Abstract
MicroRNAs (miRNAs) are small non-coding RNAs regulating post-transcriptional gene expression. They play important roles in many biological processes under physiological or pathological conditions, including development, metabolism, tumorigenesis, metastasis, and immune response. Over the past 15 years, significant insights have been gained into the roles of miRNAs in cancer. Depending on the cancer type, miRNAs can act as oncogenes, tumor suppressors, or metastasis regulators. In this review, we focus on the role of miRNAs as components of molecular networks regulating metastasis. These miRNAs, termed metastamiRs, promote or inhibit metastasis through various mechanisms, including regulation of migration, invasion, colonization, cancer stem cell properties, epithelial-mesenchymal transition, and microenvironment. Some of these metastamiRs represent attractive therapeutic targets for cancer treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
MicroRNAs (miRNAs) are small, single-stranded RNAs that are approximately 22 nucleotides in length. They downregulate the expression of genes encoding proteins or long non-coding RNAs (lncRNAs), by inhibiting mRNA translation or by promoting target RNA degradation (Fig. 1). It is estimated that miRNAs regulate more than 30% of human protein-coding genes [1].
Schematic representation of the miRNA biogenesis. In the nucleus, the primary miRNA transcript (pri-miRNA), produced by RNA polymerase II or III, is cleaved by the microprocessor complex Drosha–DGCR8. The resulting hairpin-shaped precursor (pre-miRNA) is exported from the nucleus by Exportin-5–Ran-GTP. In the cytoplasm, the RNase Dicer, paired with the double-stranded RNA-binding protein TRBP, cleaves the pre-miRNA into the miRNA duplex of mature length. The functional strand of the duplex is loaded together with Argonaute (AGO) proteins into the RNA-induced silencing complex (RISC), where it guides RISC to silence target RNAs through RNA cleavage and/or translational inhibition
The human genome produces more than 1500 miRNAs (www.mirbase.org). Mature miRNAs are generated from their hairpin-shaped precursors encoded by the genome. Figure 1 summarizes the steps of the miRNA biogenesis [2]. Most miRNA genes are transcribed by RNA polymerase II, and others are transcribed by RNA polymerase III [1]. In the nucleus, the RNase III enzyme Drosha pairs with the double-stranded RNA binding protein DGCR8 to form the microprocessor, which cleaves the primary transcripts of miRNAs (pri-miRNAs) into hairpin-shaped miRNA precursors (pre-miRNAs) [3,4,5]. Some pri-miRNAs require additional factors for efficient cleavage [2].
Subsequently, Exportin-5 transports pre-miRNAs in a RanGTP-dependent manner to the cytoplasm [6,7,8], where another type-III RNase, Dicer, cleaves the pre-miRNA into a ~22-nucleotide miRNA duplex with 5′ phosphate groups, 3′ overhangs, and in most cases, imperfect base pairing [9, 10]. The mismatches lead to degradation of one strand of the duplex, and the other strand is loaded into the RNA-induced silencing complex (RISC). The RISC binds to either perfect, or in most cases, imperfect complementary sequences in the target RNAs, such as lncRNAs or the 3′ UTR of mRNAs, leading to either inhibition of mRNA translation or degradation of the target RNAs, or both [1, 11, 12].
By targeting mRNAs or lncRNAs involved in cell proliferation, apoptosis, migration, differentiation, and so on, miRNAs play important roles in physiological and pathological processes, including development [13], metabolism [14], tumor initiation and metastasis [15,16,17], viral infection [18], and immune response [19]. Over the past 15 years, the field of cancer-implicated miRNAs has expanded dramatically, which prompted the development of miRNA therapeutics [20]. A number of studies have revealed that miRNA dysregulation plays causal roles in cancer; researchers define “oncomiRs” as miRNAs acting as oncogenes or tumor suppressors and “metastamiRs” as metastasis-regulating miRNAs.
2 MicroRNAs in cancer
Several approaches, including chromosomal analysis, miRNA microarrays, miRNA qPCR arrays, and more recently, high-throughput small RNA sequencing platforms, have been employed to generate miRNA profiles in human cancers. Numerous studies have demonstrated that miRNAs are dysregulated in cancer. In 2002, Calin et al. provided the first evidence of aberrant miRNA expression in human cancer, showing that the expression of miR-15a and miR-16 was lost in B-cell chronic lymphocytic leukemia (CLL) due to deletion of the 13q14 chromosomal locus [21]. Interestingly, this region does not encode any proteins, indicating that the miRNAs miR-15a and miR-16 may be suppressors of CLL. Indeed, mice deficient in miR-15 and miR-16 developed CLL-like disease [22]. Following this remarkable discovery, differential miRNA expression between normal tissues, primary tumor tissues, and metastases has been extensively documented. For instance, Lu et al. profiled the expression of 217 miRNAs in 334 normal tissues and tumor samples. They identified miRNAs upregulated or downregulated in cancer and found that miRNA signatures classified cancer types better than mRNAs [23].
While dysregulation of miRNA expression can be either a cause or a consequence of cancer formation, functional studies have demonstrated that miRNAs can play causal roles in tumor initiation and growth. For instance, the miR-17~92 miRNA cluster, often referred to as oncomiR-1, is overexpressed in B-cell lymphoma and colon cancer relative to the corresponding normal tissues [24]. He et al. overexpressed this miRNA cluster in fetal liver hematopoietic stem cells (HSCs) isolated from Eμ-Myc transgenic mice and injected these cells into recipient mice. Tumors derived from HSCs overexpressing the miRNA cluster were more aggressive, resulting in worse tumor-free survival and overall survival in mice [24]. These findings suggest that the miR-17~92 miRNA cluster serves as an oncogene and promotes Myc-induced lymphomagenesis. In addition, Xiao et al. reported that transgenic overexpression of the miR-17~92 cluster in mouse lymphocytes led to lymphoproliferative disease, autoimmunity, and premature death [25]. Mechanistically, this miRNA cluster inhibits the expression of PTEN (a tumor suppressor) and Bim (a pro-apoptotic protein) [25].
Another example of a miRNA serving as an oncogene is miR-21, a miRNA overexpressed in most types of cancer, including lung cancer, breast cancer, ovarian cancer, glioblastoma, leukemia, and lymphoma [26]. In mice, genetic ablation of miR-21 partially blocked non-small cell lung cancer in a K-Ras-induced tumor model, and 4- to 6-fold transgenic overexpression of miR-21 exacerbated K-Ras-induced lung tumorigenesis, although these miR-21 knockout and transgenic mice showed no obvious phenotypes [27]. In contrast, when miR-21 was overexpressed in mice at a higher level (16-fold higher than the basal level) in a tissue-specific and doxycycline-controlled manner, the animals developed pre-B-cell lymphoma, which could be reversed by turning off miR-21 expression, demonstrating that this oncomiR plays a critical role in lymphoma initiation and maintenance [28].
Some miRNAs are tumor suppressors. A well-established tumor-suppressing miRNA is let-7, the first known human miRNA [29]. Let-7 targets many oncogenes, including RAS, MYC, CDK6, HMGA2, and BACH1 [30]. Overexpression of let-7 in lung cancer cell lines downregulated RAS protein and inhibited tumor growth in xenograft mouse models [31, 32]. Another example of a tumor suppressor miRNA is the miR-34 family [33]. miR-34 is a direct transcriptional target of p53, and its overexpression can induce growth arrest in both primary and tumor-derived cell lines [34], which is mediated by its multiple targets regulating cell cycle, apoptosis, and proliferation, including CDK4, CDK6, CCNE2, CCND1, MET, and BCL2 [33, 34]. Surprisingly, genetic deletion of all mir-34 genes in mice did not increase spontaneous tumor formation [35]. However, mice with prostate-specific knockout of both miR-34 and p53 showed expansion of prostate stem cells and developed high-grade prostatic intraepithelial neoplasia and early invasive adenocarcinoma [36], indicating that mir-34 genes are bona fide tumor suppressors.
3 MicroRNAs in metastasis
Cancer metastasis is still a formidable and fatal challenge. Thus far, there are no effective therapies for treating metastatic cancer [37,38,39]. Early dissemination of metastatic seeds in some cancers encumbers metastasis prevention during cancer treatment [38, 40]. Moreover, because of the cost, time, and the lack of biomarkers to identify patients who are at high risk of metastatic disease, it is challenging to run metastasis prevention trials on patients with early-stage cancer [41]. Therefore, it is urgent to develop better prognostic markers for metastasis, novel agents and regimens that target specific abnormalities, and effective predictive markers for therapy response [42].
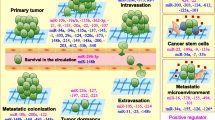
Since 2007, various mechanisms underlying miRNA-mediated regulation of metastasis have been identified, such as regulation of oncogenes, tumor suppressor genes, metastasis genes, cancer stem cell properties, epithelial-mesenchymal transition (EMT), microenvironment, and exosome secretion; moreover, miRNA biogenesis enzymes and their regulators also play functional roles in metastasis (Fig. 2). Here, we discuss examples for each mechanism, illustrating how miRNAs regulate tumor progression and metastasis.
Mechanisms by which miRNAs regulate metastasis. MetastamiRs can function through regulation of oncogenes, tumor suppressor genes, metastasis genes, cancer stem cell properties, epithelial-mesenchymal transition (EMT), microenvironment, and exosome secretion. Regulators of miRNA biogenesis also play functional roles in metastasis
3.1 MicroRNAs regulate metastasis by targeting oncogenes, tumor suppressors, or metastasis genes
MiRNA regulation of metastasis was first reported in 2007. Ma et al. identified metastasis-associated miRNAs by comparing miRNA expression between normal human mammary epithelial cells, non-metastatic breast cancer cells, and metastatic breast cancer cells. They found that overexpression of miR-10b in human breast cancer cell lines promoted lung metastasis in orthotopic xenograft models [43]. Conversely, silencing miR-10b inhibited lung metastasis in mice bearing mammary tumors formed by the 4T1 cell line [44]. This provided the first functional evidence for a metastasis-regulating miRNA. Later, a number of groups showed that miR-10b is overexpressed in highly metastatic or invasive human tumors, including metastatic breast cancer, pancreatic cancer, and glioblastoma [45]; this miRNA has also been shown to promote either tumor growth or metastasis in various cancer types [45]. Interestingly, two recent studies showed that combining miR-10b antisense inhibitors with low-dose doxorubicin led to complete, durable regression of existing lymph node metastases and distant metastases in preclinical models of metastatic breast cancer, in which combination treatment was delivered after surgical removal of the primary mammary tumor, once metastases were confirmed by bioluminescent imaging of live animals [46, 47]. These findings suggest that targeting miR-10b has potential as an anti-metastatic therapy.
Although growing numbers of miRNAs are implicated in breast cancer progression based on cell culture and xenograft models, their functions in spontaneous mammary tumors were not demonstrated by miRNA knockout approaches until Kim et al. generated miR-10b-null mice to study the role of this miRNA in vivo. Genetic deletion of miR-10b did not cause substantial developmental defects but dramatically suppressed oncogene-induced mammary tumorigenesis and metastasis in a polyomavirus middle T (PyMT) model of metastatic breast cancer [48], providing in vivo proof that miR-10b contributes to breast cancer progression. Mechanistically, miR-10b targets multiple tumor suppressor genes (e.g., TBX5 and PTEN) and metastasis suppressor genes (e.g., HOXD10), and these genes are upregulated in miR-10b-deficient mouse embryonic fibroblasts and PyMT mammary tumors, suggesting that they are physiologically relevant bona fide miR-10b targets (Fig. 3). TBX5, which is a transcription factor, activates the expression of two tumor suppressor genes, PTEN and DYRK1A [48], while the transcriptional repressor HOXD10 inhibits the expression of multiple pro-metastatic proteins, including RhoC, α3 integrin, uPAR, and MT1-MMP [49]. Moreover, miR-10b has been shown to promote proliferation by targeting NF1 (encoding neurofibromin, a negative regulator of RAS) [50] and to promote migration and invasion by targeting KLF4 [51]. Collectively, miR-10b downregulates multiple tumor suppressors or metastasis suppressors, either directly or through its direct target TBX5, ultimately contributing to tumorigenesis and metastasis (Fig. 3). Inhibition of miR-10b may provide therapeutic opportunities through reactivation of downstream tumor-suppressing and metastasis-suppressing pathways.
A second metastasis-promoting miRNA identified by Ma et al. is miR-9 [43, 52]. Initially, miR-9 was found to target CDH1 (encoding E-cadherin) and promote metastatic ability in breast cancer cells expressing this adhesion molecule [52]. Subsequently, Chen e al. found that miR-9 can also induce metastasis in E-cadherin-negative breast cancer cells, which led to the identification of leukemia inhibitory factor receptor (LIFR) as a breast cancer metastasis suppressor targeted by miR-9 [53]. Interestingly, LIFR suppresses metastasis by activating Hippo signaling, leading to phosphorylation and functional inactivation of the Hippo pathway effector YAP. Depleting LIFR in non-metastatic breast cancer cells led to massive lung metastasis in xenograft models, which was reversed by silencing of YAP; conversely, restoring LIFR expression in highly metastatic breast cancer cells suppressed lung metastasis formation, which was reversed by a non-phosphorylatable YAP mutant [53]. Notably, LIFR protein is downregulated in human breast cancer, and loss of LIFR in non-metastatic breast tumors is highly associated with poor metastasis-free, recurrence-free, and overall survival outcomes in approximately 1000 patients [53]. Recently, LIFR was also reported to suppress breast cancer bone metastasis [54] and liver cancer lung metastasis [55]. Taken together, miR-9 targets multiple metastasis suppressors.
miR-182 is another miRNA that has been shown to promote metastasis in both genetically engineered mouse models and xenograft models. miR-182 transgenic mice displayed an increase in tumor cell dissemination and lung metastasis in a sarcoma model after amputation of the tumor-bearing limb, while deletion of this miRNA in the same model inhibited sarcoma metastasis to the lung [56]. Mechanistically, miR-182 directly targets Rsu1, Mtss1, Pai1, and Timp1 to promote migration, extracellular matrix degradation, and sarcoma metastasis [56]. Moreover, miR-182 promotes melanoma metastasis by targeting MITF and FOXO3 [57].
On the other hand, some miRNAs suppress metastasis by targeting pro-metastatic genes. By profiling miRNA expression in the highly metastatic sublines of the MDA-MB-231 human breast cancer cell line, Tavazoie et al. identified miR-206, miR-335, and miR-126 as anti-metastatic miRNAs [58]. Mechanistically, miR-206 directly targets NOTCH3, leading to induction of apoptosis and inhibition of migration [59]; miR-335 targets SOX4, TNC, and PTPRN2 to inhibit tumor cell migration and invasion [58]; and miR-126 suppresses metastasis by targeting MERTK, PITPNC1, IGFBP2, and CXCL12 (encoding SDF-1α) in cancer cells. Among these miR-126 targets, PITPNC1 upregulates the expression of IGFBP2; both the cleaved MERTK ectodomain and secreted IGFBP2 promote cell migration [60]; and SDF-1α induces the recruitment of mesenchymal stem cells and inflammatory monocytes to the primary tumor environment, leading to enhanced metastasis [61].
3.2 MicroRNAs regulate metastasis by modulating cancer stem cell properties
Cancer stem cells (CSCs) are the tumor-initiating cell population, and by definition, they are responsible for establishing primary and secondary tumors [62,63,64]. MiRNAs regulating CSC properties have been shown to play a role in metastasis. For example, the level of let-7 family members is downregulated in breast CSCs and is upregulated with differentiation. Silencing let-7 expression in non-CSCs promoted self-renewal, while restoring let-7 expression in breast CSCs suppressed tumorigenesis and metastasis [65]. Mechanistically, let-7 regulates CSC properties by targeting RAS, HMGA2, and BACH1 [65,66,67]. Similarly, miR-34a is downregulated in prostate CSCs. Ectopic expression of miR-34a in prostate CSCs inhibited metastasis, while silencing of miR-34a in prostate non-CSCs enhanced tumor growth and metastasis. These effects are mediated by CD44, which is a direct miR-34 target and a CSC marker of prostate and breast cancers [68]. Another example is miR-141, a member of the miR-200 family, which is underexpressed in prostate CSCs. Overexpression of miR-141 in prostate CSCs inhibited tumor regeneration and metastasis. The metastasis-suppressing effect of miR-141 is mediated through its multiple targets, including the Rho GTPase family members (CDC42, CDC42EP3, RAC1, and ARPC5) and stem cell factors (CD44 and EZH2) [69].
3.3 MicroRNAs regulate metastasis through epithelial-mesenchymal transition
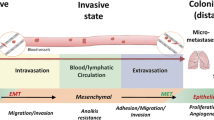
Induction of the epithelial-mesenchymal transition (EMT) in carcinoma cells facilitates tumor cell invasion and dissemination, whereas its reverse process, mesenchymal-epithelial transition (MET), promotes metastatic colonization in some cancer types [70, 71]. Certain metastamiRs regulate the EMT process. For example, miR-205 and the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141, and miR-429, with the same consensus seed sequence) have been shown to inhibit EMT and promote MET by targeting the EMT-inducing transcription factors ZEB1 and ZEB2 [72, 73]; conversely, ZEB1 directly represses the transcription of mir-200 and mir-205 genes [74,75,76] (Fig. 4). Functionally, on one hand, overexpression of miR-200 in lung cancer cells inhibited metastasis from primary tumors [77]; on the other hand, consistent with the concept that the MET process facilitates metastatic colonization, overexpression of miR-200 in the 4TO7 mouse mammary tumor cells promoted macroscopic metastasis formation in the lung and liver after these cells were injected intravenously into syngeneic mice [78, 79]. Therefore, the miR-200 miRNA family inhibits tumor invasion and metastatic dissemination at the primary sites, but enhances metastatic colonization at distant organs. Another EMT-suppressing miRNA is miR-29b, which targets several genes involved in differentiation and epithelial plasticity, including TGFB1, ITGA6, and ITGB1, and targets a network of pro-metastatic genes and microenvironmental genes, including VEGFA, ANGPTL4, LOX, MMP2, MMP9, and PDGF. Silencing miR-29 expression in breast cancer cells promoted EMT and metastasis [80].
Regulation and mechanism of action of Dicer in metastasis. Dicer’s positive regulators TAp63 and KDM6A/B suppress metastasis, and its negative regulators miR-103/107 and miR-630 promote metastasis. Downregulation of Dicer attenuates the biogenesis of miRNAs including miR-200, leading to derepression of ZEB1 and induction of EMT, invasion, and metastasis
In addition to EMT-inhibiting miRNAs, there also exist EMT-promoting miRNAs. For instance, by directly targeting the mRNA encoding E-cadherin, the pro-metastatic miRNA miR-9 promotes EMT, migration, and invasion of breast cancer cells [52]. Moreover, the miR-103/107 family targets Dicer and attenuates the biogenesis of miRNAs including miR-200, leading to induction of EMT and metastatic dissemination of otherwise non-aggressive breast cancer cells [81] (Fig. 4).
It should be noted that all EMT inducers are not functionally equal. By doing miRNA microarray analysis of mammary epithelial cells induced to undergo EMT by various transcription factors, Chen et al. identified a common set of EMT-associated miRNAs [82]: from this set, miR-205 and miR-200 are the most significantly downregulated miRNAs in EMT, while among the commonly upregulated miRNAs, miR-22 and miR-100 are capable of inducing EMT [82]. However, these two miRNAs have strikingly different functions. miR-22 targets the TET family of methylcytosine dioxygenases, leading to increased methylation of the mir-200 gene promoter and decreased expression of miR-200. Notably, in the MMTV-neu mouse model of breast cancer, transgenic overexpression of miR-22 activated EMT in the mammary tumor and induced metastasis [83]. In contrast, Chen et al. reported that miR-100 simultaneously induces EMT and inhibits tumorigenesis, migration, and invasion by targeting distinct genes [82]. This provided an example of an EMT inducer that suppresses cell movement and tumor invasion, indicating that EMT is not always associated with increased tumorigenicity, motility, and invasiveness, and that different EMT-inducing miRNAs have distinct functions depending on their specific target genes.
3.4 MicroRNAs regulate metastasis through the microenvironment
In addition to cell-autonomous effects of metastamiRs on cancer cells, miRNAs can also regulate metastasis by modulating niche cells in the tumor microenvironment. For example, miR-34a blocks osteoclastogenesis, resulting in inhibition of breast cancer bone metastasis [84]. MiR-34a knockout mice exhibited increased bone resorption and decreased bone mass, while osteoclast-specific miR-34 transgenic mice showed an opposite phenotype; moreover, ovariectomy-induced osteoporosis and bone metastasis (from breast or skin tumor cells) were lessened in these transgenic animals. Mechanistically, miR-34a directly targets transforming growth factor-β-induced factor 2 (Tgif2), a pro-osteoclastogenic protein [84] (Fig. 5). Another example is that miR-133a, miR-141, and miR-219 suppress Mitf expression in pre-osteoclasts and inhibit osteoclastogenesis, leading to reduced bone metastasis [85] (Fig. 5).
3.5 MicroRNAs regulate metastasis through exosomes
Over the past few years, tumor cell-secreted miRNAs, including exosome-transferred miRNAs, have emerged as a new mechanism mediating the tumor-stroma crosstalk and metastasis (Fig. 6a). Metastatic breast cancer cells secrete miR-10b-containing exosomes, which induce invasiveness of non-malignant mammary epithelial cells upon uptake by those cells [86] (Fig. 3). MiR-9 is secreted by tumor cells, which in turn promotes endothelial cell migration and angiogenesis through activation of the JAK-STAT signaling [87]. Thus, these two metastamiRs have a cell-non-autonomous effect in the tumor microenvironment. MiR-105 carried by exosomes is secreted by metastatic breast cancer cells. Once it enters adjacent endothelial cells, miR-105 targets the mRNA encoding the tight junction protein ZO-1 to remove the vascular endothelial barrier to metastasis. Ectopic expression of miR-105 in otherwise non-metastatic breast cancer cells induced vascular permeability and distant metastasis [88]. Similarly, exosome-mediated transfer of tumor cell-secreted miR-122 targets pyruvate kinase M2 (PKM2) in niche cells, leading to downregulation of GLUT1 expression and inhibition of glucose uptake; concurrently, the secretion of miR-122 reduces the level of this miRNA in cancer cells and promotes glucose uptake to support their growth. Both processes contribute to metastatic colonization [89].
MiRNAs regulate metastasis through exosome secretion. a Tumor cell-secreted exosomal miRNAs enter niche cells (e.g., endothelial cells) in the microenvironment to promote migration, invasion, and metastasis. b Niche cell-secreted exosomal miRNAs (e.g., astrocyte-derived exosomal miR-19a) enter metastasized tumor cells to promote metastatic outgrowth
MiRNA-containing exosomes secreted by the microenvironment, but not by tumor cells, also play an important role in metastasis. Breast cancer cells metastasized to the brain, but not to other organs, lose PTEN expression, which is restored if tumor cells leave the brain microenvironment [90]. This phenotype is caused by miR-19a, a PTEN-targeting miRNA from astrocyte-derived exosomes. The adaptive PTEN loss in brain-metastatic tumor cells activates NF-κB and AKT signaling to promote cell proliferation and inhibit apoptosis. In addition, loss of PTEN also leads to increased secretion of the chemokine CCL2, which in turn recruits IBA1-expressing myeloid cells to facilitate metastatic outgrowth of tumor cells in the brain [90] (Fig. 6b).
3.6 MicroRNA biogenesis and metastasis
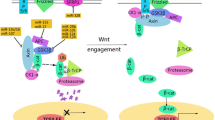
While a growing body of evidence demonstrated that specific miRNAs promote or suppress tumor metastasis, recent studies have indicated that the regulators of miRNA biogenesis also play functional roles in metastatic progression. The two key enzymes in miRNA processing, Drosha and Dicer, are downregulated in cancer, and low expression levels of these two proteins correlate with poor clinical outcomes [91, 92]. Interestingly, regulators of Dicer expression are involved in tumor metastasis, probably through modulation of metastasis-suppressing miRNAs such as miR-200 family members (Fig. 4). For instance, the miRNAs that directly target Dicer, miR-103/107 and miR-630, have been shown to promote metastasis [81, 93]. TAp63, a p53 family member, binds to the Dicer promoter and activates its expression [94]. Dicer is underexpressed in metastatic human tumors deficient in TAp63, and deletion of TAp63 in mice reduced Dicer levels in tumors and induced formation of metastases [94]. In addition, hypoxia can downregulate Dicer expression through inhibition of oxygen-dependent H3K27me3 demethylases KDM6A/B, leading to epigenetic silencing of the Dicer promoter. Subsequently, reduced miRNA biogenesis results in downregulation of miR-200 and derepression of the miR-200 target ZEB1 [95]. Whether Dicer functionally mediates the role of hypoxia in metastasis remains to be determined. AGO2 (Argonaute 2) is another miRNA biogenesis protein that may be implicated in metastasis. In response to hypoxia, AGO2 is phosphorylated by epidermal growth factor receptor (EGFR) at the Y393 residue in breast cancer cells, leading to decreased binding of AGO2 to Dicer. This mechanism has been shown to mediate EGFR-induced cancer cell survival and invasiveness [96].
4 Concluding remarks
Because of the incomplete understanding of metastasis and the lack of practical clinical trials, metastatic disease remains largely incurable. Over the past decade, significant progress has been made in understanding the functions of specific miRNAs in metastasis. Some of these metastamiRs represent attractive therapeutic targets for cancer treatment. It should be noted that the functions of miRNAs are often context-dependent and tissue type-specific. Moreover, a single miRNA (or miRNA cluster/family) may play a dual role in the invasion-metastasis cascade. As mentioned above, miR-200 family members suppress EMT, tumor invasion, and dissemination at the primary sites but promote colonization at distant anatomic sites. Therefore, the therapeutic potential of miRNA-based agents should be carefully examined in clinically relevant models and settings, before they enter clinical trials. In addition, the role and mechanisms of action of miRNA biogenesis in metastasis warrant further investigation.
Recently, the crosstalk between miRNAs and other RNAs has emerged as an intriguing research topic. In particular, competing endogenous RNAs (ceRNAs) are defined as RNAs that compete with each other through common miRNA recognition elements, including mRNAs, lncRNAs, circular RNAs, and RNAs produced from pseudogenes [97]. It has been proposed that not all miRNAs are prone to ceRNA competition and that the ceRNA effect is significant only under specific conditions, such as a low miRNA:target ratio and a high-affinity ceRNA [98, 99]. Nevertheless, functional studies have indicated that ceRNAs can regulate oncogenes and tumor suppressor genes through miRNAs. For example, PTEN expression levels can be upregulated by RNAs that share miRNA binding sites with PTEN, including the PTEN pseudogene PTENP1 [100,101,102]. It would be interesting to investigate whether ceRNAs play a role in tumor metastasis by acting as molecular decoys for miRNAs.
References
Bartel, D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297.
Winter, J., Jung, S., Keller, S., Gregory, R. I., & Diederichs, S. (2009). Many roads to maturity: microRNA biogenesis pathways and their regulation. Nature Cell Biology, 11(3), 228–234.
Gregory, R. I., Yan, K. P., Amuthan, G., Chendrimada, T., Doratotaj, B., Cooch, N., et al. (2004). The microprocessor complex mediates the genesis of microRNAs. Nature, 432(7014), 235–240.
Han, J., Lee, Y., Yeom, K. H., Kim, Y. K., Jin, H., & Kim, V. N. (2004). The Drosha-DGCR8 complex in primary microRNA processing. Genes & Development, 18(24), 3016–3027.
Han, J., Lee, Y., Yeom, K. H., Nam, J. W., Heo, I., Rhee, J. K., et al. (2006). Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell, 125(5), 887–901.
Lund, E., Guttinger, S., Calado, A., Dahlberg, J. E., & Kutay, U. (2004). Nuclear export of microRNA precursors. Science, 303(5654), 95–98.
Yi, R., Qin, Y., Macara, I. G., & Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Development, 17(24), 3011–3016.
Bohnsack, M. T., Czaplinski, K., & Gorlich, D. (2004). Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA, 10(2), 185–191.
Ha, M., & Kim, V. N. (2014). Regulation of microRNA biogenesis. Nature Reviews. Molecular Cell Biology, 15(8), 509–524.
Lin, S., & Gregory, R. I. (2015). MicroRNA biogenesis pathways in cancer. Nature Reviews. Cancer, 15(6), 321–333.
Eichhorn, S. W., Guo, H., McGeary, S. E., Rodriguez-Mias, R. A., Shin, C., Baek, D., et al. (2014). mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Molecular Cell, 56(1), 104–115.
Guo, H., Ingolia, N. T., Weissman, J. S., & Bartel, D. P. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature, 466(7308), 835–840.
Ambros, V. (2004). The functions of animal microRNAs. Nature, 431(7006), 350–355.
Massart, J., Katayama, M., & Krook, A. (2016). Micromanaging glucose and lipid metabolism in skeletal muscle: role of microRNAs. Biochimica et Biophysica Acta, 1861(12 Pt B), 2130–2138.
Rupaimoole, R., & Slack, F. J. (2017). MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nature Reviews. Drug Discovery, 16(3), 203–222.
Ma, L. (2016). MicroRNA and metastasis. Advances in Cancer Research, 132, 165–207.
Pencheva, N., & Tavazoie, S. F. (2013). Control of metastatic progression by microRNA regulatory networks. Nature Cell Biology, 15(6), 546–554.
Trobaugh, D. W., & Klimstra, W. B. (2017). MicroRNA regulation of RNA virus replication and pathogenesis. Trends in Molecular Medicine, 23(1), 80–93.
Xiao, C., & Rajewsky, K. (2009). MicroRNA control in the immune system: basic principles. Cell, 136(1), 26–36.
Ling, H., Fabbri, M., & Calin, G. A. (2013). MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nature Reviews. Drug Discovery, 12(11), 847–865.
Calin, G. A., Dumitru, C. D., Shimizu, M., Bichi, R., Zupo, S., Noch, E., et al. (2002). Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proceedings of the National Academy of Sciences of the United States of America, 99(24), 15524–15529.
Klein, U., Lia, M., Crespo, M., Siegel, R., Shen, Q., Mo, T., et al. (2010). The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell, 17(1), 28–40.
Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., et al. (2005). MicroRNA expression profiles classify human cancers. Nature, 435(7043), 834–838.
He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature, 435(7043), 828–833.
Xiao, C., Srinivasan, L., Calado, D. P., Patterson, H. C., Zhang, B., Wang, J., et al. (2008). Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nature Immunology, 9(4), 405–414.
Krichevsky, A. M., & Gabriely, G. (2009). MiR-21: a small multi-faceted RNA. Journal of Cellular and Molecular Medicine, 13(1), 39–53.
Hatley, M. E., Patrick, D. M., Garcia, M. R., Richardson, J. A., Bassel-Duby, R., van Rooij, E., et al. (2010). Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell, 18(3), 282–293.
Medina, P. P., Nolde, M., & Slack, F. J. (2010). OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature, 467(7311), 86–90.
Pasquinelli, A. E., Reinhart, B. J., Slack, F., Martindale, M. Q., Kuroda, M. I., Maller, B., et al. (2000). Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature, 408(6808), 86–89.
Balzeau, J., Menezes, M. R., Cao, S., & Hagan, J. P. (2017). The LIN28/let-7 pathway in cancer. Frontiers in Genetics, 8, 31.
Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., et al. (2005). RAS is regulated by the let-7 microRNA family. Cell, 120(5), 635–647.
Esquela-Kerscher, A., Trang, P., Wiggins, J. F., Patrawala, L., Cheng, A., Ford, L., et al. (2008). The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle, 7(6), 759–764.
He, X., He, L., & Hannon, G. J. (2007). The guardian’s little helper: microRNAs in the p53 tumor suppressor network. Cancer Research, 67(23), 11099–11101.
He, L., He, X., Lim, L. P., de Stanchina, E., Xuan, Z., Liang, Y., et al. (2007). A microRNA component of the p53 tumour suppressor network. Nature, 447(7148), 1130–1134.
Concepcion, C. P., Han, Y. C., Mu, P., Bonetti, C., Yao, E., D'Andrea, A., et al. (2012). Intact p53-dependent responses in miR-34-deficient mice. PLoS Genetics, 8(7), e1002797.
Cheng, C. Y., Hwang, C. I., Corney, D. C., Flesken-Nikitin, A., Jiang, L., Oner, G. M., et al. (2014). MiR-34 cooperates with p53 in suppression of prostate cancer by joint regulation of stem cell compartment. Cell Reports, 6(6), 1000–1007.
Brabletz, T., Lyden, D., Steeg, P. S., & Werb, Z. (2013). Roadblocks to translational advances on metastasis research. Nature Medicine, 19(9), 1104–1109.
Wan, L., Pantel, K., & Kang, Y. (2013). Tumor metastasis: moving new biological insights into the clinic. Nature Medicine, 19(11), 1450–1464.
Eccles, S. A., & Welch, D. R. (2007). Metastasis: recent discoveries and novel treatment strategies. Lancet, 369(9574), 1742–1757.
Talmadge, J. E., & Fidler, I. J. (2010). AACR centennial series: the biology of cancer metastasis: historical perspective. Cancer Research, 70(14), 5649–5669.
Steeg, P. S. (2012). Perspective: the right trials. Nature, 485(7400), S58–S59.
Sun, Y., & Ma, L. (2015). The emerging molecular machinery and therapeutic targets of metastasis. Trends in Pharmacological Sciences, 36(6), 349–359.
Ma, L., Teruya-Feldstein, J., & Weinberg, R. A. (2007). Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature, 449(7163), 682–688.
Ma, L., Reinhardt, F., Pan, E., Soutschek, J., Bhat, B., Marcusson, E. G., et al. (2010). Therapeutic silencing of miR-10b inhibits metastasis in a mouse mammary tumor model. Nature Biotechnology, 28(4), 341–347.
Ma, L. (2010). Role of miR-10b in breast cancer metastasis. Breast Cancer Research, 12(5), 210.
Yoo, B., Kavishwar, A., Ross, A., Wang, P., Tabassum, D. P., Polyak, K., et al. (2015). Combining miR-10b-targeted nanotherapy with low-dose doxorubicin elicits durable regressions of metastatic breast cancer. Cancer Research, 75(20), 4407–4415.
Yoo, B., Kavishwar, A., Wang, P., Ross, A., Pantazopoulos, P., Dudley, M., et al. (2017). Therapy targeted to the metastatic niche is effective in a model of stage IV breast cancer. Scientific Reports, 7, 45060.
Kim, J., Siverly, A. N., Chen, D., Wang, M., Yuan, Y., Wang, Y., et al. (2016). Ablation of miR-10b suppresses oncogene-induced mammary tumorigenesis and metastasis and reactivates tumor-suppressive pathways. Cancer Research, 76(21), 6424–6435.
Myers, C., Charboneau, A., Cheung, I., Hanks, D., & Boudreau, N. (2002). Sustained expression of homeobox D10 inhibits angiogenesis. The American Journal of Pathology, 161(6), 2099–2109.
Chai, G., Liu, N., Ma, J., Li, H., Oblinger, J. L., Prahalad, A. K., et al. (2010). MicroRNA-10b regulates tumorigenesis in neurofibromatosis type 1. Cancer Science, 101(9), 1997–2004.
Tian, Y., Luo, A., Cai, Y., Su, Q., Ding, F., Chen, H., et al. (2010). MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. The Journal of Biological Chemistry, 285(11), 7986–7994.
Ma, L., Young, J., Prabhala, H., Pan, E., Mestdagh, P., Muth, D., et al. (2010). MiR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nature Cell Biology, 12(3), 247–256.
Chen, D., Sun, Y., Wei, Y., Zhang, P., Rezaeian, A. H., Teruya-Feldstein, J., et al. (2012). LIFR is a breast cancer metastasis suppressor upstream of the Hippo-YAP pathway and a prognostic marker. Nature Medicine, 18(10), 1511–1517.
Johnson, R. W., Finger, E. C., Olcina, M. M., Vilalta, M., Aguilera, T., Miao, Y., et al. (2016). Induction of LIFR confers a dormancy phenotype in breast cancer cells disseminated to the bone marrow. Nature Cell Biology, 18(10), 1078–1089.
Luo, Q., Wang, C., Jin, G., Gu, D., Wang, N., Song, J., et al. (2015). LIFR functions as a metastasis suppressor in hepatocellular carcinoma by negatively regulating phosphoinositide 3-kinase/AKT pathway. Carcinogenesis, 36(10), 1201–1212.
Sachdeva, M., Mito, J. K., Lee, C. L., Zhang, M., Li, Z., Dodd, R. D., et al. (2014). MicroRNA-182 drives metastasis of primary sarcomas by targeting multiple genes. The Journal of Clinical Investigation, 124(10), 4305–4319.
Segura, M. F., Hanniford, D., Menendez, S., Reavie, L., Zou, X., Alvarez-Diaz, S., et al. (2009). Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proceedings of the National Academy of Sciences of the United States of America, 106(6), 1814–1819.
Tavazoie, S. F., Alarcon, C., Oskarsson, T., Padua, D., Wang, Q., Bos, P. D., et al. (2008). Endogenous human microRNAs that suppress breast cancer metastasis. Nature, 451(7175), 147–152.
Song, G., Zhang, Y., & Wang, L. (2009). MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. The Journal of Biological Chemistry, 284(46), 31921–31927.
Png, K. J., Halberg, N., Yoshida, M., & Tavazoie, S. F. (2011). A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature, 481(7380), 190–194.
Zhang, Y., Yang, P., Sun, T., Li, D., Xu, X., Rui, Y., et al. (2013). MiR-126 and miR-126* repress recruitment of mesenchymal stem cells and inflammatory monocytes to inhibit breast cancer metastasis. Nature Cell Biology, 15(3), 284–294.
Liu, H., Patel, M. R., Prescher, J. A., Patsialou, A., Qian, D., Lin, J., et al. (2010). Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proceedings of the National Academy of Sciences of the United States of America, 107(42), 18115–18120.
Malanchi, I., Santamaria-Martinez, A., Susanto, E., Peng, H., Lehr, H. A., Delaloye, J. F., et al. (2011). Interactions between cancer stem cells and their niche govern metastatic colonization. Nature, 481(7379), 85–89.
Al-Hajj, M., Wicha, M. S., Benito-Hernandez, A., Morrison, S. J., & Clarke, M. F. (2003). Prospective identification of tumorigenic breast cancer cells. Proceedings of the National Academy of Sciences of the United States of America, 100(7), 3983–3988.
Yu, F., Yao, H., Zhu, P., Zhang, X., Pan, Q., Gong, C., et al. (2007). Let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell, 131(6), 1109–1123.
Dangi-Garimella, S., Yun, J., Eves, E. M., Newman, M., Erkeland, S. J., Hammond, S. M., et al. (2009). Raf kinase inhibitory protein suppresses a metastasis signalling cascade involving LIN28 and let-7. The EMBO Journal, 28(4), 347–358.
Yun, J., Frankenberger, C. A., Kuo, W. L., Boelens, M. C., Eves, E. M., Cheng, N., et al. (2011). Signalling pathway for RKIP and Let-7 regulates and predicts metastatic breast cancer. The EMBO Journal, 30(21), 4500–4514.
Liu, C., Kelnar, K., Liu, B., Chen, X., Calhoun-Davis, T., Li, H., et al. (2011). The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Medicine, 17(2), 211–215.
Liu, C., Liu, R., Zhang, D., Deng, Q., Liu, B., Chao, H. P., et al. (2017). MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nature Communications, 8, 14270.
Tsai, J. H., Donaher, J. L., Murphy, D. A., Chau, S., & Yang, J. (2012). Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell, 22(6), 725–736.
Tsai, J. H., & Yang, J. (2013). Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes & Development, 27(20), 2192–2206.
Gregory, P. A., Bert, A. G., Paterson, E. L., Barry, S. C., Tsykin, A., Farshid, G., et al. (2008). The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature Cell Biology, 10(5), 593–601.
Park, S. M., Gaur, A. B., Lengyel, E., & Peter, M. E. (2008). The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes & Development, 22(7), 894–907.
Burk, U., Schubert, J., Wellner, U., Schmalhofer, O., Vincan, E., Spaderna, S., et al. (2008). A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Reports, 9(6), 582–589.
Gregory, P. A., Bracken, C. P., Smith, E., Bert, A. G., Wright, J. A., Roslan, S., et al. (2011). An autocrine TGF-beta/ZEB/miR-200 signaling network regulates establishment and maintenance of epithelial-mesenchymal transition. Molecular Biology of the Cell, 22(10), 1686–1698.
Zhang, P., Wang, L., Rodriguez-Aguayo, C., Yuan, Y., Debeb, B. G., Chen, D., et al. (2014). MiR-205 acts as a tumour radiosensitizer by targeting ZEB1 and Ubc13. Nature Communications, 5, 5671.
Gibbons, D. L., Lin, W., Creighton, C. J., Rizvi, Z. H., Gregory, P. A., Goodall, G. J., et al. (2009). Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes & Development, 23(18), 2140–2151.
Dykxhoorn, D. M., Wu, Y., Xie, H., Yu, F., Lal, A., Petrocca, F., et al. (2009). MiR-200 enhances mouse breast cancer cell colonization to form distant metastases. PLoS One, 4(9), e7181.
Korpal, M., Ell, B. J., Buffa, F. M., Ibrahim, T., Blanco, M. A., Celia-Terrassa, T., et al. (2011). Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nature Medicine, 17(9), 1101–1108.
Chou, J., Lin, J. H., Brenot, A., Kim, J. W., Provot, S., & Werb, Z. (2013). GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nature Cell Biology, 15(2), 201–213.
Martello, G., Rosato, A., Ferrari, F., Manfrin, A., Cordenonsi, M., Dupont, S., et al. (2010). A microRNA targeting dicer for metastasis control. Cell, 141(7), 1195–1207.
Chen, D., Sun, Y., Yuan, Y., Han, Z., Zhang, P., Zhang, J., et al. (2014). MiR-100 induces epithelial-mesenchymal transition but suppresses tumorigenesis, migration and invasion. PLoS Genetics, 10(2), e1004177.
Song, S. J., Poliseno, L., Song, M. S., Ala, U., Webster, K., Ng, C., et al. (2013). MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell, 154(2), 311–324.
Krzeszinski, J. Y., Wei, W., Huynh, H., Jin, Z., Wang, X., Chang, T. C., et al. (2014). MiR-34a blocks osteoporosis and bone metastasis by inhibiting osteoclastogenesis and Tgif2. Nature, 512(7515), 431–435.
Ell, B., Mercatali, L., Ibrahim, T., Campbell, N., Schwarzenbach, H., Pantel, K., et al. (2013). Tumor-induced osteoclast miRNA changes as regulators and biomarkers of osteolytic bone metastasis. Cancer Cell, 24(4), 542–556.
Singh, R., Pochampally, R., Watabe, K., Lu, Z., & Mo, Y. Y. (2014). Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Molecular Cancer, 13, 256.
Zhuang, G., Wu, X., Jiang, Z., Kasman, I., Yao, J., Guan, Y., et al. (2012). Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. The EMBO Journal, 31(17), 3513–3523.
Zhou, W., Fong, M. Y., Min, Y., Somlo, G., Liu, L., Palomares, M. R., et al. (2014). Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell, 25(4), 501–515.
Fong, M. Y., Zhou, W., Liu, L., Alontaga, A. Y., Chandra, M., Ashby, J., et al. (2015). Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nature Cell Biology, 17(2), 183–194.
Zhang, L., Zhang, S., Yao, J., Lowery, F. J., Zhang, Q., Huang, W. C., et al. (2015). Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature, 527(7576), 100–104.
Merritt, W. M., Lin, Y. G., Han, L. Y., Kamat, A. A., Spannuth, W. A., Schmandt, R., et al. (2008). Dicer, Drosha, and outcomes in patients with ovarian cancer. The New England Journal of Medicine, 359(25), 2641–2650.
Torres, A., Torres, K., Paszkowski, T., Jodlowska-Jedrych, B., Radomanski, T., Ksiazek, A., et al. (2011). Major regulators of microRNAs biogenesis Dicer and Drosha are down-regulated in endometrial cancer. Tumour Biology, 32(4), 769–776.
Rupaimoole, R., Ivan, C., Yang, D., Gharpure, K. M., Wu, S. Y., Pecot, C. V., et al. (2016). Hypoxia-upregulated microRNA-630 targets Dicer, leading to increased tumor progression. Oncogene, 35(33), 4312–4320.
Su, X., Chakravarti, D., Cho, M. S., Liu, L., Gi, Y. J., Lin, Y. L., et al. (2010). TAp63 suppresses metastasis through coordinate regulation of Dicer and miRNAs. Nature, 467(7318), 986–990.
van den Beucken, T., Koch, E., Chu, K., Rupaimoole, R., Prickaerts, P., Adriaens, M., et al. (2014). Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nature Communications, 5, 5203.
Shen, J., Xia, W., Khotskaya, Y. B., Huo, L., Nakanishi, K., Lim, S. O., et al. (2013). EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature, 497(7449), 383–387.
Salmena, L., Poliseno, L., Tay, Y., Kats, L., & Pandolfi, P. P. (2011). A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell, 146(3), 353–358.
Bosson, A. D., Zamudio, J. R., & Sharp, P. A. (2014). Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Molecular Cell, 56(3), 347–359.
Denzler, R., Agarwal, V., Stefano, J., Bartel, D. P., & Stoffel, M. (2014). Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Molecular Cell, 54(5), 766–776.
Poliseno, L., Salmena, L., Zhang, J., Carver, B., Haveman, W. J., & Pandolfi, P. P. (2010). A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature, 465(7301), 1033–1038.
Karreth, F. A., Tay, Y., Perna, D., Ala, U., Tan, S. M., Rust, A. G., et al. (2011). In vivo identification of tumor-suppressive PTEN ceRNAs in an oncogenic BRAF-induced mouse model of melanoma. Cell, 147(2), 382–395.
Tay, Y., Kats, L., Salmena, L., Weiss, D., Tan, S. M., Ala, U., et al. (2011). Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell, 147(2), 344–357.
Acknowledgements
We thank Baochau Ton for critical reading of the manuscript. The authors’ research is supported by the US National Institutes of Health grants R01CA166051 and R01CA181029, a Cancer Prevention and Research Institute of Texas grant RP150319, and a Stand Up To Cancer Innovative Research Grant.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Kim, J., Yao, F., Xiao, Z. et al. MicroRNAs and metastasis: small RNAs play big roles. Cancer Metastasis Rev 37, 5–15 (2018). https://doi.org/10.1007/s10555-017-9712-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10555-017-9712-y