Abstract
Purpose
The CD44+/CD24− cell phenotype is enriched in triple negative breast cancers, is associated with tumor invasive properties, and serves as a cell surface marker profile of breast cancer stem-like cells. Activation of Epidermal Growth Factor Receptor (EGFR) promotes the CD44+/CD24− phenotype, but the specific signaling pathway downstream of EGFR responsible for this effect is not clear. The purpose of this study was to determine the role of the MEK/ERK pathway in the expansion of CD44+/CD24− populations in TNBC cells in response to EGFR activation.
Methods
Representative TNBC cell lines SUM159PT (claudin-low) and SUM149PT (basal) were used to evaluate cell surface expression of CD44 and CD24 by flow cytometry in response to EGFR and MEK inhibition or activation. EGFR and ERK phosphorylation levels were analyzed by Western blotting. The relationship between EGFR phosphorylation and MEK activation score in basal and claudin-low tumors from the TCGA database was examined.
Results
Inhibition of ERK activation with selumetinib, a MEK1/2 inhibitor, blocked EGF-induced expansion of CD44+/CD24− populations. Sustained activation of ERK by overexpression of constitutively active MEK1 was sufficient to expand CD44+/CD24− populations in cells in which EGFR activity was blocked by either erlotinib, an EGFR kinase inhibitor, or BB-94, a metalloprotease inhibitor that prevents generation of soluble EGFR ligands. In basal and claudin-low tumors from the TCGA database, there was a positive correlation between EGFR_pY1068 and MEK activation score in tumors without genomic loss of DUSP4, a negative regulator of ERK, but not in tumors harboring DUSP4 deletion.
Conclusion
Our results demonstrate that ERK activation is a key event in EGFR-dependent regulation of CD44+/CD24− populations. Furthermore, our findings highlight the role of ligand-mediated EGFR signaling in the control of MEK/ERK pathway output in TNBC tumors without DUSP4 loss.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Epidermal Growth Factor Receptor (EGFR) is frequently overexpressed in triple negative breast cancers (TNBCs, negative for expression of estrogen receptor (ER) and progesterone receptor (PR), and lacking amplification of the human epidermal growth factor receptor-2 (HER2) gene), and its overexpression is associated with poor clinical outcomes [1,2,3]. EGFR and its downstream signaling pathways regulate many aspects of cell behavior associated with tumor growth and progression, including cell proliferation, survival, epithelial-to-mesenchymal transition (EMT), migration, invasion, and drug resistance [4,5,6,7,8,9,10,11,12,13].
The CD44+/CD24− cell surface marker profile is associated with basal-like breast tumors [14], which are largely represented by triple-negative tumors [15], EMT [16], enhanced invasiveness [17], and stem-like properties of breast cancer cells [18]. Recently, it has been reported that inhibition of EGFR signaling in SUM159PT and MDA-MB-231 TNBC cell lines using Cetuximab, an anti-EGFR blocking monoclonal antibody, reduced CD44+/CD24−/low and Aldefluor+ cell populations, decreased mammosphere formation, and partially inhibited tumor growth in vivo in mouse xenograft models [19]. These results indicated that inhibition of EGFR signaling reduced cancer stem cell (CSC) populations and suggested that anti-EGFR therapies, in combination with chemotherapy, may be more effective in eliminating CSCs compared to chemotherapy alone in some TNBC patients. It was further postulated that the reduction of CSC populations by Cetuximab was mediated through inhibition of autophagy [19]. However, while EGFR may regulate autophagy in a context-dependent manner, most of the published reports indicate that EGFR tyrosine kinase activity inhibits autophagy [13, 20,21,22,23]. Therefore, inhibition of EGFR activity with Cetuximab should lead to activation, rather than inhibition, of autophagy, and the mechanism by which EGFR would control CSC populations is not clear.
Importantly, the CSC phenotype in basal and claudin-low breast cancers was reported to be promoted by activation of two mitogen-activated protein kinase (MAPK) pathways: the extracellular signal-regulated kinase (ERK) pathway and the Jun N-terminal kinase (JNK) pathway [24]. Specifically, activation of these pathways due to genomic loss of dual specificity phosphatase 4 (DUSP4), a negative regulator of ERK1/2 and JNK1/2, expanded CSC populations in several TNBC cell lines. Conversely, enforced expression of DUSP4 in BT549 and SUM159PT cell lines reduced CD44+/CD24− populations [24]. Since ERK1 and ERK2 are downstream effectors of mitogen-activated protein kinase kinases 1 and 2 (MEK1/2) [25], which in turn are regulated by EGFR, our first goal was to determine whether EGFR activity controls the CD44+/CD24− phenotype through the MEK/ERK pathway.
The second goal of this study was to examine the role of metalloproteases in regulation of the CD44+/CD24− phenotype and the MEK/ERK pathway output in TNBC. ADAM metalloproteases release soluble ligands for EGFR, namely EGF, heparin-binding EGF (HB-EGF), amphiregulin, epiregulin, transforming growth factor α (TGF-α), or betacellulin, and act as upstream regulators of EGFR [26, 27]. Ligand-dependent activation of EGFR represents the critical first step of the transcriptional programs regulated by the MEK/ERK pathway, provided that the tumors lack genetic alterations in pathway components that would render the pathway constitutively active. While activating mutations in the EGFR/RAS/RAF/MEK/ERK pathway are rare in breast cancer, approximately 50% of TNBCs are characterized by hemi- or homozygous deletion of the DUSP4 gene, which leads to aberrant pathway activation [24, 28, 29]. Thus, TNBCs harboring DUSP4 genomic loss should be less dependent on EGFR activation. However, in the remaining ~50% of TNBCs without DUSP4 copy loss, efficient MEK/ERK pathway activation might require the function of metalloproteases, generation of soluble EGFR ligands, and ligand-dependent EGFR activation.
Here, we show that ERK1/2 activation is necessary for EGFR-induced expansion of CD44+/CD24− populations. Furthermore, we show that sustained activation of ERK1/2 by overexpression of constitutively active MEK1 is sufficient to expand CD44+/CD24− populations in cells in which EGFR activity is blocked by either erlotinib, an EGFR kinase inhibitor, or BB-94, a metalloprotease inhibitor that prevents generation of soluble EGFR ligands. These results indicate that ERK1/2 plays an essential role in ligand-dependent EGFR signaling, which promotes the CD44+/CD24− marker profile. Moreover, in basal and claudin-low tumors from the TCGA database, there is a positive correlation between EGFR_pY1068 and MEK1/2 activation score in tumors without DUSP4 loss, but not in tumors harboring DUSP4 loss. This further highlights the role of ligand-mediated EGFR signaling in regulation of the MEK/ERK pathway in TNBC tumors without DUSP4 loss. The results of our investigations may help identify biomarkers to help predict which TNBC patients are most likely to respond to EGFR inhibitors.
Methods
Reagents and antibodies
Antibodies for immunoblotting included anti-EGFR_pY1068 (#D7A5), anti-total EGFR (#D38B1), anti-ERK1/2_pT202/Y204 (#D13.14.4E), anti-total ERK1/2 (#137F5), and anti-MEK1/2 (#D1A5), all from Cell Signaling Technology. Antibodies for flow cytometry, PE-conjugated anti-CD24 (#ML5) and APC-conjugated anti-CD44 (#IM7), were from BD Biosciences and Affymetrix eBioscience, respectively.
Calculation of EGFR and MEK activation scores
The top 100 genes whose expression was most significantly changed upon stable expression of EGFR or constitutively active MEK in MCF-7 cells, compared to control MCF-7 cells adapted for long-term estrogen-independent growth [30], were retrieved from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/), using accession number GSE3542. Expression values for these EGFR- or MEK-regulated genes in CD44+/CD24− and CD44−/CD24+ subpopulations of MCF10A cells [31] were then extracted from GEO using accession number GSE15192. The EGFR and MEK scores were calculated as:
where w is the weight +1 or −1, depending on whether the gene was upregulated or downregulated in the signature, and x is the normalized gene expression level.
TCGA data mining
Expression values for MEK-regulated genes (mRNA expression z-scores, measured by Agilent microarrays) and the EGFR phosphorylation status at Y992, Y1068, and Y1173 (protein expression z-scores, measured by reverse-phase protein arrays) were retrieved from The Cancer Genome Atlas (TCGA) (Nature 2012 dataset) [32] via the cBioPortal for Cancer Genomics (http://www.cbioportal.org/public-portal/) [33, 34]. The DUSP4 copy number status was determined by the Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm. GISTIC copy numbers “−2” (a deep loss) and “−1” (a shallow loss) for DUSP4 were considered homozygous and heterozygous deletions of DUSP4, respectively. Tumors for which DUSP4 GISTIC copy numbers were ≥ 0 were assumed not to harbor DUSP4 deletion. Pearson r correlation coefficient and two-tailed P values were calculated using GraphPad Prism 6.0 software.
Cell culture, retroviral transduction, generation of stable cell lines, and immunoblotting were performed as previously described [35,36,37]. Additional experimental details are provided in Online Resource 1: Supplementary Methods.
Results
It has been previously shown that the transcriptional signature of activation of the MEK pathway is positively correlated with the CD44:CD24 mRNA ratio in the NCI-Integrative Cancer Biology Program-50 (ICBP50) panel of breast cancer cell lines [24] and in mammosphere cultures derived from primary breast tumors [24, 38]. Here, we examined the EGFR and MEK pathway activation scores in flow cytometry-sorted subpopulations of MCF10A cells expressing the CD44+/CD24− or CD44−/CD24+ marker profile [31] (data were retrieved from GEO:GSE15192). To calculate the EGFR and MEK activation scores, we used the top 100 genes whose expression was most significantly changed upon stable expression of ligand-activatable EGFR or constitutively active MEK in MCF-7 cells [30]. We determined that the CD44+/CD24− subpopulation had significantly higher EGFR and MEK activation scores than the CD44−/CD24+ subpopulation (Fig. 1a).
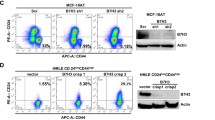
EGFR signaling modulates the CD44+/CD24− marker profile in representative TNBC cells. a EGFR and MEK pathway activation scores in CD44−/CD24+ and CD44+/CD24− subpopulations of MCF10A cells. Microarray gene expression data for these two cell populations were retrieved from GEO:GSE15192, and EGFR and MEK scores were calculated based on Ref. [30], as described in Methods. Results are shown as means from 4 determinations, ± S.E.M. b Response of SUM159PT and SUM149PT cells to sustained EGFR inhibition or activation. Cells were treated for 72 h with DMSO alone, 1 μM erlotinib, an EGFR inhibitor, dissolved in DMSO, or for 48 h with 20 ng/ml EGF and DMSO. The extent of EGFR phosphorylation at Y1068 and ERK1/2 phosphorylation at T202/Y204, total EGFR, and total ERK1/2 were analyzed by Western blotting. Tubulin is a gel-loading control. c–f Effect of EGFR inhibition or activation on the CD44 and CD24 markers. SUM159PT and SUM149PT cells were treated as in (panel b), stained with anti-CD24-PE and anti-CD44-APC antibodies, and analyzed by flow cytometry. Quadrant markers were set based on control antibody staining. Results of a representative experiment (n = 5) are shown as dot plots (c and e) or histogram analyses (d and f)
To study a potential cause-and-effect relationship between EGFR activation and cell surface expression of CD44 and CD24, and the role of MEK1/2 in this process, we utilized two representative TNBC cell lines, SUM159PT and SUM149PT, corresponding to the claudin-low and basal molecular subtypes of breast cancer, respectively [39]. We first examined the activation status of EGFR and its downstream effector ERK1/2 in these two cell lines in response to erlotinib, a specific inhibitor of EGFR tyrosine kinase activity, and to EGF, an activating ligand. Treatment for 72 h with 1 μM erlotinib caused a decrease in EGFR phosphorylation at Y1068, one of the major autophosphorylation sites in EGFR, in both cell lines. Stimulation for 48 h with 20 ng/ml EGF increased the EGFR phosphorylation signal and decreased the amount of total EGFR, leading to higher pEGFR/EGFR ratios (Fig. 1b), which is consistent with enhanced turnover of activated EGFR [40, 41]. Importantly, the extent of ERK1/2 phosphorylation decreased after erlotinib treatment and increased in the presence of EGF in both cell lines (Fig. 1b). This is notable, because SUM159PT cells, but not SUM149PT cells, contain an activating G12D mutation in the HRAS gene [42]. One of the reasons why ERK1/2 phosphorylation in SUM159PT cells was still modulated by EGFR could be the fact that the G12D mutation is heterozygous and approximately half of the HRAS protein, plus other members of the RAS family, are wild-type and amenable to regulation by external signals. Based on the results shown in Fig. 1b, we concluded that at the basal level, EGFR and ERK1/2 were partially active in both SUM159PT and SUM149PT cells, and they were further stimulated by adding EGF.
As reported previously, we found that the sizes of CD44+/CD24− populations in SUM159PT and SUM149PT cells were very different [43, 44], and amounted to ~90–95% for SUM159PT cells and ~5–10% for SUM149PT cells (Fig. 1c, e, respectively). Inhibition of the basal activation levels of EGFR with erlotinib decreased CD44+/CD24− populations in both SUM159PT and SUM149PT cells. Histogram analysis of the flow cytometry data showed a modest, but reproducible increase of CD24 expression in erlotinib-treated SUM159PT cells (Fig. 1d) and a clear increase of the CD24 staining in erlotinib-treated SUM149PT cells (Fig. 1f). Erlotinib did not have any effect on CD44 levels in SUM159PT cells, but it decreased CD44 expression in SUM149PT cells (Fig. 1d, f, respectively). These results agreed with a recently reported effect of Cetuximab on the CD44+/CD24− population in MDA-MB-231 cells [19]. EGF treatment caused a small increase of CD44+/CD24− populations in SUM159PT and SUM149PT cells (Fig. 1c and e, respectively), and these effects were statistically significant (see Figs. 2c, 3c for statistical analyses).
MEK1/2 inhibition blocks the effect of EGF on the CD44+/CD24− marker profile in SUM159PT cells. a Dose response of selumetinib, a MEK1/2 inhibitor, on the extent of ERK1/2 phosphorylation. Cells were treated with either DMSO or indicated concentrations of selumetinib. Twenty-four hours later, 20 ng/ml EGF or vehicle alone was added for an additional 48 h. Cells were analyzed by Western blotting using anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies; β1-integrin is a gel-loading control. b Cells were treated for 24 h with DMSO or 75 μM selumetinib, and then for an additional 48 h with or without 20 ng/ml EGF. Cells were stained with anti-CD24-PE and anti-CD44-APC antibodies and analyzed by flow cytometry. c Percentages of CD44+/CD24− cells are shown as mean values ± S.E.M. from three independent experiments. *P < 0.05; ***P < 0.001
MEK1/2 inhibition blocks the effect of EGF on the CD44+/CD24− marker profile in SUM149PT cells. a Dose response of selumetinib, a MEK1/2 inhibitor, on the extent of ERK1/2 phosphorylation. Cells were treated with either DMSO or indicated concentrations of selumetinib. Twenty-four hours later, 20 ng/ml EGF or vehicle alone was added for an additional 48 h. Cells were analyzed by Western blotting using anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies; β1-integrin is a gel-loading control. b Cells were treated for 24 h with DMSO or 10 μM selumetinib, and then for an additional 48 h with or without 20 ng/ml EGF. Cells were stained with anti-CD24-PE and anti-CD44-APC antibodies and analyzed by flow cytometry. c Percentages of CD44+/CD24− cells are shown as mean values ± S.E.M. from three independent experiments. *P < 0.05; **P < 0.01
To determine the role of the MEK pathway in maintaining CD44+/CD24− populations, cells were treated for 72 h with selumetinib, a MEK1/2 inhibitor. Selumetinib dose response in SUM159PT cells established that the lowest concentration of the inhibitor that entirely blocked MEK1/2-mediated phosphorylation of ERK1/2 in response to sustained EGFR activation was 75 μM (Fig. 2a). Treatment of SUM159PT cells with 75 μM selumetinib did not significantly affect cell viability (results not shown), but it dramatically decreased the CD44+/CD24− population, and this effect was not rescued by 48 h incubation with 20 ng/ml EGF (Fig. 2b, c). In SUM149PT cells, the lowest effective concentration of selumetinib was determined to be 10 μM (Fig. 3a), and this concentration had negligible effect on cell viability (results not shown). Treatment of SUM149PT cells with 10 μM selumetinib entirely eliminated the CD44+/CD24− population and, as in SUM159PT cells, this effect was not rescued by 48 h incubation with 20 ng/ml EGF (Fig. 3b, c). Thus, MEK1/2 activity is required for EGF-induced expansion of CD44+/CD24− populations.
To examine whether activation of the MEK/ERK pathway in the absence of an active EGFR is sufficient to expand CD44+/CD24− populations, we established cells with stable overexpression of a constitutively active phosphomimetic mutant of human MEK1 (MEK1-DD) [45], wild-type human MEK1 bearing an N-terminal HA-tag (HA-MEK1-WT) [46], or empty vector (EV). Overexpression of MEK1-DD or HA-MEK1-WT in SUM159PT cells was confirmed by Western blotting using anti-MEK1/2 or anti-HA-tag antibodies (Fig. 4a). The basal phosphorylation level of ERK1/2 in MEK1-DD-expressing SUM159PT cells was elevated and similar to the level of ERK1/2 phosphorylation in EGF-treated cells, validating constitutive activation of MEK1-DD (Fig. 4b). Importantly, while inhibition of EGFR with erlotinib diminished CD44+/CD24− populations in EV- and HA-MEK1-WT-expressing SUM159PT cells, erlotinib did not have any effect on the CD44+/CD24− marker profile in MEK1-DD-expressing cells (Fig. 4c, d).
Constitutively active MEK1 blocks the effect of erlotinib on CD44+/CD24− population in SUM159PT cells. a Confirmation of MEK1 overexpression. Cells stably transduced with empty vector (EV), constitutively active MEK1 (MEK1-DD), or wild-type HA-tagged MEK1 (HA-MEK1-WT) were analyzed by Western blotting using anti-MEK1 and anti-HA-tag antibodies; β1-integrin is a gel-loading control. b Verification of constitutive activation of MEK1-DD. Cells expressing EV, MEK1-DD, or HA-MEK1-WT were incubated for 30 min with or without 20 ng/ml EGF and analyzed by Western blotting using anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies. c Effect of MEK1-DD on the CD44+/CD24− marker profile. EV, MEK1-DD, or HA-MEK1-WT cells were treated with either DMSO or 1 μM erlotinib for 72 h, stained with anti-CD24-PE and anti-CD44-APC antibodies, and analyzed by flow cytometry. d Percentages of CD44+/CD24− cells are shown as mean values ± S.E.M. from two independent experiments. *P < 0.05; **P < 0.01
Similar results were obtained for SUM149PT cells with stable overexpression of EV, MEK1-DD, or HA-MEK1-WT (Fig. 5a). The basal phosphorylation level of ERK1/2 in MEK1-DD-expressing SUM149PT cells was higher than it was in EV- or HA-MEK1-WT-expressing cells and, in contrast to EV or HA-MEK1-WT cells, it was not inhibited by erlotinib (Fig. 5b), confirming the constitutive activation of MEK1-DD. Furthermore, while treatment of EV- or HA-MEK1-WT-expressing SUM149PT cells with erlotinib diminished CD44+/CD24− population, overexpression of MEK1-DD blocked the effect of erlotinib on the CD44+/CD24− population (Fig. 5c, d). Collectively, the results in Figs. 4 and 5 indicate that constitutive activation of the MEK/ERK pathway in the absence of an active EGFR was sufficient to expand CD44+/CD24− populations in SUM159PT and SUM149PT cells.
Constitutively active MEK1 blocks the effect of erlotinib on CD44+/CD24− population in SUM149PT cells. a Confirmation of MEK1 overexpression. SUM149PT cells stably transduced with empty vector (EV), constitutively active MEK1 (MEK1-DD), or wild-type HA-tagged MEK1 (HA-MEK1-WT), were analyzed by Western blotting using anti-MEK1/2 and anti-HA-tag antibodies; β1-integrin is a gel-loading control. b Verification of constitutive activation of MEK1-DD. Cells expressing EV, MEK1-DD, or HA-MEK1-WT were incubated for 24 h with DMSO or 1 μM erlotinib and analyzed by Western blotting using anti-phospho-ERK1/2 and anti-total ERK1/2 antibodies. c Effect of MEK1-DD on the CD44+/CD24− marker profile. EV, MEK1-DD, or HA-MEK1-WT cells were treated with either DMSO or 1 μM erlotinib for 72 h, stained with anti-CD24-PE and anti-CD44-APC antibodies, and analyzed by flow cytometry. d Percentages of CD44+/CD24− cells are shown as mean values ± S.E.M. from two independent experiments. *P < 0.05; ***P < 0.001
Partial activation of EGFR in SUM159PT and SUM149PT cells in the absence of the exogenously added EGF, which was also observed previously in our lab in SUM159PT cells in serum-free media [37], suggested that EGFR might have been activated by endogenously expressed ligands. All EGFR ligands are synthesized as transmembrane precursors that need to be converted to biologically active, soluble molecules via cleavage by ADAM metalloproteases [26, 27]. This raised a possibility that ADAMs may be involved in regulation of the CD44+/CD24− marker profile. This hypothesis was tested here by treatment of cells with batimastat (BB-94), a broad-spectrum metalloprotease inhibitor. BB-94 did not have a noticeable effect on cell viability or proliferation rates at the concentrations up to 30 μM (results not shown). A 48 h incubation of SUM159PT, SUM149PT, or MCF10A, a non-tumorigenic mammary epithelial cell line, with 10 μM BB-94 significantly decreased the amount of soluble amphiregulin, an EGFR ligand highly expressed in all three cell lines, in the media (Fig. 6a). Consequently, the basal phosphorylation level of EGFR was significantly reduced after BB-94 treatment (Fig. 6b). The effect of BB-94 was eliminated after 30 min stimulation with exogenous EGF (Fig. 6b), which was consistent with the fact that metalloproteases act as upstream regulators of ligand availability and EGFR activation.
Metalloprotease inhibitor batimastat (BB-94) decreases the amount of soluble amphiregulin (AREG) released from cells to the media and reduces the basal activation level of EGFR. SUM159PT, SUM149PT, or MCF10A cells were incubated for 48 h with DMSO or 10 μM BB-94. a The amount of soluble AREG in conditioned media was measured using ELISA. Shown are the mean values ± S.E.M. from three independent measurements; ***P < 0.001, ****P < 0.0001. b DMSO- or BB-94-treated cells were incubated for 30 min with or without 20 ng/ml EGF and analyzed by Western blotting using anti-phospho-EGFR or anti-total EGFR antibody; tubulin is a gel-loading control
In addition to its effect on EGFR phosphorylation, a 72-h incubation of SUM159PT cells with 10 μM BB-94 significantly inhibited the basal phosphorylation level of ERK1/2 (Fig. 7a). Importantly, CD44+/CD24− population was diminished after BB-94 treatment of EV- of HA-MEK1-WT-expressing SUM159PT cells, but not of cells expressing the constitutively active MEK1-DD (Fig. 7b, c). Similarly, BB-94-treated SUM149PT cells exhibited decreased levels of EGFR and ERK1/2 phosphorylation after 72-h treatment with 10 μM BB-94 (Fig. 8a), and BB-94 treatment reduced CD44+/CD24− populations in EV- and HA-MEK1-DD-expressing SUM149PT cells, but not in MEK1-DD-expressing cells (Fig. 8b, c). Thus, inhibition of metalloproteases, which are upstream modulators of EGFR signaling, with BB-94 exerted a similar effect to direct inhibition of EGFR tyrosine kinase activity with erlotinib.
Metalloprotease inhibitor batimastat (BB-94) reduces CD44+/CD24− population in SUM159PT cells, and this effect is blocked by the presence of constitutively active MEK1. a Effect of 72-h treatment with BB-94 on the basal activation levels of EGFR and ERK1/2. Cells were analyzed by Western blotting using anti-phospho-EGFR, anti-total EGFR, anti-phospho-ERK1/2, or anti-total ERK1/2 antibody. b Effect of BB-94 on the CD44+/CD24− marker profile, in the absence or presence of constitutively active MEK1. SUM159PT cells with stable expression of EV, MEK1-DD, or HA-MEK1-WT were treated as in (panel a), stained with anti-CD24-PE and anti-CD44-APC antibodies, and analyzed by flow cytometry. c Percentages of CD44+/CD24− cells are shown as mean values ± SEM from two independent experiments. *P < 0.05; **P < 0.01
Metalloprotease inhibitor batimastat (BB-94) reduces CD44+/CD24− population in SUM149PT cells, and this effect is blocked by the presence of constitutively active MEK1. a Effect of 72-h treatment with BB-94 on the basal activation levels of EGFR and ERK1/2. Cells were analyzed by Western blotting using anti-phospho-EGFR, anti-total EGFR, anti-phospho-ERK1/2, or anti-total ERK1/2 antibody. b Effect of BB-94 on the CD44+/CD24− marker profile, in the absence or presence of constitutively active MEK1. SUM149PT cells with stable expression of EV, MEK1-DD, or HA-MEK1-WT were treated as in (panel a), stained with anti-CD24-PE and anti-CD44-APC antibodies, and analyzed by flow cytometry. c Percentages of CD44+/CD24− cells are shown as mean values ± S.E.M. from two independent experiments. **P < 0.01; ***P < 0.001
While ligand-mediated activation of EGFR and the resulting activation of the MEK/ERK pathway may be important in some TNBCs, in other breast tumors in which ERK1/2 is hyperactive due to genomic loss of DUSP4, the status of EGFR activation should be less relevant for the MEK/ERK pathway output. To examine the relationship between EGFR activation, a transcriptional signature associated with MEK/ERK activation, and DUSP4 copy number alterations, we analyzed gene expression data for basal and claudin-low tumors (which are predominantly triple-negative) from the TCGA database [32]. EGFR phosphorylation status was available for three Tyr residues, Y992, Y1068, and Y1173, in 72 tumors. EGFR phosphorylation at Y1068 facilitates the binding of an adaptor protein GRB2 and activation of the RAS/RAF/MEK/ERK pathway, while phosphorylated Y992 and Y1173 are the major binding sites for PLCγ and SHP1 phosphatase, leading to the activation of PKC and receptor dephosphorylation, respectively [47, 48]. The MEK activation score in the 72 basal and claudin-low tumors was calculated based on the expression levels of top 100 MEK-regulated genes [30]. DUSP4 copy number data were generated by the GISTIC algorithm (see Methods).
There was a significant positive correlation (Pearson r = 0.375, P = 0.029) between EGFR_pY1068 and MEK activation score in tumors without DUSP4 deletion, but not in tumors harboring a heterozygous or homozygous deletion of DUSP4 (Table 1). EGFR_pY992 and EGFR_pY1173 were not significantly correlated with MEK activation in any tumor group. These results are consistent with the notion that metalloprotease-dependent ligand-mediated EGFR signaling plays an important role in regulation of the MEK/ERK pathway in TNBC tumors without DUSP4 loss.
Discussion
While a tumor-promoting role of EGFR signaling in TNBC has been well established, the results of EGFR-targeted therapies for TNBC, either as a monotherapy or in combination with cisplatin, [49], carboplatin [50], or Ixabepilone [51], have been disappointing. One of the reasons is the lack of specific markers predicting which patients are most likely to respond to anti-EGFR therapies [1, 2, 52,53,54,55]. Recent studies demonstrating that EGFR inhibitors reduce CSC populations in TNBC tumors [19] further highlight the importance of EGFR in the pathology of triple-negative disease and underscore the need for better markers guiding patient selection for anti-EGFR therapies. Understanding the mechanism by which EGFR activation controls CSCs is needed to maximize the clinical benefit of EGFR inhibitors in the treatment of TNBCs.
Here, we show that the MEK/ERK pathway is a key component of the signaling cascade downstream of the activated EGFR that regulates the CD44+/CD24− phenotype in SUM159PT and SUM149PT cells. MEK1/2 inhibition reduced CD44+/CD24− populations in both cell lines, and this effect was not rescued by activation of EGFR with exogenously added EGF. In contrast, overexpression of constitutively active MEK1 was sufficient to increase CD44+/CD24− populations, and this effect was not blocked by EGFR inhibition with erlotinib. Furthermore, we showed that inhibition of cell surface metalloproteases that generate soluble, bioactive ligands for EGFR diminished CD44+/CD24− populations, and metalloprotease inhibition with BB-94 was as effective as inhibition of tyrosine kinase activity of EGFR with erlotinib. Also, BB-94-mediated reduction of CD44+/CD24− populations was entirely blocked by overexpression of constitutively active MEK1. Collectively, these results demonstrate that MEK/ERK activity is both necessary and sufficient for EGFR-mediated regulation of CD44+/CD24− populations.
Our results corroborate previous studies demonstrating an important role of MEK/ERK in regulation of CSCs in breast tumors. As reported by Balko et al., activation of MAPK pathways due to DUSP4 knockdown increased mammosphere formation in MDA-MB-231 cells, and overexpression of DUSP4 in SUM159PT cells decreased mammosphere formation, reduced the CD44+/CD24− compartment, and impeded tumor growth in a mouse xenograft model [24]. Furthermore, pharmacological inhibition of MEK1/2 reduced mammosphere numbers in MDA-MB-231, BT549, and SUM159PT cell cultures and decreased the CD44+/CD24− population in MDA-MB-231 cells [24]. MEK1/2 inhibition also reduced anchorage-independent cell growth of MDA-MB-231 and SUM149PT cell lines, reversed EMT in 3D culture system, inhibited ALDH1 activity, and prevented lung metastasis in a MDA-MB-231 xenograft model [11].
While EGFR is one of the main upstream regulators the MEK/ERK pathway, and while EGFR inhibition, similarly to MEK1/2 inhibition, reduces the numbers of breast cancer cells with the stem-like phenotype [10, 19, 56], direct involvement of MEK/ERK in the EGFR-mediated control of breast CSCs has not been demonstrated before. Instead, it has been proposed that EGFR inhibition reduces CSC population in breast cancer through inhibition of autophagy [19]. This notion is supported by the observations that chloroquine, a lysotropic agent that inhibits a late step of autophagy, or knockdown of autophagy-specific genes diminished the CSC populations in TNBC cell lines [57,58,59]. The universally positive role of autophagy in CSC maintenance is, however, at odds with a report demonstrating that autophagy deficiency stabilizes the transcription factor TWIST1, promotes EMT in vitro, and tumor growth and metastasis using a A431 squamous cell carcinoma xenograft mouse model [60, 61]. Most importantly, in several cancer cell types, including breast cancer, EGFR activation suppresses autophagy [13, 20, 21]. Thus, EGFR inhibition should lead to an increase, rather than a decrease, in autophagy [22, 23], and the role of autophagy in EGFR-mediated regulation of breast CSCs needs further clarification.
The MEK/ERK pathway is hyperactivated in ~50% of TNBCs due to genomic loss of DUSP4, a negative regulator of ERK1/2. Since activating mutations in the MEK/ERK pathway components upstream of ERK1/2 are rare in breast cancer, it might be postulated that TNBCs without DUSP4 loss should rely on ligand-dependent EGFR activation to generate a sizeable ERK pathway output. Indeed, our analysis of gene expression and phosphoproteomics data from the TCGA database shows a significant positive correlation between EGFR phosphorylation at Y1068 and MEK activation score in basal and claudin-low breast cancers without DUSP4 loss, but not in basal and claudin-low breast cancers harboring DUSP4 loss. Thus, in the absence of DUSP4 loss, ligand-dependent metalloprotease-mediated activation of EGFR plays an essential role in regulating the MEK/ERK pathway output. Therefore, the efficacy of EGFR inhibitors should be greater in TNBCs containing an intact DUSP4 gene than in DUSP4-deficient TNBCs.
In summary, our results underscore the central role of the MEK/ERK signaling pathway in regulation of breast CSCs. A large body of evidence demonstrates an important role of the MEK/ERK signaling pathway in other aspects of breast cancer pathology as well. For example, inhibition of the MEK1/2 activity in estrogen receptor α (ERα)-negative breast cancer cell lines using a pharmacological inhibitor U0126 resulted in re-expression of ERα [30, 62]. More recently, increased activation of MEK/ERK in tumor cells was shown to be associated with reduced numbers of tumor-infiltrating lymphocytes in TNBC, leading to immune evasion [63]. Thus, targeting the MEK/ERK pathway may offer multiple benefits in DUSP4-deficient TNBC, whereas, EGFR inhibitors should be more efficient in DUSP4-positive TNBC.
Abbreviations
- TNBC:
-
Triple negative breast cancer
- EGF:
-
Epidermal growth factor
- EGFR:
-
Epidermal growth factor receptor
- HER2:
-
Human epidermal growth factor receptor 2
- ER:
-
Estrogen receptor
- PR:
-
Progesterone receptor
- MEK:
-
Mitogen-activated protein kinase kinase
- MAPK:
-
Mitogen-activated protein kinase
- ERK:
-
Extracellular signal-regulated kinase
- JNK:
-
Jun N-terminal kinase
- DUSP4:
-
Dual specificity phosphatase 4
- EMT:
-
Epithelial-to-mesenchymal transition
- CSCs:
-
Cancer stem cells
- FACS:
-
Fluorescence-activated cell sorting
- PE:
-
Phycoerythrin
- APC:
-
Allophycocyanin
- GEO:
-
Gene expression omnibus
- TCGA:
-
The Cancer Genome Atlas
- GISTIC:
-
Genomic identification of significant targets in cancer
References
Masuda H, Zhang D, Bartholomeusz C, Doihara H, Hortobagyi GN, Ueno NT (2012) Role of epidermal growth factor receptor in breast cancer. Breast Cancer Res Treat 136:331–345
Williams CB, Soloff AC, Ethier SP, Yeh ES (2015) Perspectives on epidermal growth factor receptor regulation in triple-negative breast cancer: ligand-mediated mechanisms of receptor regulation and potential for clinical targeting. Adv Cancer Res 127:253–281
Hsu JL, Hung MC (2016) The role of HER2, EGFR, and other receptor tyrosine kinases in breast cancer. Cancer Metastasis Rev 35:575–588
Verbeek BS, Adriaansen-Slot SS, Vroom TM, Beckers T, Rijksen G (1998) Overexpression of EGFR and c-erbB2 causes enhanced cell migration in human breast cancer cells and NIH3T3 fibroblasts. FEBS Lett 425:145–150
Mukhopadhyay P, Lakshmanan I, Ponnusamy MP, Chakraborty S, Jain M, Pai P et al (2013) MUC4 overexpression augments cell migration and metastasis through EGFR family proteins in triple negative breast cancer cells. PLoS ONE 8:e54455
Maretzky T, Evers A, Zhou W, Swendeman SL, Wong PM, Rafii S et al (2011) Migration of growth factor-stimulated epithelial and endothelial cells depends on EGFR transactivation by ADAM17. Nat Commun 2:229
Reginato MJ, Mills KR, Paulus JK, Lynch DK, Sgroi DC, Debnath J et al (2003) Integrins and EGFR coordinately regulate the pro-apoptotic protein Bim to prevent anoikis. Nat Cell Biol 5:733–740
Ahmed N, Maines-Bandiera S, Quinn MA, Unger WG, Dedhar S, Auersperg N (2006) Molecular pathways regulating EGF-induced epithelio-mesenchymal transition in human ovarian surface epithelium. Am J Physiol Cell Physiol 290:C1532–C1542
Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y et al (2007) Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res 67:9066–9076
Zhang D, LaFortune TA, Krishnamurthy S, Esteva FJ, Cristofanilli M, Liu P et al (2009) Epidermal growth factor receptor tyrosine kinase inhibitor reverses mesenchymal to epithelial phenotype and inhibits metastasis in inflammatory breast cancer. Clin Cancer Res 15:6639–6648
Bartholomeusz C, Xie X, Pitner MK, Kondo K, Dadbin A, Lee J et al (2015) MEK inhibitor Selumetinib (AZD6244; ARRY-142886) prevents lung metastasis in a triple-negative breast cancer xenograft model. Mol Cancer Ther 14:2773–2781
Wang M, Kern AM, Hulskotter M, Greninger P, Singh A, Pan Y et al (2014) EGFR-mediated chromatin condensation protects KRAS-mutant cancer cells against ionizing radiation. Cancer Res 74:2825–2834
Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G et al (2013) EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 154:1269–1284
Honeth G, Bendahl PO, Ringner M, Saal LH, Gruvberger-Saal SK, Lovgren K et al (2008) The CD44+/CD24− phenotype is enriched in basal-like breast tumors. Breast Cancer Res 10:R53
Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM (2013) Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 18:123–133
Blick T, Hugo H, Widodo E, Waltham M, Pinto C, Mani SA et al (2010) Epithelial mesenchymal transition traits in human breast cancer cell lines parallel the CD44hi/CD24lo/− stem cell phenotype in human breast cancer. J Mammary Gland Biol Neoplasia 15:235–252
Sheridan C, Kishimoto H, Fuchs RK, Mehrotra S, Bhat-Nakshatri P, Turner CH et al (2006) CD44+/CD24− breast cancer cells exhibit enhanced invasive properties: an early step necessary for metastasis. Breast Cancer Res 8:R59
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF (2003) Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100:3983–3988
Tanei T, Choi DS, Rodriguez AA, Liang DH, Dobrolecki L, Ghosh M et al (2016) Antitumor activity of Cetuximab in combination with Ixabepilone on triple negative breast cancer stem cells. Breast Cancer Res 18:6
Henson E, Chen Y, Gibson S (2017) EGFR family members’ regulation of autophagy is at a crossroads of cell survival and death in cancer. Cancers (Basel) 9:27
Chen Y, Henson ES, Xiao W, Huang D, McMillan-Ward EM, Israels SJ et al (2016) Tyrosine kinase receptor EGFR regulates the switch in cancer cells between cell survival and cell death induced by autophagy in hypoxia. Autophagy 12:1029–1046
Fung C, Chen X, Grandis JR, Duvvuri U (2012) EGFR tyrosine kinase inhibition induces autophagy in cancer cells. Cancer Biol Ther 13:1417–1424
Li X, Fan Z (2010) The epidermal growth factor receptor antibody cetuximab induces autophagy in cancer cells by downregulating HIF-1α and Bcl-2 and activating the beclin 1/hVps34 complex. Cancer Res 70:5942–5952
Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW et al (2013) Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res 73:6346–6358
Morrison DK (2012) MAP kinase pathways. Cold Spring Harb Perspect Biol 4:a011254
Blobel CP (2005) ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 6:32–43
Kataoka H (2009) EGFR ligands and their signaling scissors, ADAMs, as new molecular targets for anticancer treatments. J Dermatol Sci 56:148–153
Giltnane JM, Balko JM (2014) Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov Med 17:275–283
Mazumdar A, Poage GM, Shepherd J, Tsimelzon A, Hartman ZC, Den Hollander P et al (2016) Analysis of phosphatases in ER-negative breast cancers identifies DUSP4 as a critical regulator of growth and invasion. Breast Cancer Res Treat 158:441–454
Creighton CJ, Hilger AM, Murthy S, Rae JM, Chinnaiyan AM, El-Ashry D (2006) Activation of mitogen-activated protein kinase in estrogen receptor & #x03B1;-positive breast cancer cells in vitro induces an in vivo molecular phenotype of estrogen receptor & #x03B1;-negative human breast tumors. Cancer Res 66:3903–3911
Bhat-Nakshatri P, Appaiah H, Ballas C, Pick-Franke P, Goulet R Jr, Badve S et al (2010) SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24− phenotype. BMC Cancer 10:411
The Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490:61–70
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–404
Li H, Duhachek-Muggy S, Qi Y, Hong Y, Behbod F, Zolkiewska A (2012) An essential role of metalloprotease-disintegrin ADAM12 in triple-negative breast cancer. Breast Cancer Res Treat 135:759–769
Li H, Duhachek-Muggy S, Dubnicka S, Zolkiewska A (2013) Metalloproteinase-disintegrin ADAM12 is associated with a breast tumor-initiating cell phenotype. Breast Cancer Res Treat 139:691–703
Duhachek-Muggy S, Qi Y, Wise R, Alyahya L, Li H, Hodge J et al (2017) Metalloprotease-disintegrin ADAM12 actively promotes the stem cell-like phenotype in claudin-low breast cancer. Mol Cancer 16:32
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al (2009) Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 106:13820–13825
Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI et al (2010) Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res 12:R68
Haglund K, Dikic I (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci 125:265–275
Sorkin A, Goh LK (2009) Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res 315:396–683
Hollestelle A, Elstrodt F, Nagel JH, Kallemeijn WW, Schutte M (2007) Phosphatidylinositol-3-OH kinase or RAS pathway mutations in human breast cancer cell lines. Mol Cancer Res 5:195–201
Fillmore CM, Kuperwasser C (2008) Human breast cancer cell lines contain stem-like cells that self-renew, give rise to phenotypically diverse progeny and survive chemotherapy. Breast Cancer Res 10:R25
Keller PJ, Arendt LM, Skibinski A, Logvinenko T, Klebba I, Dong S et al (2011) Defining the cellular precursors to human breast cancer. Proc Natl Acad Sci USA 109:2772–2777
Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF et al (2007) Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129:1065–1079
Brady DC, Crowe MS, Turski ML, Hobbs GA, Yao X, Chaikuad A et al (2014) Copper is required for oncogenic BRAF signalling and tumorigenesis. Nature 509:492–496
Nyati MK, Morgan MA, Feng FY, Lawrence TS (2006) Integration of EGFR inhibitors with radiochemotherapy. Nat Rev Cancer 6:876–885
Shostak K, Chariot A (2015) EGFR and NF-κB: partners in cancer. Trends Mol Med 21:385–393
Baselga J, Gomez P, Greil R, Braga S, Climent MA, Wardley AM et al (2013) Randomized phase II study of the anti-epidermal growth factor receptor monoclonal antibody cetuximab with cisplatin versus cisplatin alone in patients with metastatic triple-negative breast cancer. J Clin Oncol 31:2586–2592
Carey LA, Rugo HS, Marcom PK, Mayer EL, Esteva FJ, Ma CX et al (2012) TBCRC 001: randomized phase II study of cetuximab in combination with carboplatin in stage IV triple-negative breast cancer. J Clin Oncol 30:2615–2623
Tredan O, Campone M, Jassem J, Vyzula R, Coudert B, Pacilio C et al (2015) Ixabepilone alone or with cetuximab as first-line treatment for advanced/metastatic triple-negative breast cancer. Clin Breast Cancer 15:8–15
Ueno NT, Zhang D (2011) Targeting EGFR in triple negative breast cancer. J Cancer 2:324–328
Costa R, Shah AN, Santa-Maria CA, Cruz MR, Mahalingam D, Carneiro BA et al (2017) Targeting Epidermal Growth Factor Receptor in triple negative breast cancer: new discoveries and practical insights for drug development. Cancer Treat Rev 53:111–119
Nakai K, Hung MC, Yamaguchi H (2016) A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am J Cancer Res 6:1609–1623
Fleisher B, Clarke C, Ait-Oudhia S (2016) Current advances in biomarkers for targeted therapy in triple-negative breast cancer. Breast Cancer 8:183–197
Farnie G, Clarke RB, Spence K, Pinnock N, Brennan K, Anderson NG et al (2007) Novel cell culture technique for primary ductal carcinoma in situ: role of Notch and epidermal growth factor receptor signaling pathways. J Natl Cancer Inst 99:616–627
Cufi S, Vazquez-Martin A, Oliveras-Ferraros C, Martin-Castillo B, Vellon L, Menendez JA (2011) Autophagy positively regulates the CD44(+) CD24(-/low) breast cancer stem-like phenotype. Cell Cycle 10:3871–3885
Choi DS, Blanco E, Kim YS, Rodriguez AA, Zhao H, Huang TH et al (2014) Chloroquine eliminates cancer stem cells through deregulation of Jak2 and DNMT1. Stem Cells 32:2309–2323
Liang DH, Choi DS, Ensor JE, Kaipparettu BA, Bass BL, Chang JC (2016) The autophagy inhibitor chloroquine targets cancer stem cells in triple negative breast cancer by inducing mitochondrial damage and impairing DNA break repair. Cancer Lett 376:249–258
Qiang L, Zhao B, Ming M, Wang N, He TC, Hwang S et al (2014) Regulation of cell proliferation and migration by p62 through stabilization of Twist1. Proc Natl Acad Sci USA 111:9241–9246
Qiang L, He YY (2014) Autophagy deficiency stabilizes TWIST1 to promote epithelial-mesenchymal transition. Autophagy 10:1864–1865
Bayliss J, Hilger A, Vishnu P, Diehl K, El-Ashry D (2007) Reversal of the estrogen receptor negative phenotype in breast cancer and restoration of antiestrogen response. Clin Cancer Res 13:7029–7036
Loi S, Dushyanthen S, Beavis PA, Salgado R, Denkert C, Savas P et al (2016) RAS/MAPK activation is associated with reduced tumor-infiltrating lymphocytes in triple-negative breast cancer: therapeutic cooperation between MEK and PD-1/PD-L1 immune checkpoint inhibitors. Clin Cancer Res 22:1499–1509
Acknowledgements
This work was supported by NIH Grant R01CA172222 to AZ. This is contribution 17-394-J from Kansas Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wise, R., Zolkiewska, A. Metalloprotease-dependent activation of EGFR modulates CD44+/CD24− populations in triple negative breast cancer cells through the MEK/ERK pathway. Breast Cancer Res Treat 166, 421–433 (2017). https://doi.org/10.1007/s10549-017-4440-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-017-4440-0