Abstract
Protein kinases are important components in oncogenic transformation of breast cancer. Evaluation of upregulated genes that codify for protein kinases could be used as biomarkers to predict clinical outcome. Gene expression and functional analyses using public datasets were performed to identify differential gene expression and functions in basal-like tumors compared with normal breast tissue. Overall survival (OS) associated with upregulated genes was explored using the KM Plotter online tool. The prognostic influence of these genes in luminal tumors and systemically untreated patients was also assessed. Of the 426 transcripts identified in basal-like tumors, 11 genes that coded for components of protein kinases were upregulated with more than a fourfold change. Regulation of cell cycle was an enriched function containing 10 of these 11 identified genes. Among them, expression of four genes, BUB1β, CDC28, NIMA, and PDZ binding kinase, were all associated with improved OS when using at least one probe in the basal-like subtype. Two genes, BUB1β and PDZ binding kinase, showed consistent association with improved OS irrespective of the gene probe used for the analysis. No association was observed for these genes with relapse-free survival. In contrast, both BUB1β and PDZ binding kinase showed worse OS in luminal tumors and in a cohort of systemically untreated patients. BUB1β and PDZ binding kinase are associated with improved OS in basal-like tumors and worse OS in luminal and untreated patients. The association with a better outcome in basal-like tumors could be due to a more favorable response to chemotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Triple negative breast cancer (TNBC) represents around 15 % of all tumors and are mainly included in the basal-like gene profiling subgroup [1]. Due to the absence of druggable molecular targets, TNBC is treated typically with cytotoxic chemotherapy [2]. The predominant mechanism of action of these anti-cancer drugs is direct DNA damage or interference with the process of mitosis [2]. Predictive markers for cytotoxic chemotherapy benefit are not available and as such, cytotoxic chemotherapy cannot be tailored. Identification of such biomarkers is therefore highly desirable.
Protein kinases play a central role in many different biological functions including proliferation, cell survival, or migration, among others [3, 4]. Proteins with kinase activity have been considered as oncogenic targets and have been used as biomarkers to select targeted agents [3–5]. In addition, protein kinases have been implicated in mechanisms of resistance [5]. In TNBC, certain kinases are important components in oncogenic transformation [6–8] and are the target of novel agents in preclinical and clinical evaluation [9–12].
Among the altered functions identified in this tumor type, proliferation is a key characteristic. Most TNBC express elevated levels of the proliferation marker Ki67 and morphologically exhibit high histological features, indicating an accelerated and continuous cell division [13]. Several protein kinases are implicated in the development of TNBC, including those that participate in the formation of the mitotic spindle during cell cycle, like polo-like kinases, Mps1/TTKs, or aurora kinases, among others; and indeed, agents targeting some of these proteins have shown preclinical activity in this breast cancer subtype [10–12].
In this article, by using an in silico approach, we identify relevant pathways and upregulated genes linked with clinical outcome in basal-like tumors. Particularly, BUB1β and PDZ binding kinase are upregulated genes involved in mitosis that are associated with better overall survival (OS) probably as they identify those tumors that respond better to chemotherapy. By contrast, expression of BUB1β and PDZ binding kinase are linked with worse outcome in luminal tumors and untreated patients.
Materials and methods
Transcriptomic and gene-set enrichment analyses
We used a public dataset (GEO DataSet accession number: GDS2250 [14]) of mRNA level data from normal breast tissue and basal-like breast tumors to identify deregulated genes. Affymetrix CEL files were downloaded and analyzed with dChip software (Dana Farber Cancer Institute, Boston, MA).
Genes with different expression values from the normal breast and basal-like breast cancer groups were obtained. Specifically, we used a fourfold change difference cutoff between both groups to identify clearly upregulated genes. We used the affymetrix software for evaluation of the volcano plot.
The list of genes was analyzed using gene-set enrichment analyses (DAVID Bioinformatics Resources 6.7) in order to identify functions of these genes. We used an adjusted p value <0.05 to select the enriched gene sets.
Outcome analyses
The KM Plotter Online Tool was used to analyze the relationship between the gene expression and patient clinical outcome in breast cancer (http://www.kmplot.com); this public database allowed us to investigate OS and relapse-free survival (RFS) [15, 16]. Information for OS were obtained from 594 patients, and RFS from 1593 patients. A cohort of systemically untreated patients including 1005 patients were also used for the analyses. Analysis was carried out as follows. First, the association between the gene of interest and outcome was explored after selection of the specific probe utilized initially. Analysis was then repeated using all probes available in the online software tool for each individual gene. Finally, in order to get insights into the association of BUB1β and PDZ binding kinase with clinical outcome, we evaluated the influence of their expression on OS in luminal tumors and systemically untreated patients.
Definitions of breast cancer subgroups reported in the online tool are as follows: triple negative (ER−/HER2−), luminal A (ESR1+/HER2−/MKI67-low), luminal B (ESR1+/HER2−/MKI67-high or ESR1+/HER2+), and basal-like (ESR1−/HER2−) breast cancers [15, 16]. In addition, the application permits to perform the analyses by using the immunohistochemical information of ER/PR and HER2 [15, 16].
All the analyses were performed independently by two authors (JPP and LDG) and reviewed by a third author (VSC). No discrepancies were observed.
Results
Transcriptomic analyses identify upregulated kinases involved in cell cycle progression
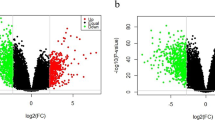
Our initial search identified 426 transcripts with a fold change greater than four between basal-like tumors (18 samples) and epithelial cells (7 samples). Of these, 17 genes coded for protein kinases; 11 of them being upregulated (Fig. 1a). Table 1 shows the Affimetrix probe sets, the gene names, and the fold change identified including p value. Volcano plot is showed in Supplementary Fig. 1. Two of the identified genes, aurora kinase A and TTK ,codify for druggable proteins.
In silico transcriptomic analyses of basal-like tumors compared with normal breast, and gene-set enrichment analyses. a Selection of upregulated genes that encodes for protein kinases with a fold change >4. b Pathway analyses. Ten out of the 11 upregulated genes are included in cell cycle regulation function
Gene-set enrichment analyses performed to identify biological functions and pathways showed that 10 of the 11 upregulated genes (91 %) participated in the regulation of cell cycle progression. Among genes not directly related to protein kinases, an additional 31 genes were associated with this function (see Fig. 1b). Supplementary Table 1 describes all genes included in the cell cycle and proliferation function as provided by DAVID Bioinformatics Resources 6.7.
Association of selected transcripts with outcome in basal-like tumors
Among the 11 upregulated genes, 4 were associated with outcome. Expression of BUB1β, CDC28, NIMA, and PDZ binding kinase were linked with improved OS (Fig. 2). These differences were statistically significant except for CDC28 which showed a non-significant association with improved outcome. Table 2 describes the functions of the evaluated genes. Repeating the analysis using all probes available for each gene confirmed the association between PDZ binding kinase and BUB1β and improved OS. The combined analyses of BUB1β and PDZ binding kinase were associated with better OS in both immunohistochemical-based subgroups (triple negative tumors), and those molecular-based subgroups (basal-like tumors) (see Supplementary Fig. 2A and B, respectively). Of note, BUB1β and PDZ were also upregulated in a subgroup of non-basal tumors from the same database (GEO DataSet accession number: GDS2250) (20 samples) as shown in Supplementary Fig. 3.
Prognostic influence of BUB1 and PDZ binding kinase in luminal tumors and systemically untreated patients
Expression of PDZ binding kinase was associated with poor OS in luminal A and luminal B tumors. BUB1β was associated with poor outcome in luminal A tumors, but in luminal B tumors, there was a non-significant association (see Fig. 3).
Similarly, expression of BUB1β and PDZ binding kinase was associated with worse OS in a cohort of systemically untreated breast cancer patients (Fig. 4).
Association with outcome of other cell cycle genes in basal-like tumors
Finally, we evaluated the association with outcome of all the 31 genes included in the cell cycle function as described in Supplementary Table 1, observing some of them linked with worse outcome (Supplementary Table 2).
Discussion
TNBC is one of the most devastating diseases as no current therapies exist beyond cytotoxic chemotherapy [2]. In this context, the identification of targets and biomarkers of response to a given treatment, including chemotherapy, is a main goal. As protein kinases have been implicated in the oncogenesis of breast cancer and are potentially druggable, we decided to evaluate the expression and prognostic relevance of this family of proteins in TNBC. A relevant finding of our study has been the identification of a substantial number of deregulated kinases which participate in cell proliferation/cell cycle regulation. This is especially interesting since it is well known that chemotherapy drugs acting on DNA/cell duplication are particularly efficient in this subtype of breast cancer.
Our study identified that BUB1β and PDZ binding kinase have a consistent association with improved OS irrespective of the gene probe used for analysis in basal-like tumors. In contrast, both BUB1β and PDZ showed worse OS in luminal tumors and in a cohort of systemically untreated patients.
The BUB1 family of genes codes for serine/threonine protein kinases that phosphorylate members of the mitotic checkpoint complex including Mad1, Mad2, BubR1, CENP-E, and PLK1 among others, and activates the spindle checkpoint [17–19]. BUB1 accumulates during G2/M and drops after mitosis [18]. PDZ binding kinase, also known as PBK, SPK, CT84, TOPK, HEL164, and Nori-3, is a serine/threonine protein kinase related to the dual-specific mitogen-activated protein kinase kinase (MAPKK) family [20]. It has been described in highly proliferative processes like the spermatogenesis and in some tumors such as lymphomas [21, 22].
Of the four genes identified as potentially prognostic, all are involved in the mitotic process. However, only BUB1β and PDZ binding kinase were associated with outcome for all the probe sets used. BUB1 and PDZ binding kinase are important in the formation of the mitotic spindle and could potentially be considered as surrogate markers of tumors where cells are highly dividing, including luminal B and triple negative tumors [17]. However, it is unclear if the expression of these genes are a better marker of proliferation compared with others, although, in our analyses, these genes provided stronger prognostication than Ki67 (data not shown).
The fact that many of these genes are linked with mitosis suggest that agents targeting kinases involved in this process could have a potential for therapeutic activity. This is the case with the remarkable preclinical activity identified for Mps1/TTK, and polo-like kinase inhibitors in triple negative breast cancer [10, 11].
The reasons for the differential prognostic value of BUB1 and PDZ binding kinase in basal and luminal tumors is unclear. One possible explanation is that as in TNBC the majority of patients are treated with chemotherapy, the improved prognosis may relate to a better response to chemotherapy. It is known that chemotherapy is more active in patients with tumors that express elevated levels of the Ki67 marker or have a high histological grade, both indicative of tumors that are highly proliferative [1].
Our study has limitations. This is an in silico evaluation of upregulated genes linked with clinical outcome in breast tumors. The fact that this is the first time that these kinases are described as upregulated and associated with outcome in different types of breast cancer warrants validation in clinical studies. The potential interaction of estrogen receptor signaling with genes coding for cell cycle and mitosis has not been explored. The analyses of outcome using the KM plotter tool have also boundaries, as it uses the median sample for dividing the samples into high- and low-expression groups. The determination of the exact cutoff value for each transcript could provide a more robust result. Finally, other proteins can be linked with outcome as some of those described in Supplementary Table 2.
We describe a set of genes in basal-like tumors that encode for mitotic checkpoint kinases and that are associated with better OS. The same genes are linked with worse outcome in luminal tumors and systemically untreated patients. The potential of these genes to predict response to chemotherapy in basal-like tumors and outcome in luminal tumors warrants further evaluation.
References
Le Du F, Eckhardt BL, Lim B, Litton JK, Moulder S, Meric-Bernstam F, Gonzalez-Angulo AM, Ueno NT (2015) Is the future of personalized therapy in triple-negative breast cancer based on molecular subtype? Oncotarget 6(15):12890–12908
Ocana A, Pandiella A (2008) Identifying breast cancer druggable oncogenic alterations: lessons learned and future targeted options. Clin Cancer Res 14(4):961–970. doi:10.1158/1078-0432.CCR-07-1630
Blume-Jensen P, Hunter T (2001) Oncogenic kinase signalling. Nature 411(6835):355–365. doi:10.1038/35077225
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) The protein kinase complement of the human genome. Science 298(5600):1912–1934. doi:10.1126/science.1075762
Ocana A, Amir E, Seruga B, Martin M, Pandiella A (2013) The evolving landscape of protein kinases in breast cancer: clinical implications. Cancer Treat Rev 39(1):68–76. doi:10.1016/j.ctrv.2012.05.004
Montero JC, Esparis-Ogando A, Re-Louhau MF, Seoane S, Abad M, Calero R, Ocana A, Pandiella A (2014) Active kinase profiling, genetic and pharmacological data define mTOR as an important common target in triple-negative breast cancer. Oncogene 33(2):148–156. doi:10.1038/onc.2012.572
Ortiz-Ruiz MJ, Alvarez-Fernandez S, Parrott T, Zaknoen S, Burrows FJ, Ocana A, Pandiella A, Esparis-Ogando A (2014) Therapeutic potential of ERK5 targeting in triple negative breast cancer. Oncotarget 5(22):11308–11318
Al-Ejeh F, Simpson PT, Saunus JM, Klein K, Kalimutho M, Shi W, Miranda M, Kutasovic J, Raghavendra A, Madore J, Reid L, Krause L, Chenevix-Trench G, Lakhani SR, Khanna KK (2014) Meta-analysis of the global gene expression profile of triple-negative breast cancer identifies genes for the prognostication and treatment of aggressive breast cancer. Oncogenesis 3:e124. doi:10.1038/oncsis.2014.41
Cuenca-Lopez MD, Serrano-Heras G, Montero JC, Corrales-Sanchez V, Gomez-Juarez M, Gascon-Escribano MJ, Morales JC, Voisin V, Nunez LE, Moris F, Bader GD, Pandiella A, Ocana A (2015) Antitumor activity of the novel multi-kinase inhibitor EC-70124 in triple negative breast cancer. Oncotarget 6(29):27923
Maire V, Nemati F, Richardson M, Vincent-Salomon A, Tesson B, Rigaill G, Gravier E, Marty-Prouvost B, De Koning L, Lang G, Gentien D, Dumont A, Barillot E, Marangoni E, Decaudin D, Roman-Roman S, Pierre A, Cruzalegui F, Depil S, Tucker GC, Dubois T (2013) Polo-like kinase 1: a potential therapeutic option in combination with conventional chemotherapy for the management of patients with triple-negative breast cancer. Cancer Res 73(2):813–823. doi:10.1158/0008-5472.CAN-12-2633
Maire V, Baldeyron C, Richardson M, Tesson B, Vincent-Salomon A, Gravier E, Marty-Prouvost B, De Koning L, Rigaill G, Dumont A, Gentien D, Barillot E, Roman-Roman S, Depil S, Cruzalegui F, Pierre A, Tucker GC, Dubois T (2013) TTK/hMPS1 is an attractive therapeutic target for triple-negative breast cancer. PLoS One 8(5):e63712. doi:10.1371/journal.pone.0063712
Maia AR, de Man J, Boon U, Janssen A, Song JY, Omerzu M, Sterrenburg JG, Prinsen MB, Willemsen-Seegers N, de Roos JA, van Doornmalen AM, Uitdehaag JC, Kops GJ, Jonkers J, Buijsman RC, Zaman GJ, Medema RH (2015) Inhibition of the spindle assembly checkpoint kinase TTK enhances the efficacy of docetaxel in a triple-negative breast cancer model. Ann Oncol. doi:10.1093/annonc/mdv293
Chu Z, Lin H, Liang X, Huang R, Tang J, Bao Y, Jiang J, Zhan Q, Zhou X (2015) Association between axillary lymph node status and Ki67 labeling index in triple-negative medullary breast carcinoma. Jpn J Clin Oncol 45(7):637–641. doi:10.1093/jjco/hyv052
Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, Liao X, Iglehart JD, Livingston DM, Ganesan S (2006) X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9(2):121–132. doi:10.1016/j.ccr.2006.01.013
Gyorffy B, Surowiak P, Budczies J, Lanczky A (2013) Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One 8(12):e82241. doi:10.1371/journal.pone.0082241
http://kmplot.com/analysis/index.php?p=service&cancer=breast. Accessed 8 Jan 2016
Li W, Lan Z, Wu H, Wu S, Meadows J, Chen J, Zhu V, Dai W (1999) BUBR1 phosphorylation is regulated during mitotic checkpoint activation. Cell Growth Differ 10(11):769–775
Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H (2008) Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol Cell 32(3):394–405. doi:10.1016/j.molcel.2008.09.017
Klebig C, Korinth D, Meraldi P (2009) Bub1 regulates chromosome segregation in a kinetochore-independent manner. J Cell Biol 185(5):841–858. doi:10.1083/jcb.200902128
Gaudet S, Branton D, Lue RA (2000) Characterization of PDZ-binding kinase, a mitotic kinase. Proc Natl Acad Sci USA 97(10):5167–5172. doi:10.1073/pnas.090102397
Zhao S, Dai J, Zhao W, Xia F, Zhou Z, Wang W, Gu S, Ying K, Xie Y, Mao Y (2001) PDZ-binding kinase participates in spermatogenesis. Int J Biochem Cell Biol 33(6):631–636
Simons-Evelyn M, Bailey-Dell K, Toretsky JA, Ross DD, Fenton R, Kalvakolanu D, Rapoport AP (2001) PBK/TOPK is a novel mitotic kinase which is upregulated in Burkitt’s lymphoma and other highly proliferative malignant cells. Blood Cells Mol Dis 27(5):825–829. doi:10.1006/bcmd.2001.0452
Acknowledgments
Instituto de Salud Carlos III (PI13/01444), ACEPAIN; Diputación de Albacete and CRIS Cancer Foundation (to AO). Ministry of Economy and Competitiveness of Spain (BFU2012-39151), the Instituto de Salud Carlos III through the Spanish Cancer Centers Network Program (RD12/0036/0003) and the Scientific Foundation of the Spanish Association Against Cancer (AECC) and the CRIS Foundation to AP. Our work is partially supported by the European Union through the FEDER program. No competing interests to declare for any author.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Bostjan Seruga receives advisory honorarium from Astellas, Sanofi and Janssen. There is no conflict of interest to declare for the rest of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2016_3720_MOESM1_ESM.ppt
Supplementary Fig. 1 Volcano plot showing genes upregulated and downregulated when comparing basal-like tumors with normal epithelial breast
10549_2016_3720_MOESM2_ESM.ppt
Supplementary Fig. 2 Association of the combined analyses of BUB1β and PDZ binding kinase with overall survival in triple negative tumors (panel A) and basal like tumors (panel B)
10549_2016_3720_MOESM3_ESM.ppt
Supplementary Fig. 3 Expression of BUB1β and PDZ binding kinase in non-basal tumors compared with normal epithelial breast
10549_2016_3720_MOESM4_ESM.pdf
Supplementary Table 1 Genes included in the function of cell cycle and proliferation as reported in the gene set enrichment analyses performed with DAVID Bioinformatics Resources 6.7
10549_2016_3720_MOESM5_ESM.ppt
Supplementary Table 2 Association of the 31 genes included in the cell cycle function with outcome (overall survival and relapse free survival) in basal-like tumors
Rights and permissions
About this article
Cite this article
Ocaña, A., Pérez-Peña, J., Díez-González, L. et al. Transcriptomic analyses identify association between mitotic kinases, PDZ-binding kinase and BUB1, and clinical outcome in breast cancer. Breast Cancer Res Treat 156, 1–8 (2016). https://doi.org/10.1007/s10549-016-3720-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-016-3720-4