Abstract
Recent studies in patients with breast cancer suggest the immune microenvironment influences response to therapy. We aimed to evaluate the relationship between growth rates of tumors in common spontaneous mammary tumor models and immune biomarkers evaluated in the tumor and blood. TgMMTV-neu and C3(1)-Tag transgenic mice were followed longitudinally from birth, and MPA–DMBA-treated mice from the time of carcinogen administration, for the development of mammary tumors. Tumor-infiltrating CD4+ and CD8+ T-cells, FOXP3+ T-regulatory cells, and myeloid-derived suppressor cells were assessed by flow cytometry. Serum cytokines were evaluated in subsets of mice. Fine needle aspirates of tumors were collected and RNA was isolated to determine levels of immune and proliferation markers. Age of tumor onset and kinetics of tumor growth were significantly different among the models. Mammary tumors from TgMMTV-neu contained a lower CD8/CD4 ratio than that of other models (p < 0.05). MPA–DMBA-induced tumors contained a higher percentage of FOXP3+ CD4+ T-cells (p < 0.01) and MDSC (p < 0.001) compared with the other models. Individuals with significantly slower tumor growth demonstrated higher levels of Type I serum cytokines prior to the development of lesions compared to those with rapid tumor growth. Moreover, the tumors of animals with more rapid tumor growth demonstrated a significant increase in the expression of genes associated with Type II immunity than those with slower-progressing tumors. These data provide a foundation for the development of in vivo models to explore the relationship between endogenous immunity and response to standard therapies for breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer growth and progression is heterogeneous and is influenced by the tumor microenvironment. For successful translation of new breast cancer therapies from in vivo models to the clinic, pre-clinical models should reflect the same heterogeneity and diverse tumor-infiltrating cell types as human disease. Many mouse mammary tumor models have been designed to mimic the genetic alterations found in human breast cancer and have been used to better understand cancer development, prevention, and response to therapy [1, 2]. Importantly, these models are immune competent and develop tumors spontaneously over longer periods of time, allowing for the influx of numerous infiltrating immune cell populations which may impact tumor growth and response to therapy.
There are many similarities between certain murine mammary tumor models and human breast cancer including similar pathologic progression from hyperplasia, to carcinoma in situ, to invasive disease [3–5]. Genomic signatures classifying human breast cancer subtypes have also been reproduced in several mouse mammary tumor models. Tumors from the transgenic mouse, TgMMTV-neu, demonstrate genetic patterns similar to human luminal B breast cancers, and C3(1)-Tag mice have a similar gene expression pattern as human basal-like breast cancers [1]. A 7,12-dimethylbenz[a]anthracene (DMBA) chemically induced mammary tumor displays a more heterogeneous genetic expression pattern overlapping both basal-like and luminal human breast cancer subtypes and is hormone receptor positive [6].
Although selected mouse mammary tumors have been genetically characterized, the natural history of tumor initiation and progression, frequency, and localization of primary tumors and metastases, and characterization of the infiltrating T-cells in the tumor microenvironment has not been extensively studied across a population of animals. Studies presented here provide a detailed phenotype of tumor growth and progression in three common models of mammary tumors and identify distinct immune phenotypes that may influence tumor growth.
Methods and materials
Mouse mammary tumor models
TgMMTV-neu mice [strain name: FVB/N-Tg(MMTVneu)202Mul/J, strain #002376] were purchased from the Jackson Laboratory and maintained under strict inbreeding conditions [7]. C3(1)-Tag mice [strain name: FVB-Tg(C3-1-TAg)cJeg/Jeg] male mice (provided by Dr. Jeff Green, NCI) were maintained by breeding to FVB/nJ parental females (Jackson Laboratory, strain #001800) [8]. All animals were housed in a specific pathogen-free facility at the University of Washington. To induce tumors in the MPA–DMBA model, FVB/nJ parental mice were treated with 15 mg of Medroxyprogesterone 17-acetate (MPA) (Sigma) subcutaneously at 6–8 weeks of age, followed by four weekly doses of 1 mg 7,12-Dimethylbenz[a]anthracene (DMBA) (Sigma) by oral gavage [6, 9]. All procedure were done in accordance with the University of Washington Institutional Animal Care and Use Committee guidelines.
Polymerase chain reaction
All C3(1)-Tag mice were tested for the SV40 Tag transgene at 3–4 weeks of age and all nontumor-bearing TgMMTV-neu mice were assessed for the presence of the neu transgene. Genomic DNA was prepared by the “HotSHOT” method from tail or ear tissue samples [10]. Lyophilized primers, for SV40, neu, or nonspecific internal control (Integrated DNA Technologies), were resuspended to a final concentration of 20 µM using sterile water (Supplemental Table S1). PCR reactions were performed using GoTaq Green Master Mix 2X (Promega), according to manufacturer’s instructions for a 25-µl reaction. After preparation, samples were placed in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems) initialized at 94 °C for 3 m, then run for 35 cycles first at 94 °C for 30 s and followed by 55 °C for 30 s, and finally at 72 °C for 60 s. A final extension cycle of 72 °C for 2 m was performed, and the samples were held at 4–10 °C until analysis. Products were run in a Tris–acetate-EDTA (TAE) buffer (in house) on a 1.5 % agarose gel (Genesee Scientific) stained with ethidium bromide (VWR).
Assessment of tumor development and growth rate
TgMMTV-neu and C3(1)-Tag mice were enrolled into an observational study at the time of birth. Sixty-nine TgMMTV-neu, 57 C3(1)-Tag, and 19 MPA–DMBA tumor-induced mice were available for analysis. Two C3(1)-Tag mice were excluded from all analyses described below due to the development of chondral abnormalities resulting in abnormally large pinnae and other defects [8]. Age of tumor onset was calculated as the ([date of first palpable tumor observation]—[mouse date of birth]), ±2 days. Mice were observed for tumor development two to three times per week, by the same operator, from 6 weeks of age until sacrifice. Tumor volumes were calculated from raw measurements by the standard volume calculation for an ellipsoid: [(length) × (width) × (depth) × (π/6)] and reported as mm3. If a mouse developed more than one tumor, tumors were tracked and measured individually. Mice were sacrificed by CO2 asphyxiation once tumor(s) reached a cumulative volume greater than 1,000 mm3, if a tumor became ulcerated, or at 1 year of age, regardless of the presence or absence of palpable mammary tumors. Mice that died without clinically palpable tumors within 47 weeks of age in TgMMTV-neu or 24 weeks of age in C3(1)-Tag mice were excluded from evaluation (n = 7 TgMMTV-neu, n = 12 C3(1)-Tag). This time frame was determined by calculating [Mean age of tumor onset + (2 × Standard Deviation of tumor onset)]. Tumor growth rates were calculated by determining the change in volume between subsequent measurements and dividing by the number of days between the measurements, resulting in a rate value of mm3/day. The tumor kinetics of each mouse was plotted by volume (mm3) per day.
Histologic evaluation of metastatic disease and primary tumors
The brain, liver, lung, and femur were collected in 10 % neutral buffered formalin, and select primary tumors were harvested and frozen in OCT media (VWR). Formalin-fixed samples were processed routinely, embedded in paraffin, stained with hematoxylin and eosin (H&E), and examined by a board-certified veterinary pathologist (PMT) for histologic evidence of metastasis. Intravenous (IV) and parenchymal (outside of vascular system) foci of carcinoma were noted. Frozen samples were sectioned, stained with H&E, and primary mammary tumors were histologically confirmed and classified [11].
Immunohistochemical staining was performed on the Leica Bond Automated Immunostainer using Leica Bond solutions and EDTA antigen retrieval with antibodies as listed in Supplemental Table 1. Murine antibodies anti-SV40 large T antigen and c-erbB2/HER2/neu (Biocare Medical), and Mouse on Mouse HRP-Polymer kits were used to document tissue of origin (BioCare Medical “PromARK” Cat No. MM510G). Secondary detection was performed using Bond Polymer Refine Detection kit (Leica Biosystems, Cat No. DS9800) followed by DAB substrate detection. The sections were counter-stained with Mayer Hematoxylin solution (Newcomer Supply, Cat No. 1202).
Flow cytometric evaluation of immune infiltrates
The immune phenotype of splenocytes and tumor-infiltrating lymphocytes (TIL) were analyzed in all the three models by flow cytometry. TILs were isolated as previously described [12]. Splenocytes were processed for analysis according to our previously published methods [13]. For FOXP3 and CD4/CD8 evaluation, 1 µg anti-mouse CD16/CD32 (BD Pharmingen) was used to block nonspecific binding for 30 min at room temperature; then 0.4 µg each of PE anti-CD3 (BD Pharmingen, clone #145-2C11), PE/Cy5 anti-CD4 (BioLegend, clone #GK1.5), and PE-Cy7 anti-CD8 (eBioscience, clone #53-6.7) were added. After overnight permeabilization of the cells, 1 µg anti-FOXP3 Alexa488 (eBioscience, clone #FJK.16 s) was added for 30 min. For myeloid-derived suppressor cell (MDSC) evaluation, 0.4 µg of GR-1 (BD Pharmingen, clone #RB6-8C5) and anti-mouse CD11b were added and incubated for 30 min at room temperature (eBioscience clone #M1/70). After the appropriate antibodies were applied to each single cell suspension of tumor or spleen for 30 min at room temperature, the stained cells were acquired using FACSCanto flow cytometer (BD Bioscience) and analyzed using FlowJo software (Tree Star Inc.). Results are reported as mean ± SEM of the total percentage of a cell population or ratio of cell quantities, as indicated.
Fine needle aspirates and real-time PCR
mRNA was made from fine needle aspirates (FNA) of TgMMTV-neu mouse tumors growing at different rates and obtained when all tumors were of volumes ranging from 120–300 mm3. RNA was isolated using the RNAqueous-4PCR kit (Ambion). RNA quality was evaluated by both electroporating the RNA in formaldehyde loading dye (evaluating for 28S and 18S ribosomal RNA bands) and by UV absorbance quantification (A 260/A 280 ratio between 1.8 and 2.1) using the nanodrop spectrometer (Thermoscientific). cDNA was generated from 500 ng of RNA using Superscript III transcriptase with oligo DT primers (Life Technologies). Real-time PCR was performed in 384-well thin-walled PCR plates using an ABI Prism 7900 HT (Applied Biosystems) under the following conditions: initial extension 50 °C for 2 min and denaturation 95 °C for 15 s; then 40 cycles of denaturation 95 °C for 15 s; and annealing/extension 60 °C for 1 min. Amplification was performed using the TaqMan Gene Expression master mix (Invitrogen) and array primers (Applied Biosystems), listed in Supplemental Table S1. Data analysis was performed using SDS 2.4.1 (Applied Biosystems). mRNA expression levels were normalized to mouse β-actin using the ΔCT method. \(\hbox{Level of expression}\,=\,2^{-(C{\rm t}\,{\text{target}} - C{{\rm t}}\,{\beta}{\text{-actin}})}\). Ct is the cycle threshold at which the fluorescence signal crosses an arbitrary value.
Serum cytokine analysis
Mouse sera were collected from female TgMMTV-neu mice as previously described [14]. Sera from 3–4 time points immediately prior to tumor development were analyzed for mice with significantly faster tumor progression compared to those with significantly slower tumor progression within a population (n = 8 mice). Thirty-two cytokines (Supplemental Table 2) were measured in duplicate using a Milliplex kit (Millipore) on a Luminex instrument (Qiagen) according to the manufacturer’s instruction.
Statistical analysis
Graphs, linear regression analysis, and ANOVA comparisons were completed using GraphPad Prism v5.03 software. A one-way ANOVA with Tukey’s post-test was used for comparisons of one variable between the three models, and a Two-Way ANOVA with Bonferroni’s post-test was used for grouped comparisons between the three models. A Student’s unpaired t test was used in rtPCR analysis, with a Welch’s correction applied when appropriate. Significance was considered at p < 0.05 for all statistical tests.
Results
Mouse mammary tumor models demonstrated significant differences in the age of tumor onset
The mean age of spontaneous tumor development across the models was variable. The mean age for the development of palpable tumor in the TgMMTV-neu mice was 35.1 ± 5.9 weeks (95 % CI: 33.6, 36.6) (Fig. 1A). The C3(1)-Tag mice developed tumors at a mean age of 18.5 ± 2.83 weeks (95 % CI: 17.7, 19.2) (Fig. 1B). Tumor development in the MPA–DMBA-induced model occurred at 10.3 ± 2.97 weeks after MPA administration (95 % CI: 8.91, 11.8) (Fig. 1C). The TgMMTV-neu mice had significantly later-onset tumors compared to both C3(1)-Tag and MPA–DMBA mice (p < 0.001). In addition, the age at tumor onset of C3(1)-Tag mice was significantly later than the time-to-tumor onset in the MPA–DMBA-induced model (p < 0.001). The age range of tumor onset in C3(1)-Tag mice was more uniform than that of the TgMMTV-neu strain; the majority of C3(1)-Tag mice developed tumors ± 5.7 weeks of the mean onset age, whereas the majority of TgMMTV-neu mice developed tumors ± 11.8 weeks of the mean onset age. All MPA–DMBA-induced mice developed tumors within 16 weeks of MPA administration (Fig. 1C). Of note, 7 of 69 TgMMTV-neu (10 %) and 1 of 57 C3(1)-Tag (2 %) did not develop palpable tumors during the observation period, despite the documented presence of the appropriate transgene (Fig. 1A, B).
Mouse mammary tumor models demonstrated significant differences in the age of tumor onset. Shown is the number of mice, y-axis, at age in weeks at first detection of palpable tumor (black bars) or sacrifice with no detectable tumor (white bars), x-axis, for models, A TgMMTV-neu (n = 69), B C3(1)-Tag (n = 57), and C MPA–DMBA induced (n = 19) as measured from administration of MPA (day 0)
Marked heterogeneity in the rate of tumor growth is observed between genetically engineered mouse mammary tumor models and is not associated with age of tumor onset
TgMMTV-neu primary tumors progressed more slowly, with an average growth rate of 73.6 mm3/week (Fig. 2A), compared to C3(1)-Tag tumors, 150.8 mm3/week (p < 0.01) (Fig. 2B), and MPA–DMBA-induced mice, 163.9 mm3/week (p < 0.01) (Fig. 2C). There was no significant difference in the rates of tumor growth between the MPA–DMBA-induced lesions and the C3(1)-Tag tumors (p = 0.73). A linear regression analysis comparing median growth rate with age of tumor onset in each of the mouse models demonstrated that age of tumor onset is not associated with rate of tumor growth within these populations; TgMMTV-neu; p = 0.286, R 2 = 0.0190 (Fig. 2D), C3(1)-Tag; p = 0.274, R 2 = 0.0221 (Fig. 2E), and MPA–DMBA; p = 0.268, R 2 = 0.0715 (Fig. 2F). Further, the rate of tumor growth was not associated with date of birth in the two spontaneous models (p = 0.88 TgMMTV-neu, p = 0.68 C3(1)-Tag). The association of tumor growth rate with date of birth was not evaluated in the MPA–DMBA model, as all mice were commercially provided and had the same date of birth reported.
Marked heterogeneity in the rate of tumor growth is observed between genetically engineered mouse mammary tumor models and is not associated with age of tumor onset. Shown is the median tumor growth rate (mm3/week), y-axis, by individual animal ranked from the lowest (1) to the highest rate (n), y-axis, for, A TgMMTV-neu (n = 62), B C3(1)-Tag (n = 56), and C MPA–DMBA induced (n = 19). Median growth rate (mm3/week), y-axis, related to age at tumor onset, x-axis for D TgMMTV-neu, E C3(1)-Tag, and F MPA–DMBA-induced mammary tumors. Solid lines show linear regression of median growth rate on age of tumor onset
Tumor growth rates can be significantly different within a specific mouse mammary tumor model
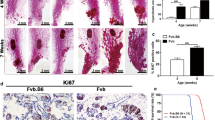
The rates of growth, calculated from tumor onset through sacrifice, between individual animals within a mouse model were variable. The growth rates between the slowest 10 % and fastest growing 10 % of tumors for TgMMTV-neu (Fig. 3A, B) and C3(1)-Tag (Fig. 3C, D) differed significantly (p = 0.0016 and p < 0.0001 respectively). In the TgMMTV-neu FNAs, we observed significant differences in tumor gene expressions between mice eventually categorized as having slower versus faster growing tumors for Cyclin D1 (p = 0.0105) and SatB1 (p = 0.0012) with higher expression of these proliferation markers being found in rapidly progressing disease (Fig. 3E, F). No significant difference was found in Ki67 (Supplemental Fig. S1A).
Tumor growth rate can be significantly different within a specific mouse mammary tumor model. Shown are the kinetics of tumor growths in volume (mm3), y-axis, from the day of tumor onset, x-axis for the most rapid 10 % (solid line, n = 6 per model) and the slowest growing 10 % (dotted line, n = 6 per model) of mammary tumors in A TgMMTV-neu and C C3(1)-Tag mouse models. Median growth rates (mm3/week), y-axis of these same mice are shown by group, x-axis for B TgMMTV-neu and D C3(1)-Tag. ∆CT, y-axis, for n = 5 fast and n = 5 slow-growing tumors, x-axis, from TgMMTV-neu FNA samples measured for E Cyclin D1 and F SatB1. Samples represent tumor volumes of 120–300 mm3 upon collection from mice. *p < 0.05, **p < 0.01
Models vary significantly in the number of mammary tumors, but have a similar incidence and pattern of metastasis
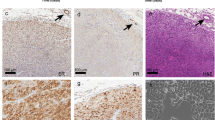
In the TgMMTV-neu model, 10 % of mice did not develop palpable mammary tumors, 49 % of TgMMTV-neu mice developed one palpable mammary tumor, 19 % developed two tumors, and 22 % developed more than three palpable tumors during their lifespan (Fig. 4A). C3(1)-Tag mice had significantly more tumors than TgMMTV-neu mice (p < 0.001) and MPA–DMBA-induced mice (p < 0.01), with 2 % of mice developing no palpable tumor; 28 % developing one tumor; 24 % two tumors; and 45 % three or more mammary tumors in their lifespan (Fig. 4A). The majority of MPA–DMBA-induced mice (70 %) developed only one primary tumor, whereas 15 % developed two tumors and 15 % three or more tumors in their lifespan (Fig. 4A). No statistical differences in intravascular (IV) and parenchymal metastases were noted in the models. The lung was the predominant metastatic site in all three mouse models, with 28 % IV metastases and 3 % parenchymal metastases in TgMMTV-neu, 13 % IV and 7 % parenchymal in C3(1)-Tag, and 6 % IV and 17 % parenchymal in MPA–DMBA-induced mice (Fig. 4B and Supplemental Table 3). Mammary origin of the metastatic foci was confirmed via immunohistochemical staining and morphology (Supplementary Fig. S2). As DMBA is systemically administered, other non-mammary organs developed primary neoplastic lesions including the lung (11 %) and stomach (5 %). The animal with the suspected gastric carcinoma had a small IV and parenchymal foci of undifferentiated carcinoma, and the site of origin could not be determined by morphology but was excluded as primary lung by prosurfactant-c immunohistochemistry.
Models vary significantly in the number of mammary tumors, but have a similar incidence and pattern of metastasis. Number of mammary tumors at the time of sacrifice, y-axis; 0 (white), 1 (light grey), 2 (dark grey), or more than 3 (black) for, x-axis, A TgMMTV-neu (n = 69), C3(1)-Tag (n = 57), and MPA–DMBA induced (n = 19). B Percent incidence of tumor in lung, y-axis, for TgMMTV-neu (n = 29), C3(1)-Tag (n = 30), and MPA–DMBA induced (n = 18) by origin and location, x-axis; metastatic mammary IV (white), metastatic mammary parenchymal (light grey), primary lung parenchymal (dark grey) or metastatic poorly differentiated carcinoma, favoring gastric origin (black). **p < 0.01, ****p < 0.0001
Mouse mammary tumor models demonstrate distinct tumor immune phenotypes
We observed a significantly lower CD8+/CD4+ ratio of CD3+ cells in the TgMMTV-neu model compared to both the MPA–DMBA-induced model (p < 0.001) and the C3(1)-Tag model (p < 0.05). There was significantly higher CD8+/CD4+ ratio in the MPA–DMBA-induced model than in the C3(1)-Tag mice (p < 0.05) (Fig. 5A). No significant difference was found in the CD8+/CD4+ ratio of CD3+ in the spleens of the three models (Fig. 5B). Increased percentages of FOXP3+ Treg cells have been found in several types of cancers [15–17]. The MPA–DMBA-induced mouse model demonstrated a significantly higher % of FOXP3+ CD4+ cells compared to TgMMTV-neu (p < 0.01) and C3(1)-Tag mice (p < 0.01) (Fig. 5C). No difference in the FOXP3+ CD4+ cells were detected between the C3(1)-Tag and TgMMTV-neu tumors or in the spleens of all three models (Fig. 5C, D). MDSC frequently accumulate as tumor burden increases resulting in immune suppression [18]. Although there were no significant differences in MDSC in the spleen of the three mouse models (Fig. 5F), we observed significantly higher MDSC levels in the tumors of MPA–DMBA-induced mice compared to the TgMMTV-neu (p < 0.001) and the C3(1)-Tag mice (p < 0.01) (Fig. 5E). There was no significant difference in MDSC between the TgMMTV-neu and C3(1)-Tag mice. A representative result from flow cytometry analyses in the three mouse models is shown in Supplementary Fig. S3.
Mouse mammary tumor models demonstrate distinct tumor immune phenotypes. CD4+/CD8+ ratio of CD3+, y-axis, in A tumors and B spleens; % FOXP3+CD4+ of all CD4+, y-axis, in C tumors and D spleens; %Gr1+CD11b+ of total cells, y-axis, in E tumors and F spleens; shown for TgMMTV-neu (n = 12), C3(1)-Tag (n = 11), and MPA–DMBA induced (n = 7), x-axis. *p < 0.05, **p < 0.01, ***p < 0.001
In addition, an analysis of immune infiltrates in spleen and TIL as related to tumor volume was performed. The only positive relationship was found in the C3(1)-Tag model. The total CD4+ T-cell number (p = 0.038, R 2 = 0.397) (Supplemental Fig. S4.A) and level of MDSC (p = 0.029, R 2 = 0.426) (Supplemental Fig. S4.B) proved to be significantly higher with greater tumor volumes. Immune infiltrates in spleen and TIL were not significantly correlated with tumor volume for any cell type studied in the other models (all p > 0.05).
Significant elevation of Type I serum cytokines and tumor expression of genes associated with Type II immunity define slow and rapid progressors, respectively
Several Th1 cytokines, including IL-2 (mean ± SE: slow 0.6 ± 0.2, vs. rapid, 0.1 ± 0.1; p = 0.017), TNF-a (3.7 ± 0.2 vs. 2.9 ± 0.3; p = 0.038), and M-CSF (7.0 ± 0.7 vs. 4.7 ± 0.8; p = 0.044) were significantly elevated in mice with slower-progressing tumors than those with rapidly progressing tumors. Two chemo-attractant cytokines, macrophage inflammatory protein 2 (MIP-2; 102.9 ± 11.0 vs. 55.1 ± 8.5; p = 0.002) and monocyte chemotactic protein-1 (MCP-1; 11.7 ± 1.0 vs. 7.5 ± 0.9; p = 0.005) were also significantly higher in slower-growing tumors. In contrast, IL-1-a was significantly decreased in the slower-progressing compared to rapidly progressing tumors (416.5 ± 71.7 vs. 642.3 ± 63.6; p = 0.027) (Supplemental Table 2). PCR analysis demonstrated significantly higher expression of immunosuppressive markers TGFβ (p < 0.01) and Gata3 (p < 0.05) in the more rapidly progressing tumors (Supplemental Fig. S1B, C). In addition, there was a trend toward greater expression of FOXP3 in the fast compared to slow-growing tumors (p = 0.099) (Supplemental Fig. S1D).
Discussion
We evaluated the natural evolution of spontaneous mouse mammary tumors in three commonly used murine models of breast cancer: TgMMTV-neu, C3(1)-Tag, and MPA–DMBA-induced mammary tumors from in the FVB/N strain. The tumor growths were heterogenic both within and across models. The most rapidly progressive tumors within a single model demonstrated significant upregulation of genes associated with proliferation and Type II immunity compared to slower-progressing lesions, whereas Type I cytokines were elevated in the serum of individuals with slower-progressing tumors. Finally, the levels and phenotype of tumor-infiltrating immune cells differed significantly between models.
Mice engineered to develop cancer from a limited series of genetic alterations have been thought not to adequately reflect the heterogeneity of human malignancy. Recent studies have demonstrated the complex heterogeneity of the genomic landscape within individual cancer patients. A detailed genomic analysis of human renal cell carcinomas revealed significant heterogeneity of mutations across several samples taken from the same lesion within a single individual [19]. Analyses evaluating mammary tumors which arose in the MMTV-Myc model have demonstrated significant genetic heterogeneity and histologic differences in tumors derived from the same breeding colony [20]. A recent investigation reported that the genetic profiles of primary tumors, residual tumors remaining after chemotherapy, and lung metastases were quite different from each other in the MMTV-PyMT mammary tumor model [21]. The varied kinetics of tumor growth we observed within the transgenic models would suggest that there is a genetic or environmental heterogeneity influencing tumor progression. These models may be useful for assessing response to therapies in both primary lesions and metastasis. The longer growth rate we observed could potentially allow for the assessment of the development of resistance to novel therapies.
With standard approaches for therapeutic modeling in mice, xenographs in immunodeficient mice or syngeneic implants, tumors do not have robust immune infiltrates. Investigations have demonstrated that tumor cell lines implanted subcutaneously into a syngeneic host have significantly fewer infiltrating immune cells than tumors that arise spontaneously in genetically engineered mice [22]. Recent studies suggest that immune cell infiltrates, present in breast cancers, affect prognosis as well as response to treatment. In an evaluation of over 1,000 patients, the percent of intratumoral lymphocytes was an independent predictor for the development of a pathologic complete response [23]. Moreover, high levels of infiltrating CD4+FOXP3+ Tregs are associated with a poor clinical outcome in breast cancer [24]. Several subsequent studies have validated that specific tumor immune infiltrates significantly impact disease-free and overall survival [25–27]. The strongest evidence for a beneficial role of immune infiltration in human breast cancer is seen in ER-negative tumors with increased CD8+ TILs conferring a significantly improved prognosis and superior response to therapy rivaling that of ER-positive tumors [27–29]. Similar to what is seen in the human, the intratumoral CD8/CD4 ratio is greater in C3(1)-Tag tumors, which are similar to triple-negative breast tumors, than in the TgMMTV-neu lesions, which are similar to luminal B breast tumors (p < 0.05). The importance of CD8+ immune infiltration on prognosis is less apparent in hormone receptor-positive disease; however the presence of increased FOXP3+ T-cells predicts a worse prognosis [24]. The hormone receptor-positive model we studied had the highest levels of both FOXP3+ and MDSCs. Gene expression data obtained from low-volume tumors suggest that immune regulation may play a role in modulating the diverse growth rates observed in these murine mammary tumors. The significant variation in both adaptive and innate immune cell infiltrates between models may provide a tool for addressing the role of immunity in modulating clinical response to standard and experimental therapies.
Spontaneous mammary tumor models offer several benefits in translational oncology: the heterogeneity of disease more closely mirrors cancer patients and the slower rate of disease progression and multiplicity of lesions provides a unique model system to address the roles of drug selection and immune modulation in treatment response or failure. Studies, such as the one described here, should provide a baseline for the development of experimental designs for the evaluation of the role of immunity in influencing the response to breast cancer therapy.
References
Herschkowitz JI, Simin K, Weigman VJ, Mikaelian I, Usary J, Hu Z, Rasmussen KE, Jones LP, Assefnia S, Chandrasekharan S, Backlund MG, Yin Y, Khramtsov AI, Bastein R, Quackenbush J, Glazer RI, Brown PH, Green JE, Kopelovich L, Furth PA, Palazzo JP, Olopade OI, Bernard PS, Churchill GA, Van Dyke T, Perou CM (2007) Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol 8(5):R76. doi:10.1186/gb-2007-8-5-r76
Usary J, Zhao W, Darr D, Roberts PJ, Liu M, Balletta L, Karginova O, Jordan J, Combest A, Bridges A, Prat A, Cheang MC, Herschkowitz JI, Rosen JM, Zamboni W, Sharpless NE, Perou CM (2013) Predicting drug responsiveness in human cancers using genetically engineered mice. Clin Cancer Res 19(17):4889–4899. doi:10.1158/1078-0432.CCR-13-0522
Shoushtari AN, Michalowska AM, Green JE (2007) Comparing genetically engineered mouse mammary cancer models with human breast cancer by expression profiling. Breast Dis 28:39–51
Vargo-Gogola T, Rosen JM (2007) Modelling breast cancer: one size does not fit all. Nat Rev Cancer 7(9):659–672. doi:10.1038/nrc2193
Van Dyke T, Jacks T (2002) Cancer modeling in the modern era: progress and challenges. Cell 108(2):135–144
Lanari C, Lamb CA, Fabris VT, Helguero LA, Soldati R, Bottino MC, Giulianelli S, Cerliani JP, Wargon V, Molinolo A (2009) The MPA mouse breast cancer model: evidence for a role of progesterone receptors in breast cancer. Endocr Relat Cancer 16(2):333–350. doi:10.1677/ERC-08-0244
Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ (1992) Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA 89(22):10578–10582
Maroulakou IG, Anver M, Garrett L, Green JE (1994) Prostate and mammary adenocarcinoma in transgenic mice carrying a rat C3(1) simian virus 40 large tumor antigen fusion gene. Proc Natl Acad Sci USA 91(23):11236–11240
Aldaz CM, Liao QY, LaBate M, Johnston DA (1996) Medroxyprogesterone acetate accelerates the development and increases the incidence of mouse mammary tumors induced by dimethylbenzanthracene. Carcinogenesis 17(9):2069–2072
Truett GE, Heeger P, Mynatt RL, Truett AA, Walker JA, Warman ML (2000) Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 29(1):52–54
Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wakefield LM, Ward JM, Green JE (2000) The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 19(8):968–988
Knutson KL, Dang Y, Lu H, Lukas J, Almand B, Gad E, Azeke E, Disis ML (2006) IL-2 immunotoxin therapy modulates tumor-associated regulatory T cells and leads to lasting immune-mediated rejection of breast cancers in neu-transgenic mice. J Immunol 177(1):84–91
Park KH, Gad E, Goodell V, Dang Y, Wild T, Higgins D, Fintak P, Childs J, Dela Rosa C, Disis ML (2008) Insulin-like growth factor-binding protein-2 is a target for the immunomodulation of breast cancer. Cancer Res 68(20):8400–8409. doi:10.1158/0008-5472.CAN-07-5891
Mao J, Ladd J, Gad E, Rastetter L, Johnson MM, Marzbani E, Childs JS, Lu H, Dang Y, Broussard E, Stanton SE, Hanash SM, Disis ML (2014) Mining the pre-diagnostic antibody repertoire of TgMMTV-neu mice to identify autoantibodies useful for the early detection of human breast cancer. J Transl Med 12:121. doi:10.1186/1479-5876-12-121
Woo EY, Chu CS, Goletz TJ, Schlienger K, Yeh H, Coukos G, Rubin SC, Kaiser LR, June CH (2001) Regulatory CD4(+)CD25(+) T cells in tumors from patients with early-stage non-small cell lung cancer and late-stage ovarian cancer. Cancer Res 61(12):4766–4772
Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen L, Zou W (2004) Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 10(9):942–949. doi:10.1038/nm1093
Wang RF, Peng G, Wang HY (2006) Regulatory T cells and toll-like receptors in tumor immunity. Semin Immunol 18(2):136–142. doi:10.1016/j.smim.2006.01.008
Ostrand-Rosenberg S (2010) Myeloid-derived suppressor cells: more mechanisms for inhibiting antitumor immunity. Cancer Immunol Immunother 59(10):1593–1600. doi:10.1007/s00262-010-0855-8
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366(10):883–892. doi:10.1056/NEJMoa1113205
Andrechek ER, Cardiff RD, Chang JT, Gatza ML, Acharya CR, Potti A, Nevins JR (2009) Genetic heterogeneity of Myc-induced mammary tumors reflecting diverse phenotypes including metastatic potential. Proc Natl Acad Sci USA 106(38):16387–16392. doi:10.1073/pnas.0901250106
Franci C, Zhou J, Jiang Z, Modrusan Z, Good Z, Jackson E, Kouros-Mehr H (2013) Biomarkers of residual disease, disseminated tumor cells, and metastases in the MMTV-PyMT breast cancer model. PLoS ONE 8(3):e58183. doi:10.1371/journal.pone.0058183
Lu H, Knutson KL, Gad E, Disis ML (2006) The tumor antigen repertoire identified in tumor-bearing neu transgenic mice predicts human tumor antigens. Cancer Res 66(19):9754–9761. doi:10.1158/0008-5472.CAN-06-1083
Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, von Torne C, Weichert W, Engels K, Solbach C, Schrader I, Dietel M, von Minckwitz G (2010) Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol 28(1):105–113. doi:10.1200/JCO.2009.23.7370
Bates GJ, Fox SB, Han C, Leek RD, Garcia JF, Harris AL, Banham AH (2006) Quantification of regulatory T cells enables the identification of high-risk breast cancer patients and those at risk of late relapse. J Clin Oncol 24(34):5373–5380. doi:10.1200/JCO.2006.05.9584
Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, de Azambuja E, Quinaux E, Di Leo A, Michiels S, Piccart MJ, Sotiriou C (2013) Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J Clin Oncol 31(7):860–867. doi:10.1200/JCO.2011.41.0902
Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR (2011) Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J Clin Oncol 29(15):1949–1955. doi:10.1200/JCO.2010.30.5037
Baker K, Lachapelle J, Zlobec I, Bismar TA, Terracciano L, Foulkes WD (2011) Prognostic significance of CD8+ T lymphocytes in breast cancer depends upon both oestrogen receptor status and histological grade. Histopathology 58(7):1107–1116. doi:10.1111/j.1365-2559.2011.03846.x
Liu S, Lachapelle J, Leung S, Gao D, Foulkes WD, Nielsen TO (2012) CD8+ lymphocyte infiltration is an independent favorable prognostic indicator in basal-like breast cancer. Breast cancer Res BCR 14(2):R48. doi:10.1186/bcr3148
Lee HJ, Seo JY, Ahn JH, Ahn SH, Gong G (2013) Tumor-associated lymphocytes predict response to neo adjuvant chemotherapy in breast cancer patients. J Breast Cancer 16(1):32–39. doi:10.4048/jbc.2013.16.1.32
Acknowledgments
The authors wish to thank the staff of the University of Washington Comparative Pathology Program/Histology and Imaging Core Research Laboratory especially Brian Johnson, Erin McCarty, and Cara Appel for their contributions to slide production as well as histochemical and immunohistochemical staining. This work was supported by the National Cancer Institute, U01 CA141539, the Department of Defense Grant, W81XWH-11-1-0760, and the National Cancer Institute contract, N01-CN-53300/WA#10. Mary L. Disis was supported by The Athena Distinguished Professorship for Breast Cancer Research.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standards
All experiments performed for this manuscript comply with the current laws of the country in which they were performed.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ekram Gad and Lauren Rastetter have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gad, E., Rastetter, L., Slota, M. et al. Natural history of tumor growth and immune modulation in common spontaneous murine mammary tumor models. Breast Cancer Res Treat 148, 501–510 (2014). https://doi.org/10.1007/s10549-014-3199-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-3199-9