Abstract
Purpose
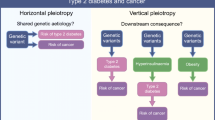
Type 2 diabetes (T2D) has been reported to be associated with an elevated risk of breast cancer. It is unclear, however, whether this association is due to shared genetic factors.
Methods
We constructed a genetic risk score (GRS) using risk variants from 33 known independent T2D susceptibility loci and evaluated its relation to breast cancer risk using the data from two consortia, including 62,328 breast cancer patients and 83,817 controls of European ancestry. Unconditional logistic regression models were used to derive adjusted odds ratios (ORs) and 95 % confidence intervals (CIs) to measure the association of breast cancer risk with T2D GRS or T2D-associated genetic risk variants. Meta-analyses were conducted to obtain summary ORs across all studies.
Results
The T2D GRS was not found to be associated with breast cancer risk, overall, by menopausal status, or for estrogen receptor positive or negative breast cancer. Three T2D associated risk variants were individually associated with breast cancer risk after adjustment for multiple comparisons using the Bonferroni method (at p < 0.001), rs9939609 (FTO) (OR 0.94, 95 % CI = 0.92–0.95, p = 4.13E−13), rs7903146 (TCF7L2) (OR 1.04, 95 % CI = 1.02–1.06, p = 1.26E−05), and rs8042680 (PRC1) (OR 0.97, 95 % CI = 0.95–0.99, p = 8.05E−04).
Conclusions
We have shown that several genetic risk variants were associated with the risk of both T2D and breast cancer. However, overall genetic susceptibility to T2D may not be related to breast cancer risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Globally, approximately 382 million people currently live with diabetes, and this number may rise to 592 million by 2035 [1]. Type 2 diabetes (T2D) accounts for over 90 % of all diabetes cases [2]. Breast cancer is the most common cancer among women in many countries, including the USA [3]. Many epidemiological studies have linked T2D to increased breast cancer risk [4–8]. Recent meta-analyses have shown a more than 20 % increase in risk of breast cancer among women with T2D compared to women without the disease [9–12]. T2D and breast cancer share some risk factors, including obesity in postmenopausal women and physical inactivity [13]. Elevated levels of circulating C-peptide and insulin-like growth factor-1, biomarkers related to insulin resistance, have also been associated with increased breast cancer risk [14, 15]. It remains unclear, however, whether the link between these two diseases is due to shared lifestyle risk factors or intrinsic etiology such as genetic susceptibility. Understanding how genetic variants related to T2D risk influence breast cancer risk may provide insights into the nature of the T2D–breast cancer relationship.
Recent genome-wide association studies (GWASs) have identified approximately 50 genetic variants associated with T2D risk. Some of these reported T2D-related genetic variants have been studied in relation to the risk of several cancers, including cancers of the pancreas [16], colon/rectum [17, 18], and prostate [19]. The influence of these variants on breast cancer risk, however, has not been adequately studied. To date, only two studies have evaluated the association of a subset of these T2D-related genetic variants with breast cancer risk [20, 21]. Both studies reported a null association, which may be due to small study size and low study power.
In this analysis, using data from two consortia including 62,328 breast cancer cases and 83,817 controls of women of European ancestry, we evaluated T2D-related genetic variants reported to date in relation to breast cancer risk. By constructing a T2D-related genetic risk score (T2D GRS) and evaluating its association with breast cancer risk, we tested the hypothesis that, overall, the alleles that increase T2D risk may also increase breast cancer risk. We also tested the hypothesis that certain T2D-related genetic variants may be associated with breast cancer risk.
Methods
Study population
Included in this analysis were 62,328 breast cancer cases and 83,817 controls of women of European ancestry recruited either in the 39 studies (Online Resource Table 1) that participated in the Breast Cancer Association Consortium (BCAC), a part of the Collaborative Oncological Gene-Environment Study (COGS), or in the 11 studies (Online Resource Table 2) that are included in the Discovery, Biology, and Risk of Inherited Variants in Breast Cancer (DRIVE) project of Genetic Associations and Mechanism in Oncology (GAME-ON). From the BCAC, we included individual-level data for 46,325 breast cancer cases and 42,482 controls. The DRIVE project included 16,003 breast cases and 41,335 controls; however, only summary statistics for the association between T2D-related risk variants and breast cancer risk were available, and thus, these summary statistics were used in our study. The study samples and participant data, including demographics and the traditional risk factors for breast cancer, were collected in each contributing study.
Single-nucleotide polymorphism (SNP) selection
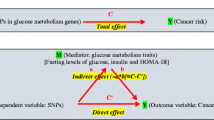
We searched for all reported genetic risk variants associated with T2D in European ancestry populations at a genome-wide significance level (p < 5 × 10−8, trait ‘Type 2 diabetes’ or ‘Type 2 diabetes and other traits’) using the US National Human Genome Resource Institute (NHGRI) Catalog of Published Genome-Wide Association Studies (GWASs Catalog, accessed 19 November 2012, at http://www.genome.gov/gwastudies). Fifty SNPs representing 33 independent loci (linkage disequilibrium (LD) R 2 < 0.1) were identified (Fig. 1).
Genetic risk score construction
The genetic risk scores were calculated in 46,325 cases and 42,482 controls included in the BCAC. At each of the 33 independent loci, we selected the SNP with the lowest p value for association with T2D reported in GWASs to represent the locus in constructing the T2D GRS. Using these 33 SNPs, a weighted T2D GRS was constructed as a measure of the overall association of genetic risk variants with T2D. In the BCAC, 11 SNPs were directly genotyped and 22 were imputed with imputation quality threshold of R 2 > 0.5. The T2D GRS was created as \(\sum\nolimits_{i}^{33} {w_{i} {\text{SNP}}_{i} }\), where \(w_{i}\) is the logarithm of the odds ratio (OR) of the ith SNP with T2D reported from previous GWAS and \({\text{SNP}}_{i}\) is the number of risk alleles carried by a given subject on the ith SNP. We hypothesized that the risk allele for T2D would be associated with increased risk of breast cancer. The 33 individual T2D risk variants identified from the NHGRI GWAS catalog are presented in Online Resource Table 3.
Genotyping
In the BCAC, genotype data were obtained either from direct genotyping with a custom Illumina iSelect genotyping array (iCOGS) that contains 211,155 SNPs [22] or from imputation with the 1,000 Genomes Project Phase I integrated variant set (version 3, March 2012 release) as the Ref. [23], using the program IMPUTE2 [24]. Details of the studies that participated in the BCAC and the methodology used by the BCAC and iCOGS have been published elsewhere [22] and can also be found on the iCOGS Web site (http://ccge.medschl.cam.ac.uk/research/consortia/icogs/).
In the DRIVE project, genotype data were obtained either from direct genotyping using Illumina or Affymetrix arrays (Online Resource Table 2) or from imputation with the HapMap version 2 CEU panel (Utah residents of Northern and Western European ancestry) as a reference, using the program MACH v1.0 or IMPUTE [24]. Details of the studies that participated in DRIVE were described in previously published papers [22, 25–28] or on the GAME-ON Web site (http://gameon.dfci.harvard.edu).
Statistical analysis
We evaluated the association between the T2D GRS and breast cancer risk using individual-level data from 46,325 breast cancer cases and 42,482 controls of European ancestry who participated in BCAC studies. Demographic characteristics and known breast cancer risk factors were summarized by case–control status using mean and standard deviation (SD) for continuous variables or frequency with percentage for categorical variables. Differences between cases and controls were compared using the Wilcoxon rank-sum test (continuous variables) or the Chi-square test (categorical variables). To assess the association between the T2D GRS and breast cancer risk factors, we used control data and calculated the mean and SD of the T2D GRS by comparison groups for each categorical variable; the difference was tested by the Wilcoxon rank-sum test. For continuous variables, the Pearson’s correlations were measured. To account for potential population stratification within our study population, genetic ancestry was estimated by principal component (PC) analysis using EIGENSTRAT software [29] on 37,000 uncorrelated SNPs (including those selected as ancestry informative markers) on the chip. The mean value of the genomic inflation factor (λ) was 1.01 for the participating studies when PCs were included in the regression models, indicating little evidence of population stratification [22]. For all analyses, the top eight PCs were included in all regression models. For the LMBC study, the study-specific principal component was further adjusted. To assess the association between the T2D GRS and breast cancer risk, we first fitted unconditional logistic regression models adjusting for age and PCs within each of the 39 contributing studies individually and recorded the β coefficients with standard errors for T2D GRS quintiles (relative to the first quintile). We then conducted a meta-analysis on the results from these 39 studies using both fixed-effect and mixed-effect models. The odds ratios (ORs) with 95 % confidence intervals (CIs) from the fixed-effects model are reported in Table 1, as are further analyses by estrogen receptor (ER) status, menopausal status, age group (<50 vs. ≥50 years), and body mass index (BMI, <25 vs. ≥25 kg/m2).
We also used the SNP-set Kernel Association Test (SKAT) to evaluate whether any SNP in the T2D-associated SNP set may be related to breast cancer risk without making the assumption that the alleles that increase T2D risk may also increase breast cancer risk [30]. To evaluate the association of each individual SNP (per copy of risk allele) with breast cancer risk, we used individual-level data from the BCAC (46,325 cases and 42,482 controls) and summary results data from DRIVE (16,003 cases and 41,335 controls). We first estimated allelic OR for each SNP for each BCAC study with adjustment similar to that in the analyses for the association of T2D GRS with breast cancer risk described above and then combined the results across all BCAC studies with results from DRIVE using the inverse-variance meta-analysis with a fixed-effect model. Both consortium-specific results and combined results are reported in Table 2. For individual SNP analyses, statistical significance was considered after adjusting for multiple comparisons using the Bonferroni method (0.05/33). For all other analyses, statistical significance was considered at a two-sided 5 % level unless stated otherwise. All analyses were conducted using R version 3.0.3 [31].
Results
Among the 88,807 BCAC participants studied, on average, cases were slightly older than controls (57.8 vs. 54.9 years, p < 0.001) and entered menopause at a younger age (48.5 vs. 48.7 years, p < 0.01), as shown in Online Resource Table 4. More cases than controls were postmenopausal (69.3 vs. 68.1 %, p < 0.01) or had a first-degree family history of breast cancer (27.7 vs. 11.2 %, p < 0.01). Among postmenopausal women, cases and controls had comparable BMI (p = 0.62). Among controls, the T2D GRS was positively correlated with BMI (postmenopausal women, Pearson r = 0.018, p = 0.03) and inversely correlated with age at menarche (Pearson r = −0.021, p < 0.01). For other categorical variables examined, the mean T2D GRS values were virtually identical across different statuses (Online Resource Table 4, right columns).
Overall, the T2D GRS was not found to be associated with breast cancer risk (p for trend = 0.69, Table 1). No significant results were observed in analyses stratified by ER status (p for trend = 0.74 and 0.47 for ER+ and ER− breast cancer, respectively), menopausal status (p for trend = 0.74 and 0.93 for premenopausal and postmenopausal women, respectively), age group (p for trend = 0.74 and 0.62 for age <50 and age ≥50 years, respectively), or BMI group (p for trend = 0.64 and 0.64 for BMI < 25 and BMI ≥ 25, respectively). Meta-analysis using mixed-effect models gave similar results (data not shown). In a sensitivity analysis, which included only the 11 directly genotyped SNPs and 14 imputed SNPs with imputation R 2 > 0.9, similar results were observed (Online Resource Table 5).
Using SKATs and without making the assumption that the alleles that increase T2D risk also increase breast cancer risk, we found evidence for potential association for some of the T2D-related SNPs with breast cancer risk (p = 3.95E−10). Of the 33 independent SNPs investigated, seven were nominally associated with breast cancer risk using BCAC data alone (Table 2). Of these, the risk allele for T2D in four SNPs was associated with a reduced risk of breast cancer. After adjusting for multiple comparisons, the association for two SNPs, rs7903146 (TCF7L2, OR 1.04, 95 % CI = 1.02–1.07, p = 1.20E−04) and rs9939609 (FTO, OR 0.93, 95 % CI = 0.91–0.95, p = 3.63E−12), remained statistically significant, and both associations were replicated in DRIVE. SNP rs8042680 (PRC1) was related to breast cancer risk in the BCAC at p = 0.02 and in DRIVE at p = 6.18E−3; meta-analyses of these data yielded a significant association after adjusting for multiple comparisons (OR 0.97, 95 % CI = 0.99–0.99, p = 8.05E−4).
Discussion
In this large study, we investigated the association of 33 independent T2D-related genetic variants with breast cancer risk individually and in combination (through the use of our GRS). Generally, we found no association between T2D GRS and risk of breast cancer overall or by ER status. Of the 33 T2D-associated SNPs investigated in this study, three showed a significant association with breast cancer risk after adjusting for multiple comparisons: rs9939609 (FTO), rs7903146 (TCF7L2), and rs8042680 (PRC1). Although this study does not provide any evidence for an overall association of T2D susceptibility and breast cancer risk, it does show that some T2D-associated SNPs may be related to breast cancer risk.
It has been hypothesized that the association between T2D and breast cancer may be mediated through insulin resistance and hyperinsulinemia [32]. T2D and breast cancer share some lifestyle risk factors, including obesity in postmenopausal women and physical inactivity. Indeed, it has been shown previously that the observed association between these two diseases may be, in part, due to residual confounding by BMI [33]. With a very large sample size, our study suggests that overall genetic susceptibility to T2D was not related to breast cancer risk, indicating that the previously observed association between T2D and breast cancer risk may be largely due to shared lifestyle risk factors. Our finding for a null association between T2D GRS and breast cancer risk is supported by two previous studies that investigated this association. In one of these studies, Chen et al. [20] investigated 18 T2D-related SNPs among 503 European ancestry cases and 633 controls from the multiethnic cohort and PAGE studies. In the other study, Hou et al. [21] pooled data for 25 genotyped and 15 imputed T2D-related SNPs from seven studies and investigated this association among 1,142 European ancestry cases and 1,137 European ancestry controls. Neither study reported a significant association between T2D GRS and overall breast cancer risk. However, these two studies had evaluated a smaller set of T2D risk variants than the current study and the sample size in both studies was substantially smaller than the current study, and thus, the statistical power in these two previous studies was low. For example, for a given SNP with a minor allele frequency of 0.3, the current study had 99.6 % power to detect an OR of 1.05 at a type I error rate of 0.05, while the previous studies had <15 % power to detect an OR of 1.05.
We identified three T2D risk variants that were associated with breast cancer risk. SNPs in strong correlation with each of these three variants have recently been identified in GWAS to be associated with breast cancer risk. SNP rs9939609 (FTO) located in region 16q12.2 and rs7903146 (TCF7L2) located in region 10q25.2 are in perfect LD (R 2 = 1) with rs17817449 and rs7904519, respectively, which were identified in relation to breast cancer risk in a GWAS conducted using BCAC data [22]. SNP rs8042680 (PRC1) is in strong LD with rs2290203 (R 2 = 0.59, 9,270 bp apart) that was recently identified as a risk variant for breast cancer in a GWAS conducted in East Asian women [34]. Interestingly, the T2D-risk allele of rs9939609 and rs8042680 is associated with a decreased risk of breast cancer. Though studies have suggested that TCF7L2 may associate with breast cancer through the Wnt/β-catenin pathway [35, 36], the exact mechanisms underlying these associations are unclear. Further studying these genes may uncover additional insights into the biology and genetics that link the risk of breast cancer and T2D.
The sample size for our study was very large. When comparing subjects in T2D GRS Q5 to those in Q1, our study had 80 % power to detect an OR for breast cancer risk as low as 1.06 (or 0.94) at 5 % type I error rate. Our study showed that the association between T2D GRS and breast cancer risk should be very small, if it exists. The GRS used in our study was constructed using SNPs with established association with T2D, as demonstrated convincingly in previous GWAS, and thus, this GRS should have a clear association with T2D. Indeed, using the resources from the Nashville Breast Health Study [37], we showed that this GRS was related to T2D in a dose–response manner (p for trend < 0.01, Online Resource Table 6). However, there are some potential limitations of our study. The T2D treatment information was not available for the study, preventing us from conducting an in-depth evaluation of the potential influence of T2D treatment on the association of T2D risk variants with breast cancer risk. To reduce potential influence of T2D treatment, we conducted an analysis among younger patients (<50 years old) who are less likely to have T2D diagnosis than the older age group. This analysis showed similar results in younger and older groups (Table 2), indicating that the influence of T2D treatment on the association of T2D risk variants with breast cancer risk should be small. Approximately two-thirds of the SNPs used to construct the T2D GRS were not directly genotyped. We imputed these SNPs using 1,000 Genomes Project data as the reference. The imputation quality was high. In a sensitivity analysis, we constructed an alternate T2D GRS using only the 11 directly genotyped SNPs and the 14 imputed SNPs which had almost perfect quality (R 2 > 0.9). This T2D GRS is highly correlated with the T2D GRS used in our primary analysis (Pearson’s r = 0.93), and using the alternate T2D GRS did not change the results appreciably. Since we started this project, 14 new genetic loci for T2D have been identified. Unfortunately, we don’t have any data for these 14 new loci for our study. However, the strength of the association of T2D risk is much weaker for these newly identified variants than the 33 variants identified previously and included in our study. Therefore, we believe that including these variants would not change the conclusion of this study. Finally, all participants in this study are of European ancestry, possibility affecting the generalizability of our study findings to other populations.
In conclusion, our study found no apparent association between a polygenetic score constructed using the known T2D risk variants identified to date in GWAS and breast cancer risk among women of European ancestry. It is possible that the previously reported association between these two diseases could be due to shared lifestyle risk factors for T2D and breast cancer, providing support for lifestyle modification as an effective prevention strategy to reduce the risk of both T2D and breast cancer. Our finding of significant associations of three T2D risk variants with breast cancer suggests a potential link of certain shared genetic and biological pathways for these common diseases.
References
Federation ID (2013) IDF diabetes atlas, 6th edn. International Diabetes Federation, Brussels
Alberti KG, Zimmet PZ (1998) Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabetic Med 15(7):539–553. doi:10.1002/(SICI)1096-9136(199807)15:7<539:AID-DIA668>3.0.CO;2-S
Siegel R, Ma J, Zou Z, Jemal A (2014) Cancer statistics, 2014. CA Cancer J Clin 64(1):9–29. doi:10.3322/caac.21208
Talamini R, Franceschi S, Favero A, Negri E, Parazzini F, La Vecchia C (1997) Selected medical conditions and risk of breast cancer. Br J Cancer 75(11):1699–1703
Weiderpass E, Gridley G, Persson I, Nyren O, Ekbom A, Adami HO (1997) Risk of endometrial and breast cancer in patients with diabetes mellitus. Int J Cancer J Int du cancer 71(3):360–363
Wideroff L, Gridley G, Mellemkjaer L, Chow WH, Linet M, Keehn S, Borch-Johnsen K, Olsen JH (1997) Cancer incidence in a population-based cohort of patients hospitalized with diabetes mellitus in Denmark. J Natl Cancer Inst 89(18):1360–1365
Michels KB, Solomon CG, Hu FB, Rosner BA, Hankinson SE, Colditz GA, Manson JE, Nurses’ Health S (2003) Type 2 diabetes and subsequent incidence of breast cancer in the Nurses’ Health Study. Diabetes Care 26(6):1752–1758
Lipscombe LL, Goodwin PJ, Zinman B, McLaughlin JR, Hux JE (2006) Diabetes mellitus and breast cancer: a retrospective population-based cohort study. Breast Cancer Res Treat 98(3):349–356. doi:10.1007/s10549-006-9172-5
Xue F, Michels KB (2007) Diabetes, metabolic syndrome, and breast cancer: a review of the current evidence. Am J Clin Nutr 86(3):s823–s835
Larsson SC, Mantzoros CS, Wolk A (2007) Diabetes mellitus and risk of breast cancer: a meta-analysis. Int J Cancer J Int du Cancer 121(4):856–862. doi:10.1002/ijc.22717
Hardefeldt PJ, Edirimanne S, Eslick GD (2012) Diabetes increases the risk of breast cancer: a meta-analysis. Endocr Relat Cancer 19(6):793–803. doi:10.1530/ERC-12-0242
Boyle P, Boniol M, Koechlin A, Robertson C, Valentini F, Coppens K, Fairley LL, Boniol M, Zheng T, Zhang Y, Pasterk M, Smans M, Curado MP, Mullie P, Gandini S, Bota M, Bolli GB, Rosenstock J, Autier P (2012) Diabetes and breast cancer risk: a meta-analysis. Br J Cancer 107(9):1608–1617. doi:10.1038/bjc.2012.414
Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, Yee D (2010) Diabetes and cancer: a consensus report. CA Cancer J Clin 60(4):207–221. doi:10.3322/caac.20078
Yang G, Lu G, Jin F, Dai Q, Best R, Shu XO, Chen JR, Pan XY, Shrubsole M, Zheng W (2001) Population-based, case-control study of blood C-peptide level and breast cancer risk. Cancer Epidemiol Biomarkers Prev 10(11):1207–1211
Endogenous H, Breast Cancer Collaborative G, Key TJ, Appleby PN, Reeves GK, Roddam AW (2010) Insulin-like growth factor 1 (IGF1), IGF binding protein 3 (IGFBP3), and breast cancer risk: pooled individual data analysis of 17 prospective studies. Lancet Oncol 11(6):530–542. doi:10.1016/S1470-2045(10)70095-4
Pierce BL, Austin MA, Ahsan H (2011) Association study of type 2 diabetes genetic susceptibility variants and risk of pancreatic cancer: an analysis of PanScan-I data. Cancer Causes Control CCC 22(6):877–883. doi:10.1007/s10552-011-9760-5
Cheng I, Caberto CP, Lum-Jones A, Seifried A, Wilkens LR, Schumacher FR, Monroe KR, Lim U, Tiirikainen M, Kolonel LN, Henderson BE, Stram DO, Haiman CA, Le Marchand L (2011) Type 2 diabetes risk variants and colorectal cancer risk: the Multiethnic Cohort and PAGE studies. Gut 60(12):1703–1711. doi:10.1136/gut.2011.237727
Sainz J, Rudolph A, Hoffmeister M, Frank B, Brenner H, Chang-Claude J, Hemminki K, Forsti A (2012) Effect of type 2 diabetes predisposing genetic variants on colorectal cancer risk. J Clin Endocrinol Metab 97(5):E845–E851. doi:10.1210/jc.2011-2565
Machiela MJ, Lindstrom S, Allen NE, Haiman CA, Albanes D, Barricarte A, Berndt SI, Bueno-de-Mesquita HB, Chanock S, Gaziano JM, Gapstur SM, Giovannucci E, Henderson BE, Jacobs EJ, Kolonel LN, Krogh V, Ma J, Stampfer MJ, Stevens VL, Stram DO, Tjonneland A, Travis R, Willett WC, Hunter DJ, Le Marchand L, Kraft P (2012) Association of type 2 diabetes susceptibility variants with advanced prostate cancer risk in the Breast and Prostate Cancer Cohort Consortium. Am J Epidemiol 176(12):1121–1129. doi:10.1093/aje/kws191
Chen F, Wilkens LR, Monroe KR, Stram DO, Kolonel LN, Henderson BE, Le Marchand L, Haiman CA (2011) No association of risk variants for diabetes and obesity with breast cancer: the Multiethnic Cohort and PAGE studies. Cancer Epidemiol Biomarkers Prev 20(5):1039–1042. doi:10.1158/1055-9965.EPI-11-0135
Hou N, Zheng Y, Gamazon ER, Ogundiran TO, Adebamowo C, Nathanson KL, Domchek SM, Rebbeck TR, Simon MS, John EM, Hennis A, Nemesure B, Wu SY, Leske MC, Ambs S, Niu Q, Zhang J, Pierce B, Cox NJ, Olopade OI, Huo D (2012) Genetic susceptibility to type 2 diabetes and breast cancer risk in women of European and African ancestry. Cancer Epidemiol Biomarkers Prev 21(3):552–556. doi:10.1158/1055-9965.EPI-11-0979
Michailidou K, Hall P, Gonzalez-Neira A, Ghoussaini M, Dennis J, Milne RL, Schmidt MK, Chang-Claude J, Bojesen SE, Bolla MK, Wang Q, Dicks E, Lee A, Turnbull C, Rahman N, Breast, Ovarian Cancer Susceptibility C, Fletcher O, Peto J, Gibson L, Dos Santos Silva I, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Czene K, Irwanto A, Liu J, Waisfisz Q, Meijers-Heijboer H, Adank M, Hereditary B, Ovarian Cancer Research Group N, van der Luijt RB, Hein R, Dahmen N, Beckman L, Meindl A, Schmutzler RK, Muller-Myhsok B, Lichtner P, Hopper JL, Southey MC, Makalic E, Schmidt DF, Uitterlinden AG, Hofman A, Hunter DJ, Chanock SJ, Vincent D, Bacot F, Tessier DC, Canisius S, Wessels LF, Haiman CA, Shah M, Luben R, Brown J, Luccarini C, Schoof N, Humphreys K, Li J, Nordestgaard BG, Nielsen SF, Flyger H, Couch FJ, Wang X, Vachon C, Stevens KN, Lambrechts D, Moisse M, Paridaens R, Christiaens MR, Rudolph A, Nickels S, Flesch-Janys D, Johnson N, Aitken Z, Aaltonen K, Heikkinen T, Broeks A, Veer LJ, van der Schoot CE, Guenel P, Truong T, Laurent-Puig P, Menegaux F, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Zamora MP, Perez JI, Pita G, Alonso MR, Cox A, Brock IW, Cross SS, Reed MW, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, Henderson BE, Schumacher F, Le Marchand L, Andrulis IL, Knight JA, Glendon G, Mulligan AM, kConFab I, Australian Ovarian Cancer Study G, Lindblom A, Margolin S, Hooning MJ, Hollestelle A, van den Ouweland AM, Jager A, Bui QM, Stone J, Dite GS, Apicella C, Tsimiklis H, Giles GG, Severi G, Baglietto L, Fasching PA, Haeberle L, Ekici AB, Beckmann MW, Brenner H, Muller H, Arndt V, Stegmaier C, Swerdlow A, Ashworth A, Orr N, Jones M, Figueroa J, Lissowska J, Brinton L, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Network G, Radice P, Peterlongo P, Manoukian S, Bonanni B, Devilee P, Tollenaar RA, Seynaeve C, van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Durda K, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Bogdanova NV, Antonenkova NN, Dork T, Kristensen VN, Anton-Culver H, Slager S, Toland AE, Edge S, Fostira F, Kang D, Yoo KY, Noh DY, Matsuo K, Ito H, Iwata H, Sueta A, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Shu XO, Lu W, Gao YT, Cai H, Teo SH, Yip CH, Phuah SY, Cornes BK, Hartman M, Miao H, Lim WY, Sng JH, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Shen CY, Hsiung CN, Wu PE, Ding SL, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Blot WJ, Signorello LB, Cai Q, Zheng W, Deming-Halverson S, Shrubsole M, Long J, Simard J, Garcia-Closas M, Pharoah PD, Chenevix-Trench G, Dunning AM, Benitez J, Easton DF (2013) Large-scale genotyping identifies 41 new loci associated with breast cancer risk. Nat Genetics 45 (4):353–361, 361e351-352. doi:10.1038/ng.2563
Michailidou K, Beesley J, Lindstrom S, Canisius S, Dennis J, Lush MJ, Maranian MJ, Bolla MK, Wang Q, Shah M, Perkins BJ, Czene K, Eriksson M, Darabi H, Brand JS, Bojesen SE, Nordestgaard BG, Flyger H, Nielsen SF, Rahman N, Turnbull C, Bocs, Fletcher O, Peto J, Gibson L, Dos-Santos-Silva I, Chang-Claude J, Flesch-Janys D, Rudolph A, Eilber U, Behrens S, Nevanlinna H, Muranen TA, Aittomaki K, Blomqvist C, Khan S, Aaltonen K, Ahsan H, Kibriya MG, Whittemore AS, John EM, Malone KE, Gammon MD, Santella RM, Ursin G, Makalic E, Schmidt DF, Casey G, Hunter DJ, Gapstur SM, Gaudet MM, Diver WR, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Berg CD, Chanock SJ, Figueroa J, Hoover RN, Lambrechts D, Neven P, Wildiers H, van Limbergen E, Schmidt MK, Broeks A, Verhoef S, Cornelissen S, Couch FJ, Olson JE, Hallberg E, Vachon C, Waisfisz Q, Meijers-Heijboer H, Adank MA, van der Luijt RB, Li J, Liu J, Humphreys K, Kang D, Choi JY, Park SK, Yoo KY, Matsuo K, Ito H, Iwata H, Tajima K, Guenel P, Truong T, Mulot C, Sanchez M, Burwinkel B, Marme F, Surowy H, Sohn C, Wu AH, Tseng CC, Van Den Berg D, Stram DO, Gonzalez-Neira A, Benitez J, Zamora MP, Perez JI, Shu XO, Lu W, Gao YT, Cai H, Cox A, Cross SS, Reed MW, Andrulis IL, Knight JA, Glendon G, Mulligan AM, Sawyer EJ, Tomlinson I, Kerin MJ, Miller N, kConFab I, Group A, Lindblom A, Margolin S, Teo SH, Yip CH, Taib NA, Tan GH, Hooning MJ, Hollestelle A, Martens JW, Collee JM, Blot W, Signorello LB, Cai Q, Hopper JL, Southey MC, Tsimiklis H, Apicella C, Shen CY, Hsiung CN, Wu PE, Hou MF, Kristensen VN, Nord S, Alnaes GI, Nbcs, Giles GG, Milne RL, McLean C, Canzian F, Trichopoulos D, Peeters P, Lund E, Sund M, Khaw KT, Gunter MJ, Palli D, Mortensen LM, Dossus L, Huerta JM, Meindl A, Schmutzler RK, Sutter C, Yang R, Muir K, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Hartman M, Miao H, Chia KS, Chan CW, Fasching PA, Hein A, Beckmann MW, Haeberle L, Brenner H, Dieffenbach AK, Arndt V, Stegmaier C, Ashworth A, Orr N, Schoemaker MJ, Swerdlow AJ, Brinton L, Garcia-Closas M, Zheng W, Halverson SL, Shrubsole M, Long J, Goldberg MS, Labreche F, Dumont M, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Grip M, Brauch H, Hamann U, Bruning T, Network G, Radice P, Peterlongo P, Manoukian S, Bernard L, Bogdanova NV, Dork T, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Devilee P, Tollenaar RA, Seynaeve C, Van Asperen CJ, Jakubowska A, Lubinski J, Jaworska K, Huzarski T, Sangrajrang S, Gaborieau V, Brennan P, McKay J, Slager S, Toland AE, Ambrosone CB, Yannoukakos D, Kabisch M, Torres D, Neuhausen SL, Anton-Culver H, Luccarini C, Baynes C, Ahmed S, Healey CS, Tessier DC, Vincent D, Bacot F, Pita G, Alonso MR, Alvarez N, Herrero D, Simard J, Pharoah PP, Kraft P, Dunning AM, Chenevix-Trench G, Hall P, Easton DF (2015) Genome-wide association analysis of more than 120,000 individuals identifies 15 new susceptibility loci for breast cancer. Nat Genetics 47 (4):373–380. doi:10.1038/ng.3242
Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR (2012) Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet 44(8):955–959. doi:10.1038/ng.2354
Garcia-Closas M, Couch FJ, Lindstrom S, Michailidou K, Schmidt MK, Brook MN, Orr N, Rhie SK, Riboli E, Feigelson HS, Le Marchand L, Buring JE, Eccles D, Miron P, Fasching PA, Brauch H, Chang-Claude J, Carpenter J, Godwin AK, Nevanlinna H, Giles GG, Cox A, Hopper JL, Bolla MK, Wang Q, Dennis J, Dicks E, Howat WJ, Schoof N, Bojesen SE, Lambrechts D, Broeks A, Andrulis IL, Guenel P, Burwinkel B, Sawyer EJ, Hollestelle A, Fletcher O, Winqvist R, Brenner H, Mannermaa A, Hamann U, Meindl A, Lindblom A, Zheng W, Devillee P, Goldberg MS, Lubinski J, Kristensen V, Swerdlow A, Anton-Culver H, Dork T, Muir K, Matsuo K, Wu AH, Radice P, Teo SH, Shu XO, Blot W, Kang D, Hartman M, Sangrajrang S, Shen CY, Southey MC, Park DJ, Hammet F, Stone J, Veer LJ, Rutgers EJ, Lophatananon A, Stewart-Brown S, Siriwanarangsan P, Peto J, Schrauder MG, Ekici AB, Beckmann MW, Dos Santos Silva I, Johnson N, Warren H, Tomlinson I, Kerin MJ, Miller N, Marme F, Schneeweiss A, Sohn C, Truong T, Laurent-Puig P, Kerbrat P, Nordestgaard BG, Nielsen SF, Flyger H, Milne RL, Perez JI, Menendez P, Muller H, Arndt V, Stegmaier C, Lichtner P, Lochmann M, Justenhoven C, Ko YD, Gene EI, breast CN, Muranen TA, Aittomaki K, Blomqvist C, Greco D, Heikkinen T, Ito H, Iwata H, Yatabe Y, Antonenkova NN, Margolin S, Kataja V, Kosma VM, Hartikainen JM, Balleine R, kConFab I, Tseng CC, Berg DV, Stram DO, Neven P, Dieudonne AS, Leunen K, Rudolph A, Nickels S, Flesch-Janys D, Peterlongo P, Peissel B, Bernard L, Olson JE, Wang X, Stevens K, Severi G, Baglietto L, McLean C, Coetzee GA, Feng Y, Henderson BE, Schumacher F, Bogdanova NV, Labreche F, Dumont M, Yip CH, Taib NA, Cheng CY, Shrubsole M, Long J, Pylkas K, Jukkola-Vuorinen A, Kauppila S, Knight JA, Glendon G, Mulligan AM, Tollenaar RA, Seynaeve CM, Kriege M, Hooning MJ, van den Ouweland AM, van Deurzen CH, Lu W, Gao YT, Cai H, Balasubramanian SP, Cross SS, Reed MW, Signorello L, Cai Q, Shah M, Miao H, Chan CW, Chia KS, Jakubowska A, Jaworska K, Durda K, Hsiung CN, Wu PE, Yu JC, Ashworth A, Jones M, Tessier DC, Gonzalez-Neira A, Pita G, Alonso MR, Vincent D, Bacot F, Ambrosone CB, Bandera EV, John EM, Chen GK, Hu JJ, Rodriguez-Gil JL, Bernstein L, Press MF, Ziegler RG, Millikan RM, Deming-Halverson SL, Nyante S, Ingles SA, Waisfisz Q, Tsimiklis H, Makalic E, Schmidt D, Bui M, Gibson L, Muller-Myhsok B, Schmutzler RK, Hein R, Dahmen N, Beckmann L, Aaltonen K, Czene K, Irwanto A, Liu J, Turnbull C, Familial Breast Cancer S, Rahman N, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Australian Breast Cancer Tissue Bank I, Olswold C, Slager S, Pilarski R, Ademuyiwa F, Konstantopoulou I, Martin NG, Montgomery GW, Slamon DJ, Rauh C, Lux MP, Jud SM, Bruning T, Weaver J, Sharma P, Pathak H, Tapper W, Gerty S, Durcan L, Trichopoulos D, Tumino R, Peeters PH, Kaaks R, Campa D, Canzian F, Weiderpass E, Johansson M, Khaw KT, Travis R, Clavel-Chapelon F, Kolonel LN, Chen C, Beck A, Hankinson SE, Berg CD, Hoover RN, Lissowska J, Figueroa JD, Chasman DI, Gaudet MM, Diver WR, Willett WC, Hunter DJ, Simard J, Benitez J, Dunning AM, Sherman ME, Chenevix-Trench G, Chanock SJ, Hall P, Pharoah PD, Vachon C, Easton DF, Haiman CA, Kraft P (2013) Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat genetics 45 (4):392–398, 398e391-392. doi:10.1038/ng.2561
Ghoussaini M, Fletcher O, Michailidou K, Turnbull C, Schmidt MK, Dicks E, Dennis J, Wang Q, Humphreys MK, Luccarini C, Baynes C, Conroy D, Maranian M, Ahmed S, Driver K, Johnson N, Orr N, dos Santos Silva I, Waisfisz Q, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Netherlands Collaborative Group on Hereditary B, Ovarian C, Hall P, Czene K, Irwanto A, Liu J, Nevanlinna H, Aittomaki K, Blomqvist C, Meindl A, Schmutzler RK, Muller-Myhsok B, Lichtner P, Chang-Claude J, Hein R, Nickels S, Flesch-Janys D, Tsimiklis H, Makalic E, Schmidt D, Bui M, Hopper JL, Apicella C, Park DJ, Southey M, Hunter DJ, Chanock SJ, Broeks A, Verhoef S, Hogervorst FB, Fasching PA, Lux MP, Beckmann MW, Ekici AB, Sawyer E, Tomlinson I, Kerin M, Marme F, Schneeweiss A, Sohn C, Burwinkel B, Guenel P, Truong T, Cordina-Duverger E, Menegaux F, Bojesen SE, Nordestgaard BG, Nielsen SF, Flyger H, Milne RL, Alonso MR, Gonzalez-Neira A, Benitez J, Anton-Culver H, Ziogas A, Bernstein L, Dur CC, Brenner H, Muller H, Arndt V, Stegmaier C, Familial Breast Cancer S, Justenhoven C, Brauch H, Bruning T, Gene Environment Interaction of Breast Cancer in Germany N, Wang-Gohrke S, Eilber U, Dork T, Schurmann P, Bremer M, Hillemanns P, Bogdanova NV, Antonenkova NN, Rogov YI, Karstens JH, Bermisheva M, Prokofieva D, Khusnutdinova E, Lindblom A, Margolin S, Mannermaa A, Kataja V, Kosma VM, Hartikainen JM, Lambrechts D, Yesilyurt BT, Floris G, Leunen K, Manoukian S, Bonanni B, Fortuzzi S, Peterlongo P, Couch FJ, Wang X, Stevens K, Lee A, Giles GG, Baglietto L, Severi G, McLean C, Alnaes GG, Kristensen V, Borrensen-Dale AL, John EM, Miron A, Winqvist R, Pylkas K, Jukkola-Vuorinen A, Kauppila S, Andrulis IL, Glendon G, Mulligan AM, Devilee P, van Asperen CJ, Tollenaar RA, Seynaeve C, Figueroa JD, Garcia-Closas M, Brinton L, Lissowska J, Hooning MJ, Hollestelle A, Oldenburg RA, van den Ouweland AM, Cox A, Reed MW, Shah M, Jakubowska A, Lubinski J, Jaworska K, Durda K, Jones M, Schoemaker M, Ashworth A, Swerdlow A, Beesley J, Chen X, kConFab I, Australian Ovarian Cancer Study G, Muir KR, Lophatananon A, Rattanamongkongul S, Chaiwerawattana A, Kang D, Yoo KY, Noh DY, Shen CY, Yu JC, Wu PE, Hsiung CN, Perkins A, Swann R, Velentzis L, Eccles DM, Tapper WJ, Gerty SM, Graham NJ, Ponder BA, Chenevix-Trench G, Pharoah PD, Lathrop M, Dunning AM, Rahman N, Peto J, Easton DF (2012) Genome-wide association analysis identifies three new breast cancer susceptibility loci. Nat Genet 44(3):312–318. doi:10.1038/ng.1049
Siddiq A, Couch FJ, Chen GK, Lindstrom S, Eccles D, Millikan RC, Michailidou K, Stram DO, Beckmann L, Rhie SK, Ambrosone CB, Aittomaki K, Amiano P, Apicella C, Australian Breast Cancer Tissue Bank I, Baglietto L, Bandera EV, Beckmann MW, Berg CD, Bernstein L, Blomqvist C, Brauch H, Brinton L, Bui QM, Buring JE, Buys SS, Campa D, Carpenter JE, Chasman DI, Chang-Claude J, Chen C, Clavel-Chapelon F, Cox A, Cross SS, Czene K, Deming SL, Diasio RB, Diver WR, Dunning AM, Durcan L, Ekici AB, Fasching PA, Familial Breast Cancer S, Feigelson HS, Fejerman L, Figueroa JD, Fletcher O, Flesch-Janys D, Gaudet MM, Consortium G, Gerty SM, Rodriguez-Gil JL, Giles GG, van Gils CH, Godwin AK, Graham N, Greco D, Hall P, Hankinson SE, Hartmann A, Hein R, Heinz J, Hoover RN, Hopper JL, Hu JJ, Huntsman S, Ingles SA, Irwanto A, Isaacs C, Jacobs KB, John EM, Justenhoven C, Kaaks R, Kolonel LN, Coetzee GA, Lathrop M, Le Marchand L, Lee AM, Lee IM, Lesnick T, Lichtner P, Liu J, Lund E, Makalic E, Martin NG, McLean CA, Meijers-Heijboer H, Meindl A, Miron P, Monroe KR, Montgomery GW, Muller-Myhsok B, Nickels S, Nyante SJ, Olswold C, Overvad K, Palli D, Park DJ, Palmer JR, Pathak H, Peto J, Pharoah P, Rahman N, Rivadeneira F, Schmidt DF, Schmutzler RK, Slager S, Southey MC, Stevens KN, Sinn HP, Press MF, Ross E, Riboli E, Ridker PM, Schumacher FR, Severi G, Dos Santos Silva I, Stone J, Sund M, Tapper WJ, Thun MJ, Travis RC, Turnbull C, Uitterlinden AG, Waisfisz Q, Wang X, Wang Z, Weaver J, Schulz-Wendtland R, Wilkens LR, Van Den Berg D, Zheng W, Ziegler RG, Ziv E, Nevanlinna H, Easton DF, Hunter DJ, Henderson BE, Chanock SJ, Garcia-Closas M, Kraft P, Haiman CA, Vachon CM (2012) A meta-analysis of genome-wide association studies of breast cancer identifies two novel susceptibility loci at 6q14 and 20q11. Hum Mol Genet 21(24):5373–5384. doi:10.1093/hmg/dds381
Ahsan H, Halpern J, Kibriya MG, Pierce BL, Tong L, Gamazon E, McGuire V, Felberg A, Shi J, Jasmine F, Roy S, Brutus R, Argos M, Melkonian S, Chang-Claude J, Andrulis I, Hopper JL, John EM, Malone K, Ursin G, Gammon MD, Thomas DC, Seminara D, Casey G, Knight JA, Southey MC, Giles GG, Santella RM, Lee E, Conti D, Duggan D, Gallinger S, Haile R, Jenkins M, Lindor NM, Newcomb P, Michailidou K, Apicella C, Park DJ, Peto J, Fletcher O, dos Santos Silva I, Lathrop M, Hunter DJ, Chanock SJ, Meindl A, Schmutzler RK, Muller-Myhsok B, Lochmann M, Beckmann L, Hein R, Makalic E, Schmidt DF, Bui QM, Stone J, Flesch-Janys D, Dahmen N, Nevanlinna H, Aittomaki K, Blomqvist C, Hall P, Czene K, Irwanto A, Liu J, Rahman N, Turnbull C, Familial Breast Cancer S, Dunning AM, Pharoah P, Waisfisz Q, Meijers-Heijboer H, Uitterlinden AG, Rivadeneira F, Nicolae D, Easton DF, Cox NJ, Whittemore AS (2014) A genome-wide association study of early-onset breast cancer identifies PFKM as a novel breast cancer gene and supports a common genetic spectrum for breast cancer at any age. Cancer Epidemiol Biomarkers Prev 23(4):658–669. doi:10.1158/1055-9965.EPI-13-0340
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8):904–909. doi:10.1038/ng1847
Wu MC, Kraft P, Epstein MP, Taylor DM, Chanock SJ, Hunter DJ, Lin X (2010) Powerful SNP-set analysis for case-control genome-wide association studies. Am J Hum Genet 86(6):929–942. doi:10.1016/j.ajhg.2010.05.002
Team RC (2014) R: A language and environment for statistical computing. R foundation for Statistical computing, Vienna
Giovannucci E, Harlan DM, Archer MC, Bergenstal RM, Gapstur SM, Habel LA, Pollak M, Regensteiner JG, Yee D (2010) Diabetes and cancer: a consensus report. Diabetes Care 33(7):1674–1685. doi:10.2337/dc10-0666
La Vecchia C, Giordano SH, Hortobagyi GN, Chabner B (2011) Overweight, obesity, diabetes, and risk of breast cancer: interlocking pieces of the puzzle. Oncologist 16(6):726–729. doi:10.1634/theoncologist.2011-0050
Cai Q, Zhang B, Sung H, Low SK, Kweon SS, Lu W, Shi J, Long J, Wen W, Choi JY, Noh DY, Shen CY, Matsuo K, Teo SH, Kim MK, Khoo US, Iwasaki M, Hartman M, Takahashi A, Ashikawa K, Matsuda K, Shin MH, Park MH, Zheng Y, Xiang YB, Ji BT, Park SK, Wu PE, Hsiung CN, Ito H, Kasuga Y, Kang P, Mariapun S, Ahn SH, Kang HS, Chan KY, Man EP, Iwata H, Tsugane S, Miao H, Liao J, Nakamura Y, Kubo M, Consortium DG-O, Delahanty RJ, Zhang Y, Li B, Li C, Gao YT, Shu XO, Kang D, Zheng W (2014) Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat Genetics 46 (8):886–890. doi:10.1038/ng.3041
Brown AM (2001) Wnt signaling in breast cancer: have we come full circle? Breast Cancer Res 3(6):351–355
Howe LR, Brown AM (2004) Wnt signaling and breast cancer. Cancer Biol Ther 3(1):36–41
Fu Z, Deming SL, Fair AM, Shrubsole MJ, Wujcik DM, Shu XO, Kelley M, Zheng W (2011) Well-done meat intake and meat-derived mutagen exposures in relation to breast cancer risk: the Nashville Breast Health Study. Breast Cancer Res Treat 129(3):919–928. doi:10.1007/s10549-011-1538-7
Acknowledgments
We thank all the individuals who took part in these studies and all the researchers, study staff, clinicians and other healthcare providers, technicians, and administrative staff who have enabled this work to be carried out. In particular, we thank: Andrew Berchuck (OCAC); Rosalind A. Eeles, Ali Amin Al Olama, Zsofia Kote-Jarai, Sara Benlloch (PRACTICAL); Antonis Antoniou, Lesley McGuffog, Ken Offit (CIMBA); Joe Dennis, Andrew Lee, Ed Dicks, Craig Luccarini, and the staff of the Centre for Genetic Epidemiology Laboratory; Javier Benitez, Anna Gonzalez-Neira, and the staff of the CNIO genotyping unit; Jacques Simard, Daniel C. Tessier, Francois Bacot, Daniel Vincent, Sylvie LaBoissière, Frederic Robidoux, and the staff of the McGill University and Génome Québec Innovation Centre; Sune F. Nielsen, Borge G. Nordestgaard, and the staff of the Copenhagen DNA laboratory; and Julie M. Cunningham, Sharon A. Windebank, Christopher A. Hilker, Jeffrey Meyer, and the staff of Mayo Clinic Genotyping Core Facility; Maggie Angelakos, Judi Maskiell, Gillian Dite (ABCFS), Ellen van der Schoot, Femake Atsma (Sanquin Bloodbank); Emiel Rutgers, Senno Verhoef, Frans Hogervorst, the Thai Ministry of Public Health (MOPH); Dr. Prat Boonyawongviroj (former Permanent Secretary of MOPH); Dr. Pornthep Siriwanarungsan (Department Director-General of Disease Control); Michael Schrauder, Matthias Rübner, Sonja Oeser, Silke Landrith, Eileen Williams, Elaine Ryder-Mills, Kara Sargus, Niall McInerney, Gabrielle Colleran, Andrew Rowan, Angela Jones, Christof Sohn, Andeas Schneeweiß, and Peter Bugert (the Danish Breast Cancer Group); Núria Álvarez; the CTS Steering Committee (including Leslie Bernstein, James Lacey, Sophia Wang, Huiyan Ma, Yani Lu, and Jessica Clague DeHart at the Beckman Research Institute of the City of Hope; Dennis Deapen, Rich Pinder, Eunjung Lee and Fred Schumacher at the University of Southern California; Pam Horn-Ross, Peggy Reynolds, and David Nelson at the Cancer Prevention Institute of California; and Hannah Park at the University of California Irvine); Hartwig Ziegler; Sonja Wolf; Volker Hermann; the GENICA network [Dr. Margarete Fischer-Bosch-Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Germany; (HB, Wing-Yee Lo, Christina Justenhoven), Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany (Yon-Dschun Ko, Christian Baisch), Institute of Pathology, University of Bonn, Germany (Hans-Peter Fischer), Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum (DKFZ) Heidelberg, Germany (UH), Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), Germany (Thomas Brüning, Beate Pesch, Sylvia Rabstein, Anne Lotz), Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Germany (Volker Harth)]; Tuomas Heikkinen; Irja Erkkilä; Kirsimari Aaltonen; Karl von Smitten; Natalia Antonenkova; Peter Hillemanns; Hans Christiansen; Eija Myöhänen; Helena Kemiläinen; Heather Thorne; Eveline Niedermayr; the AOCS Management Group (D Bowtell, G Chenevix-Trench, A deFazio, D Gertig, A Green, P Webb); the ACS Management Group (A Green, P Parsons, N Hayward, P Webb, D Whiteman); the LAABC data collection team, especially Annie Fung and June Yashiki; Gilian Peuteman; Dominiek Smeets; Thomas Van Brussel; Kathleen Corthouts; Nadia Obi; Judith Heinz; Sabine Behrens; Ursula Eilber; Muhabbet Celik; Til Olchers; the Mayo Clinic Breast Cancer Patient Registry; Martine Tranchant; Marie-France Valois; Annie Turgeon; Lea Heguy; Phuah Sze Yee; Peter Kang; Kang In Nee; Shivaani Mariapun; Yoon Sook-Yee; Daphne Lee; Teh Yew Ching; Nur Aishah Mohd Taib; Meeri Otsukka; Kari Mononen; Teresa Selander; Nayana Weerasooriya; OFBCR staff: E Krol-Warmerdam, J Molenaar, J Blom; Louise Brinton; Neonila Szeszenia-Dabrowska; Beata Peplonska; Witold Zatonski; Pei Chao; Michael Stagner; Petra Bos; Jannet Blom; Ellen Crepin; Anja Nieuwlaat; Annette Heemskerk; the Erasmus MC Family Cancer Clinic; Sue Higham; Simon Cross; Helen Cramp; Dan Connley; Sabapathy Balasubramanian; Ian Brock; the Eastern Cancer Registration and Information Centre; the SEARCH and EPIC teams; Michael Kerin, Nicola Miller, Niall McInerney, Gabrielle Colleran (BIGGS), and Pierre Kerbrat; Patrick Arveux; Romuald Le Scodan; Yves Raoul; Pierre Laurent-Puig; Claire Mulot (CECILE), Hartwig Ziegler, Sonja Wolf, Volker Hermann, Christa Stegmaier and Katja Butterbach (ESTHER), Taru A. Muranen (HEBCS), Natalia Antonenkova, Peter Hillemanns, Hans Christiansen and Johann H. Karstens (HMBCS), Gilian Peuteman, Dominiek Smeets, Thomas Van Brussel and Kathleen Corthouts (LMBC), Petra Seibold, Judith Heinz, Nadia Obi, Alina Vrieling, Sabine Behrens, Ursula Eilber, Muhabbet Celik, Til Olchers and Stefan Nickels (MARIE). MBCSG wish to thank Paolo Radice, Bernard Peissel, and Daniela Zaffaroni of the Fondazione IRCCS Istituto Nazionale dei Tumori (INT); Bernardo Bonanni, Monica Barile, and Irene Feroce of the Istituto Europeo di Oncologia (IEO) and Loris Bernard and the personnel of the Cogentech Cancer Genetic Test Laboratory. Cancer Council Victoria acknowledges the Traditional Owners of the land and waters throughout Victoria and pays respect to them, their culture and their Elders past, present, and future. We would like to thank Martine Tranchant (Cancer Genomics Laboratory, CHU de Québec Research Center), Marie-France Valois, Annie Turgeon, and Lea Heguy (McGill University Health Center, Royal Victoria Hospital; McGill University) for DNA extraction, sample management, and skillful technical assistance. J.S. is Chairholder of the Canada Research Chair in Oncogenetics. OBCS thanks Arja Jukkola-Vuorinen, Mervi Grip, Saila Kauppila, Kari Mononen, and Meeri Otsukka for data collection and sample preparation. Craig Luccarini; Don Conroy; Caroline Baynes; Kimberley Chua; the Ohio State University Human Genetics Sample Bank; and Robert Pilarski. Data on SCCS cancer cases used in this publication were provided by the: Alabama Statewide Cancer Registry; Kentucky Cancer Registry, Lexington, KY; Tennessee Department of Health, Office of Cancer Surveillance; Florida Cancer Data System; North Carolina Central Cancer Registry, North Carolina Division of Public Health; Georgia Comprehensive Cancer Registry; Louisiana Tumor Registry; Mississippi Cancer Registry; South Carolina Central Cancer Registry; Virginia Department of Health, Virginia Cancer Registry; and Arkansas Department of Health, Cancer Registry. DFBBCS thank Muriel Adank for selecting the samples and Margreet Ausems, Christi van Asperen, Senno Verhoef, and Rogier van Oldenburg for providing samples from their Clinical Genetic centers. We thank Pascal Arp, Mila Jhamai, Marijn Verkerk, Lizbeth Herrera, and Marjolein Peters for their help in creating the GWAS database, and Karol Estrada and Maksim V. Struchalin for their support in creation and analysis of imputed data. The authors are grateful to the study participants, the staff from the Rotterdam Study, and the participating general practitioners and pharmacists.
Financial Support
The work conducted for this project at the Vanderbilt Epidemiology Center is supported in part by NIH Grant R37CA070867 and endowment funds for the Ingram Professorship and Anne Potter Wilson Chair in Medicine. BCAC is funded by Cancer Research UK (C1287/A10118, C1287/A12014) and by the European Community’s Seventh Framework Programme under Grant agreement no 223175 (HEALTH-F2–2009-223175) (COGS). Meetings of the BCAC have been funded by the European Union COST programme (BM0606). Genotyping of the iCOGS array was funded by the European Union (HEALTH-F2-2009-223175), Cancer Research UK (C8197/A16565 and C1287/A10710), the Canadian Institutes of Health Research for the ‘CIHR Team in Familial Risks of Breast Cancer’ program and the Ministry of Economic Development, Innovation and Export Trade of Quebec (PSR-SIIRI-701). Additional support for the iCOGS infrastructure was provided by the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065, and 1U19 CA148112—the GAME-ON initiative), the Department of Defense (W81XWH-10-1-0341), Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The Australia, California, and Ontario sites of the Breast Cancer Family Registry were supported by Grant UM1 CA164920 from the National Cancer Institute (USA). The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the USA Government or the BCFR. The ABCFS (Australia site of the BCFR) was also supported by the National Health and Medical Research Council of Australia, the New South Wales Cancer Council, the Victorian Health Promotion Foundation (Australia) and the Victorian Breast Cancer Research Consortium. John L. Hopper is a National Health and Medical Research Council (NHMRC) Senior Principal Research Fellow and M.C.S. is a NHMRC Senior Research Fellow. Work at the OFBCR (Ontario site of the BCFR) was also supported by the Canadian Institutes of Health Research ‘CIHR Team in Familial Risks of Breast Cancer’ program. The ABCS study was supported by the Dutch Cancer Society [Grants NKI 2007-3839; 2009 4363] and BBMRI-NL, which is a Research Infrastructure financed by the Dutch government (NWO 184.021.007). The work of the BBCC was partly funded by ELAN-Fond of the University Hospital of Erlangen. The BBCS is funded by Cancer Research UK and Breakthrough Breast Cancer and acknowledges NHS funding to the NIHR Biomedical Research Centre, and the National Cancer Research Network (NCRN). Elinor J. Sawyer is supported by NIHR Comprehensive Biomedical Research Centre, Guy’s & St. Thomas’ NHS Foundation Trust in partnership with King’s College London, UK. Core funding to the Wellcome Trust Centre for Human Genetics was provided by the Wellcome Trust (090532/Z/09/Z). Ian Tomlinson is supported by the Oxford Biomedical Research Centre. The BSUCH study was supported by the Dietmar-Hopp Foundation, the Helmholtz Society, and the German Cancer Research Center (DKFZ). The CECILE study was funded by Fondation de France, Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Agence Nationale de Sécurité Sanitaire (ANSES), and Agence Nationale de la Recherche (ANR). The CGPS was supported by the Chief Physician Johan Boserup and Lise Boserup Fund, the Danish Medical Research Council, and Herlev Hospital. The CNIO-BCS was supported by the Genome Spain Foundation, the Red Temática de Investigación Cooperativa en Cáncer, and grants from the Asociación Española Contra el Cáncer and the Fondo de Investigación Sanitario (PI11/00923 and PI081120). The Human Genotyping-CEGEN Unit, CNIO is supported by the Instituto de Salud Carlos III. The CTS was initially supported by the California Breast Cancer Act of 1993 and the California Breast Cancer Research Fund (contract 97-10500) and is currently funded through the National Institutes of Health (R01 CA77398). Collection of cancer incidence data was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885. HAC receives support from the Lon V Smith Foundation (LVS39420). The ESTHER study was supported by a grant from the Baden Württemberg Ministry of Science, Research and Arts. Additional cases were recruited in the context of the VERDI study, which was supported by a grant from the German Cancer Aid (Deutsche Krebshilfe). The GENICA was funded by the Federal Ministry of Education and Research (BMBF) Germany Grants 01KW9975/5, 01KW9976/8, 01KW9977/0, and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum (IPA), as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus Bonn, Germany. The HEBCS was supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, the Nordic Cancer Union, and the Sigrid Juselius Foundation. The HMBCS was supported by the Rudolf Bartling Foundation. Financial support for KARBAC was provided through the regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institute, the Stockholm Cancer Foundation, and the Swedish Cancer Society. The KBCP was financially supported by the special Government Funding (EVO) of Kuopio University Hospital grants, Cancer Fund of North Savo, the Finnish Cancer Organizations, the Academy of Finland, and by the strategic funding of the University of Eastern Finland. kConFab is supported by grants from the National Breast Cancer Foundation, the NHMRC, the Queensland Cancer Fund, the Cancer Councils of New South Wales, Victoria, Tasmania and South Australia, and the Cancer Foundation of Western Australia. The kConFab Clinical Follow Up Study was funded by the NHMRC (145684, 288704, 454508). Kelly-Anne Phillips is a National Breast Cancer Foundation Fellow (Australia). Financial support for the AOCS was provided by the United States Army Medical Research and Materiel Command (DAMD17-01-1-0729), the Cancer Council of Tasmania and Cancer Foundation of Western Australia, and the NHMRC (199600). Georgia Chenevix-Trench and P.W. are supported by the NHMRC. LMBC is supported by the ‘Stichting tegen Kanker’ (232-2008 and 196-2010). Diether Lambrechts is supported by the FWO and the KULPFV/10/016-SymBioSysII and by a ERC consolidator grant. The MARIE study was supported by the Deutsche Krebshilfe e.V. [70-2892-BR I, 106332, 108253, 108419], the Hamburg Cancer Society, the German Cancer Research Center, and the Federal Ministry of Education and Research (BMBF) Germany [01KH0402]. MBCSG is supported by grants from the Italian Association for Cancer Research (AIRC) and by funds from the Italian citizens who allocated a 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori, according to Italian laws (INT-Institutional strategic projects ‘5 × 1,000’). The MCBCS was supported by the NIH Grants (CA122340, CA128978) and a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), the Breast Cancer Research Foundation, and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. MCCS cohort recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC Grants 209057, 251553, and 504711 and by infrastructure provided by Cancer Council Victoria. The MEC was supported by NIH Grants CA63464, CA54281, CA098758, and CA132839. The work of MTLGEBCS was supported by the Quebec Breast Cancer Foundation, the Canadian Institutes of Health Research for the ‘CIHR Team in Familial Risks of Breast Cancer’ program—Grant No CRN-87521 and the Ministry of Economic Development, Innovation and Export Trade—Grant No PSR-SIIRI-701. The NBCS was supported by grants from the Norwegian Research council (155218/V40, 175240/S10 to A.L.B.D., FUGE-NFR 181600/V11 to V.N.K. and a Swizz Bridge Award to A.L.B.D.). The NBHS was supported by NIH Grant R01CA100374. Biological sample preparation was conducted the Survey and Biospecimen Shared Resource, which is supported by P30 CA68485. OBCS was supported by the Academy of Finland (Grant Number 250083, 122715 and Center of Excellence Grant Number 251314), the Finnish Cancer Foundation, the Sigrid Juselius Foundation, the University of Oulu, the University of Oulu Support Foundation and the special Governmental EVO funds for Oulu University Hospital-based research activities. This OFBCR was supported by grant UM1 CA164920 from the National Cancer Institute. The content of this manuscript does not necessarily reflect the views or policies of the National Cancer Institute or any of the collaborating centers in the Breast Cancer Family Registry (BCFR), nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government or the BCFR. The ORIGO study was supported by the Dutch Cancer Society (RUL 1997-1505) and the Biobanking and Biomolecular Resources Research Infrastructure (BBMRI-NL CP16). The PBCS was funded by Intramural Research Funds of the National Cancer Institute, Department of Health and Human Services, USA. The pKARMA study was supported by Märit and Hans Rausings Initiative Against Breast Cancer. The RBCS was funded by the Dutch Cancer Society (DDHK 2004-3124, DDHK 2009-4318). The SASBAC was supported by funding from the Agency for Science, Technology and Research of Singapore (A*STAR), the US National Institute of Health (NIH), and the Susan G. Komen Breast Cancer Foundation. KC was financed by the Swedish Cancer Society (5128-B07-01PAF). The SBCS was supported by Yorkshire Cancer Research S305PA, S299, and S295. SEARCH is funded by a programme grant from Cancer Research UK (C490/A10124) and supported by the UK National Institute for Health Research Biomedical Research Centre at the University of Cambridge. SKKDKFZS is supported by the DKFZ. The SZBCS was supported by Grant PBZ_KBN_122/P05/2004. The TNBCC was supported by: a Specialized Program of Research Excellence (SPORE) in Breast Cancer (CA116201), a grant from the Breast Cancer Research Foundation, a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation, the Stefanie Spielman Breast Cancer fund and the OSU Comprehensive Cancer Center, DBBR (a CCSG Share Resource by National Institutes of Health Grant P30 CA016056), the Hellenic Cooperative Oncology Group research Grant (HR R_BG/04) and the Greek General Secretary for Research and Technology (GSRT) Program, Research Excellence II, the European Union (European Social Fund—ESF), and Greek national funds through the Operational Program ‘Education and Lifelong Learning’ of the National Strategic Reference Framework (NSRF)—ARISTEIA. The UKBGS is funded by Breakthrough Breast Cancer and the Institute of Cancer Research (ICR). ICR acknowledges NHS funding to the NIHR Biomedical Research Centre. The DFBBCS GWAS was funded by the Netherlands Organisation for Scientific Research (NWO) as part of a ZonMw/VIDI Grant number 91756341. The generation and management of GWAS genotype data for the Rotterdam Study (control samples) are supported by the Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study is funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, Z., Wen, W., Michailidou, K. et al. Association of genetic susceptibility variants for type 2 diabetes with breast cancer risk in women of European ancestry. Cancer Causes Control 27, 679–693 (2016). https://doi.org/10.1007/s10552-016-0741-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10552-016-0741-6