the lower chamber before culturing for 16 h. The cells in the upper chamber (2 × 105 cells) were then placed in 4% paraformaldehyde for 20 min and mixed with 0.5% hematoxylin and eosin for 10 min. The percentage of cell migration in each well was determined in a blinded manner.
Cell counting kit-8 (CCK-8) assay
The cell counting kit-8 (CCK-8) assay kit (KeyGen, Nanjing, China) was used, and chondrocytes (2 × 103 cells/well) were stimulated with 10 μg SMSC-Exos and SMSC-155-5p-Exos (10 × 1011 particles/mL) with or without miR-155-5p inhibitor and Runx2. After 6 h, the cells were placed in 96-well plates and cultured at 37 °C in 5% CO2. Each sample was assayed three times. Cell viability was recorded at 0, 24, 48, 72, and 96 h. The optical density (OD) was examined on a Multiskan GO microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) at 450 nm.
Histology and immunohistochemistry
Cartilage tissue was mixed with 10% neutral buffered formalin overnight and 30% (v/v) buffered formic acid for decalcification. Then, the samples were dehydrated and integrated in paraffin, followed by generation of 5 μm serial sections. Immunohistochemistry was conducted as reported (Liu et al. 2018) to detect cartilage matrix proteins, such as CoII, and apoptotic proteins, such as P65. Immunohistochemical images were randomly taken and assessed by two to three pathologists. The OARSI score was calculated by two to three blinded pathologists to assess the progression of disease (Tao et al. 2017).
Statistical analysis
We used SPSS 17.0 (IBM, Armonk, NY, USA) and GraphPad Prism 8.0 software (GraphPad Software, Inc., San Diego, CA), and the results are shown as the mean ± standard deviation. p < 0.05 indicated statistical significance (Student’s t test).
Results
Isolation and characterization of SMSC Exos
SMSCs were from the synovial membrane, and the cells had a spindle-like shape after the culture density reached 80–90% (Fig. 1a). Similar to previously described Exos, the purified Exos were between 100 and 120 nm in size and exhibited a typical cup shape, as shown by electron microscopy (Fig. 1b). As shown in Fig. 1c, the expression of the markers CD63 and CD81 was abundant in the exosomal fractions but was not detected in whole-cell lysates. In addition, the riboprotein marker TFIIB was not expressed in the Exos (Fig. 1c). These results confirmed that Exos were successfully isolated. To explore whether the SMSC Exos could be taken up by chondrocytes, we cultured SMSCs with green fluorescent lipophilic dye, and chondrocytes were cultured for 12 h with Exos taken from marked SMSCs. Green-marked Exos were detected in the perinuclear region of the chondrocytes (Fig. 1d), which confirmed that the Exos were internalized.
Isolation and identification of SMSC-Exos. a The cell morphology of the SMSCs is spindle-like (scale bar: 50 μm). b The morphology of Exos was examined by transmission electron microscopy (scale bar: 100 nm). c Exo surface markers (CD81, CD63) and the riboprotein marker TFIIB were detected by Western blot analysis. d Green-labeled Exos were absorbed by chondrocytes, and the nuclei of chondrocytes were stained by DAPI (scale bar: 100 μm). The results are representative of three independent experiments
The SMSC-Exos promote proliferation and migration but inhibit apoptosis in osteoarthritic chondrocytes
To explore the effect of the SMSC-Exos in OA, we used SMSC-Exos at a concentration of 10 × 1011 particles/mL and incubated them with osteoarthritic chondrocytes for 36 h. Then, we performed a series of in vitro assays, including proliferation assays, Transwell migration assays, and Western blot analyses. Our results showed that the SMSC-Exos significantly increased osteoarthritic chondrocyte proliferation (Fig. 2a), promoted cell migration (Fig. 2b), and suppressed the apoptosis of osteoarthritic chondrocytes (Fig. 2c, d). However, the SMSC-Exos had no effect on ECM secretion in osteoarthritic chondrocytes (Fig. 2e). These data indicated that Exos derived from SMSCs can improve the damage to osteoarthritic chondrocytes in vitro.
The effect of the SMSC-Exos on osteoarthritic chondrocytes. a CCK-8 assays were used to detect proliferation with or without SMSC-Exo treatment in osteoarthritic chondrocytes. b Transwell assays were performed to examine the migration with or without SMSCs-Exo treatment in osteoarthritic chondrocytes (scale bar: 100 μm). c,d Flow cytometry and Western blotting were used to analyze apoptosis in osteoarthritic chondrocytes with or without SMSC-Exo treatment. e ECM secretion of osteoarthritic chondrocytes with or without SMSC-Exo treatment was examined using Western blot analysis. The results are representative of three independent experiments; *p < 0.05, **p < 0.01; ns, no significance
miRNA expression profiles in the synovial membrane between OA patients and healthy individuals and screening of potential functional miRNAs
Small RNA sequencing was conducted to identify differentially expressed miRNAs in the synovial membrane between OA patients and healthy individuals. Considering the relatively small sample size and the heterogeneity in the samples, we arbitrarily assigned a fold threshold > 2 and p < 0.05, without multiple detection and correction, as the cut-off point. We found that 52 miRNAs were differentially expressed in the synovial membranes between the OA patients and healthy individuals (Fig. 3a). We selected the top 20 miRNAs with the most significant downregulation of expression between the two groups and performed RT-qPCR analysis to verify the difference in expression of these genes (Fig. 3b). According to the PCR results, we identified the 4 miRNAs that had the most obvious downregulation and further verified them in the Exo fractions by RT-qPCR. These results showed that miR-155-5p was the most abundant miRNA in the SMSC-Exos (Fig. 3c). We next constructed overexpression vectors for these four miRNAs and investigated their effects on apoptosis and ECM secretion of osteoarthritic chondrocytes. Western blot analysis indicated that miR-155-5p inhibited apoptosis significantly (Fig. 3d) while increasing ECM secretion of osteoarthritic chondrocytes (Fig. 3e). The miR-155-5p expression in SMSC-Exos in normal synovial tissue was higher than that in the osteoarthritic SMSC-Exos (Fig. 3f). To obtain Exos containing high levels of miR-155-5p, we isolated Exos from the SMSC-155-5p-Exos. Compared with SMSC-Exos, miR-155-5p expression increased more than 60-fold in the SMSC-155-5p-Exos (Fig. 3g). The expression of miR-155-5p in the chondrocytes treated with the SMSC-155-5p-Exos increased nearly 40 times compared with that in the untreated chondrocytes (Fig. 3h).
Screening of the potential functional miRNAs in osteoarthritic chondrocytes. a Differential expression of miRNAs in the synovial membrane between OA patients and healthy individuals. b RT-qPCR analysis was used to verify the difference in expression of the miRNAs. c Four of the most obviously downregulated miRNAs were further verified in SMSC-Exos by PCR analysis. d,e Western blot analysis was performed to detect the expression of proteins related to apoptosis and ECM secretion in osteoarthritic chondrocytes transfected with miRNAs. f miR-155-5p expression in the SMSC-Exos from controls and OA patients was compared with RT-qPCR analysis. g RT-qPCR analysis was used to detect miR-155-5p expression in the SMSC-Exos and the SMSC-155-5p-Exos. h RT-qPCR analysis was used to detect miR-155-5p expression in the chondrocytes treated with the SMSC-Exos or the SMSC-155-5p-Exos. The results are representative of three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001
Exosomal miR-155-5p increases proliferation, migration, and ECM secretion and attenuates apoptosis in osteoarthritic chondrocytes
To explore the role of miR-155-5p in the SMSC-155-5p-Exos, we used a miR-155-5p inhibitor, which was transferred into osteoarthritic chondrocytes via the SMSC-155-5p-Exos. We found that the SMSC-155-5p-Exos were more effective than the SMSC-Exos in promoting the proliferation and migration of osteoarthritic chondrocytes, and the SMSC-155-5p-Exos also promoted ECM secretion. However, the effects of the SMSC-155-5p-Exos on osteoarthritic chondrocyte proliferation, migration, and ECM secretion were partially reversed by the miR-155-5p inhibitor (Fig. 4a, b, and e). In addition, the effects of the SMSC-155-5p-Exos on osteoarthritic chondrocyte apoptosis were much stronger than those of the SMSC-Exos. The inhibitory effect of the SMSC-155-5p-Exos on osteoarthritic chondrocyte apoptosis was blocked by the miR-155-5p inhibitor (Fig. 4c, d).
Effect of miR-155-5p in osteoarthritic chondrocytes. a CCK-8 assays were used to detect proliferation of osteoarthritic chondrocytes transfected with the SMSC-Exos and the SMSC-155-5p-Exos with or without the miR-155-5p inhibitor. b Transwell assays were performed to examine the migration of osteoarthritic chondrocytes transfected with the SMSC-Exos and the SMSC-155-5p-Exos with or without the miR-155-5p inhibitor (scale bar: 100 μm). c,d Flow cytometry and Western blotting were used to analyze apoptosis in osteoarthritic chondrocytes transfected with the SMSC-Exos and the SMSC-155-5p-Exos with or without the miR-155-5p inhibitor. e The ECM-related protein expression of SOX9 and CoII was examined using Western blot analysis in osteoarthritic chondrocytes transfected with the SMSC-Exos and the SMSC-155-5p-Exos with or without the miR-155-5p inhibitor. The results are representative of three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001
Runx2 is a target of miR-155-5p which is negatively relative with miR-155-5p
Previously, miRNAs were found to be related to transcription factors; the predicted mRNAs that interact with miR-155-5p were analyzed by TargetScan and miRBase software, and we predicted that Runx2 was a potential target of miR-155-5p. As shown in Fig. 5a, miR-155-5p has one potential complementary sequence with Runx2. A dual-luciferase reporter assay of HEK 293 cells was performed. After transfection with the miR-155-5p mimic, the relative luciferase activity was significantly decreased while the relative luciferase activity increased after transfection with the miR-155-5p inhibitor in the WT transfection group. A comparison of the relative luciferase activity between the two groups showed no significant difference after the UTR-mut group was transfected with the miR-155-5p mimic or miR-155-5p inhibitor (Fig. 5b). To further investigate the results, we performed Western blot analysis to assess the expression of Runx2. As shown in Fig. 5c, Runx2 function was inhibited when miR-155-5p was overexpressed, whereas inhibition of miR-155-5p upregulated the protein levels of Runx2. In addition, Runx2 expression was increased in osteoarthritic synovial tissues, and an analysis showed that miR-155-5p is negatively related to Runx2 in the osteoarthritic synovial membrane (Fig. 5d). We also explored whether the functions of the SMSC-155-5p-Exos are mediated by upregulation of Runx2. CCK-8 assays indicated that the proliferative effect of the SMSC-155-5p-Exos on osteoarthritic chondrocytes was blocked by overexpressing Runx2 (Fig. 5e). Transwell assays indicated that the antimigratory effects of the SMSC-155-5p-Exos in osteoarthritic chondrocytes were abolished by Runx2 overexpression (Fig. 5f). In addition, flow cytometry and Western blot analysis indicated that enhanced Runx2 blocked the anti-apoptotic effects of the SMSC-155-5p-Exos in osteoarthritic chondrocytes (Fig. 5g, h). Furthermore, overexpression of Runx2 partially reversed the SMSC-155-5p-Exo-mediated promotion of ECM secretion (Fig. 5i).
Runx2 is a target of miR-155-5p and is negatively correlated with miR-155-5p. a TargetScan and miRBase software were used to predict the potential targets of miR-155-5p. b A dual-luciferase reporter assay was performed in HEK 293 cells to confirm the relationship between Runx2 and miR-155-5p. c Western blot analysis was performed to detect Runx2 expression with miR-155-5p mimic or inhibitor treatment. d RT-qPCR was used to detect the expression of Runx2 and miR-155-5p in the osteoarthritic synovial membrane, and the relationship of Runx2 and miR-155-5p was analyzed. e–i Osteoarthritic chondrocytes transfected with SMSC-Exos and SMSC-155-5p-Exos with or without the miR-155-5p inhibitor were used to detect proliferation (e), migration (f), apoptosis (scale bar: 100 μm) (g,h), and ECM secretion (i). The results are representative of three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significance
The SMSC-155-5p-Exos prevent OA
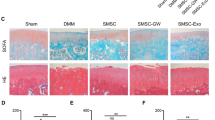
To explore the role of Exos in prevention of OA, we constructed a mouse model of OA. The staining results of femoral condyle sections indicated that caspase-3 activity was increased in the OA model but could be significantly inhibited by both the SMSC-Exos and the SMSC-155-5p-Exos. Consistent with the results of in vitro experiments, decreased CoII expression in the mouse model of OA was not elevated in the SMSC-Exos group but increased significantly in the SMSC-155-5p-Exo group (Fig. 6a, b). Moreover, the decrease in chondrocyte number and the increase in OARSI scores in the mouse model of OA were reversed by the SMSC-Exos and the SMSC-155-5p-Exos, and the ability of the SMSC-155-5p-Exos to prevent OA was more significant than that of the SMSC-Exos (Fig. 6c, d). These data indicated that Exos with or without some special modifications may have an important role in clinical applications.
The SMSC-155-5p-Exos prevents OA. a,b Photomicrographs of femoral condyle sections of mice with OA were stained and performed to measure the expression of caspase-3 and CoII (scale bar: 100 μm). c,d Statistical results of the chondrocytes and the result of statistical analysis of OARSI score are shown in each group. The results are representative of three independent experiments; *p < 0.05, **p < 0.01, ***p < 0.001; ns, no significance
Discussion
Exos, as an important paracrine mechanism, are critical in intercellular communication (Wu et al. 2019). This study confirmed that Exos derived from SMSCs attenuate OA by enhancing proliferation and ECM secretion and attenuating apoptosis. We also showed that expression of miR-155-5p was corrected in OA via microarray analysis of synovial tissue in OA patients; the overexpression of miR-155-5p exerted the same effect as the SMSC-Exos on osteoarthritic chondrocytes by negatively regulating Runx2 expression. Further analysis demonstrated that Exos derived from miR-155-5p-overexpressing SMSCs had a more significant effect on OA than the SMSC-Exos.
Recently, numerous reports on miRNAs in OA have been published and have increased interest in the pathophysiological process of OA (Tsai et al. 2017; Mao et al. 2017). An increasing number of miRNAs have been confirmed to be aberrantly expressed in OA (Papanagnou et al. 2016). To explore the expression and effect of miRNAs in OA, we used microarray and RT-qPCR analysis to screen differentially expressed miRNAs, such as miR-155-5p, in osteoarthritic synovial tissue compared with normal synovial tissue. Previous studies have indicated that miR-155-5p can control cell proliferation and apoptosis in many diseases (Zhao et al. 2019; Al-Haidari et al. 2017). The overexpression of miR-155-5p could enhance osteoarthritic chondrocyte proliferation, migration, and ECM secretion and inhibit apoptosis. Recent studies have examined how specific miRNAs affect cartilage function and the occurrence of OA.
Previous studies have reported that miRNAs interact with proteins to regulate protein function or directly localize in cellular compartments to regulate cellular functions (Shi et al. 2019; Sun et al. 2018). This study has indicated that miR-155-5p and Runx2 can interact through binding sites, which negatively regulates the expression of Runx2. Chondrocyte proliferation and differentiation were shown to be regulated by noncoding RNA. For example, miR-204 could bind the 3′ untranslated region (UTR) of the Runx2 gene and influence its expression in mesenchymal progenitor cells, chondrocytes, and osteoblasts (Zhang et al. 2011). In this study, we demonstrated that Runx2 is upregulated in osteoarthritic synovial tissue and is negatively associated with miR-155-5p. After miR-155-5p treatment, the related indicators of OA were significantly improved, and the expression of Runx2 was also decreased. In addition, we found that overexpression of Runx2 could significantly reverse the effect of miR-155-5p. Therefore, we infer that Runx2 is critical for the attenuation of OA-associated injury by miR-155-5p.
SMSCs have a strong ability to proliferate, do not show a decrease in viability after passaging, and are easy to isolate and extract (Guo et al. 2016). In this study, we found that SMSC-derived Exos can be easily extracted in large quantities. Although Exos derived from SMSCs could strongly promote chondrocyte proliferation and migration and inhibit apoptosis, they had no effect on ECM synthesis and secretion of osteoarthritic chondrocytes. Interestingly, SMSC-155-5p-Exo treatment of chondrocytes not only had significant effects on proliferation and migration and inhibition of apoptosis but also promoted the secretion of the ECM-related proteins CoII and SOX9. The secretion of miR-155-5p in the SMSC-155-5p-Exos was nearly 40 times higher than that in the SMSC-Exos. This finding indicates that the influence of Exos on ECM secretion in chondrocytes is mainly mediated by the overexpression of miR-155-5p. These results indicate that Runx2, a miR-155-5p target gene, blocks the effect of the SMSC-155-5p-Exos on proliferation, migration, ECM secretion, and apoptosis in osteoarthritic chondrocytes. Therefore, using SMSC-155-5p-Exos will be a convenient and reliable strategy, and SMSC-155-5p-Exos may be a suitable therapeutic agent for the treatment of OA.
In conclusion, by targeting Runx2, miR-155-5p promoted chondrocyte proliferation, migration, and ECM secretion and inhibited apoptosis. Furthermore, we found that the SMSC-155-5p-Exos alleviated OA-associated injury and promoted cartilage regeneration, which may be explored as a new strategy for OA therapy.
References
Al-Haidari AA, Syk I, Thorlacius H. Mir-155-5p positively regulates ccl17-induced colon cancer cell migration by targeting rhoa. Oncotarget. 2017;8:14887–96.
Bornes TD, Adesida AB, Jomha NM. Mesenchymal stem cells in the treatment of traumatic articular cartilage defects: a comprehensive review. Arthritis Research & Therapy. 2014;16:432.
Clarke SP, Poulis N, Moreton BJ, Walsh DA, Lincoln NB. Evaluation of a group acceptance commitment therapy intervention for people with knee or hip osteoarthritis: a pilot randomized controlled trial. Disabil Rehabil. 2017;39:663–70.
Guo SC, Tao SC, Yin WJ, Qi X, Sheng JG, Zhang CQ. Exosomes from human synovial-derived mesenchymal stem cells prevent glucocorticoid-induced osteonecrosis of the femoral head in the rat. Int J Biol Sci. 2016;12:1262–72.
Hausburg MA, Frederick ED, McNair P, Schwappach J, Banton KL, Roshon M, et al. Clinically relevant redifferentiation of fibroblast-like chondrocytes into functional chondrocytes by the low molecular weight fraction of human serum albumin. Clin Exp Rheumatol. 2018;36:891–5.
Li Q, Chen W, Song M, Chen W, Yang Z, Yang A. Weighted gene co-expression network analysis and prognostic analysis identifies hub genes and the molecular mechanism related to head and neck squamous cell carcinoma. Cancer Biol Ther. 2019a;206:1–10.
Li S, Ma F, Pang X, Tang B, Lin L. Synthesis of chondroitin sulfate magnesium for osteoarthritis treatment. Carbohydr Polym. 2019b;212:387–94.
Liu Y, Lin L, Zou R, Wen C, Wang Z, Lin F. Msc-derived exosomes promote proliferation and inhibit apoptosis of chondrocytes via lncrna-klf3-as1/mir-206/git1 axis in osteoarthritis. Cell Cycle. 2018;17:2411–22.
Mao G, Zhang Z, Huang Z, Chen W, Huang G, Meng F, et al. MicroRNA-92a-3p regulates the expression of cartilage-specific genes by directly targeting histone deacetylase 2 in chondrogenesis and degradation. Osteoarthr Cartil. 2017;25:521–32.
Mao G, Hu S, Zhang Z, Wu P, Zhao X, Lin R, et al. Exosomal mir-95-5p regulates chondrogenesis and cartilage degradation via histone deacetylase 2/8. J Cell Mol Med. 2018;22:5354–66.
Neybecker P, Henrionnet C, Pape E, Mainard D, Galois L, Loeuille D, et al. In vitro and in vivo potentialities for cartilage repair from human advanced knee osteoarthritis synovial fluid-derived mesenchymal stem cells. Stem Cell Res Ther. 2018;9:329.
Owens C, Conaghan PG. Improving joint pain and function in osteoarthritis. The Practitioner. 2016;260:17–20.
Papanagnou P, Stivarou T, Tsironi M. The role of miRNAs in common inflammatory arthropathies: osteoarthritis and gouty arthritis. Biomolecules. 2016;6:44.
Qi H, Liu DP, Xiao DW, Tian DC, Su YW, Jin SF. Exosomes derived from mesenchymal stem cells inhibit mitochondrial dysfunction-induced apoptosis of chondrocytes via p38, ERK, and AKT pathways. In Vitro Cell Dev Biol Anim. 2019;55:203–10.
Rai A, Greening DW, Chen M, Xu R, Ji H, Simpson RJ. Exosomes derived from human primary and metastatic colorectal cancer cells contribute to functional heterogeneity of activated fibroblasts by reprogramming their proteome. Proteomics. 2018;14:e1800148.
Reisbig NA, Pinnell E, Scheuerman L, Hussein H, Bertone AL. Synovium extra cellular matrices seeded with transduced mesenchymal stem cells stimulate chondrocyte maturation in vitro and cartilage healing in clinically-induced rat-knee lesions in vivo. PLoS One. 2019;14:e0212664.
Shi Z, She K, Li H, Yuan X, Han X, Wang Y. Microrna-454 contributes to sustaining the proliferation and invasion of trophoblast cells through inhibiting nodal/alk7 signaling in pre-eclampsia. Chem Biol Interact. 2019;298:8–14.
Solmi M, Koyanagi A, Thompson T, Fornaro M, Correll CU, Veronese N. Network analysis of the relationship between depressive symptoms, demographics, nutrition, quality of life and medical condition factors in the osteoarthritis initiative database cohort of elderly North-American adults with or at risk for osteoarthritis—corrigendum. Epidemiol Psychiatric Sci. 2019;29:e14.
Sun M, Zhai M, Zhang N, Wang R, Liang H, Han Q, et al. MicroRNA-148b-3p is involved in regulating hypoxia/reoxygenation-induced injury of cardiomyocytes in vitro through modulating sirt7/p53 signaling. Chem Biol Interact. 2018;296:211–9.
Sun M, Hussain S, Hu Y, Yan J, Min Z, Lan X, et al. Maintenance of sox9 stability and ECM homeostasis by selenium-sensitive prmt5 in cartilage. Osteoarthr Cartil. 2019;27:932–44.
Tao SC, Yuan T, Zhang YL, Yin WJ, Guo SC, Zhang CQ. Exosomes derived from mir-140-5p-overexpressing human synovial mesenchymal stem cells enhance cartilage tissue regeneration and prevent osteoarthritis of the knee in a rat model. Theranostics. 2017;7:180–95.
Tsai CH, Liu SC, Wang YH, Su CM, Huang CC, Hsu CJ, et al. Osteopontin inhibition of mir-129-3p enhances il-17 expression and monocyte migration in rheumatoid arthritis. Biochim Biophys Acta Gen Subj. 2017;1861:15–22.
Wang K, Li Y, Dai Y, Han L, Zhu Y, Xue C, et al. Peptides from Antarctic krill (Euphausia superba) improve osteoarthritis via inhibiting hif-2alpha-mediated death receptor apoptosis and metabolism regulation in osteoarthritic mice. J Agric Food Chem. 2019;67:3125–33.
Wu J, Li H, Xie H, Wu X, Lan P. The malignant role of exosomes in the communication among colorectal cancer cell, macrophage and microbiome. Carcinogenesis. 2019;40:601–10.
Zhang Y, Xie RL, Croce CM, Stein JL, Lian JB, van Wijnen AJ, et al. A program of microRNAs controls osteogenic lineage progression by targeting transcription factor runx2. Proc Natl Acad Sci U S A. 2011;108:9863–8.
Zhao XS, Han B, Zhao JX, Tao N, Dong CY. Mir-155-5p affects Wilms' tumor cell proliferation and apoptosis via targeting creb1. Eur Rev Med Pharmacol Sci. 2019;23:1030–7.
Funding
This research was supported by the National Natural Science Foundation of China (81873990, 81873991, and 81672238), the Jiangsu Provincial Medical Youth Talent (QNRC2016751), and the Natural Science Foundation of Jiangsu Province (BK20180001).
Author information
Authors and Affiliations
Contributions
Guarantor of integrity of the entire study: Zhirong Wang
Study concepts: Zhirong Wang
Study design: Zhirong Wang
Literature search: Yuefeng Hao, Dechun Geng
Clinical studies: Kai Yan, Gaoran Ge
Experimental studies: Kai Yan, Gaoran Ge
Data acquisition: Di Zhang, Jiaxiang Bai, Xiaobin Guo, Jing Zhou
Data analysis: Tianpeng Xu, Menglei Xu, Xiao Long
Statistical analysis: Yuefeng Hao, Dechun Geng
Manuscript preparation: Zhirong Wang, Kai Yan
Manuscript editing: Zhirong Wang, Kai Yan
Manuscript review: Zhirong Wang
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they do not have conflicts of interests.
Ethical approval
This study was approved by the relevant Ethics Committee.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Z., Yan, K., Ge, G. et al. Exosomes derived from miR-155-5p–overexpressing synovial mesenchymal stem cells prevent osteoarthritis via enhancing proliferation and migration, attenuating apoptosis, and modulating extracellular matrix secretion in chondrocytes. Cell Biol Toxicol 37, 85–96 (2021). https://doi.org/10.1007/s10565-020-09559-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-020-09559-9