Abstract
Increasing evidence suggests that dysfunction of histone lysine demethylase is associated with abnormal chromatin remodeling and gene silencing, contributing to breast tumorigenesis. In silico analysis shows that the newly identified histone demethylase lysine-specific demethylase 2 is highly expressed in breast cancer, especially in invasive tumors. However, it is currently unknown how LSD2 regulates chromatin remodeling and gene expression regulation in breast cancer. Using short hairpin RNA, we stably knocked down LSD2 (LSD2-KD) in MDA-MB-231 breast cancer cells. LSD2-KD led to accumulation of H3K4me1/2 without changing methylation levels of other key histone lysine residues, suggesting that LSD2 acts as a bona fide H3K4 demethylase in breast cancer cells. LSD2-KD resulted in decreased colony formation and attenuated global DNA methylation in MDA-MB-231 cells. Additionally, treatment with the DNMT inhibitor, 5-aza-deoxycytidine (DAC), synergistically increased mRNA expression of aberrantly silenced genes important in breast cancer development, including PR, RARβ, ERα, SFRP1, SFRP2, and E-cadherin in LSD2-KD cells. Furthermore, LSD2-KD cells are more susceptible to cell death than scramble controls, and combined treatment with tranylcypromine, an LSD2 inhibitor, and DAC resulted in synergistic growth inhibition of breast cancer cells. DNMT inhibition by DAC in LSD2-KD cells led to internucleosomal DNA fragmentation, enhanced PARP cleavage and increased sub-G1 apoptotic cell population. These results demonstrate an important role for LSD2 in regulation of DNA methylation and gene silencing in breast cancer, and suggest that inhibition of LSD2 in combination with DNA methyltransferase inhibition represents a novel approach for epigenetic therapy of breast cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The groundbreaking discovery that histones can be demethylated highlights the pervasive and dynamic nature of post-translational modifications of chromatin. The reversibility of histone methylation by histone demethylases provides a novel and promising target for therapeutic intervention. The amine oxidase family of histone demethylases consists of LSD1 (KDM1A or AOF2) and LSD2 (KDM1B or AOF1). Both are FAD-dependent and capable of demethylating mono- and di-methylated lysine 4 of histone H3 (H3K4me1/2) [1–3].
While LSD1 and LSD2 share 33 % amino acid sequence homology in the amine oxidase domain [3] and target the same enzymatic substrate (H3K4me1/2), recent evidence indicates distinct functions for each. LSD2 lacks a tower domain which LSD1 uses for protein–protein interactions. LSD2 also contains an N-terminal CW-type zinc finger domain which is absent in LSD1 [3]. LSD1 is largely associated with promoter regions of genes [4], while LSD2 associates more with coding regions [5]. LSD2 does not bind histone deacetylases (HDACs) or the REST corepressor 1, while LSD1 is known to bind both [3, 4, 6]. Therefore, LSD2 and LSD1 have distinct functions. However, the precise role of LSD2 in breast cancer tumorigenesis has not been fully investigated.
A recent study has reported that LSD2 is needed for de novo DNA methylation during embryonic development [7]. Abnormal DNA methylation is frequently associated with dysregulated histone activity that contributes to aberrant gene silencing in breast tumors and is accompanied by poor prognosis [8]. Silencing of important genes such as the nuclear receptors estrogen receptor, progesterone receptor, and retinoid acid receptor β (RARβ), and tumor suppressor genes such as E-cadherin (CDH1) and secreted frizzled related proteins promote breast cancer progression and resistance to targeted therapy [4, 9–12].
LSD1 inhibition also leads to reexpression of silenced genes, and significantly attenuates breast cancer cell growth [12–14]. Crosstalk between HDACs and other key chromatin modifiers is supported by our recent work showing that LSD1 interacts with HDACs to regulate gene expression and breast cancer cell growth [15]. On the basis of these recent findings, we addressed the role of LSD2 in breast cancer and its crosstalk with other chromatin modifiers. We examined the functional link between LSD2 and different key epigenetic enzymes in chromatin remodeling, gene transcription, and therapeutic response. We demonstrate for the first time that LSD2-KD leads to increased susceptibility to DAC and enhanced reexpression of silenced genes.
Materials and methods
Reagents and cell culture conditions
Cells were maintained in DMEM (Corning cellgro, Tewksbury, MA) with 5 % fetal bovine serum (complete media), and 400 μg/ml G418 at 37 °C in a humidified atmosphere with 5 % CO2. Experiments were performed in complete media without G418. Live cell images were taken on a Zeiss Axiovert 40C using a Moticam 2 digital camera and Motic Images Plus 2.0 acquisition software.
LSD2 shRNA treatment and stable cell line generation
MDA-MB-231 or MCF7 cells were transfected with one of 4 short hairpin RNA (shRNA) or a scramble shRNA (SA Biosciences, Valencia, CA) using Attractene (SA Biosciences). Cells were then plated in 10 cm dishes at low density in media containing G418 (800 μg/ml for MDA-MB-231, and 600 μg/ml for MCF7) for selection. Single-cell colonies were tested for LSD2 knockdown by quantitative PCR. Two colonies expressing the lowest LSD2 mRNA levels were developed from shRNA 2 and shRNA 3 and used throughout this manuscript.
Western blotting
Nuclear extracts were prepared using the NE-PER Kit (Thermo Scientific, Rockford, IL), separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted using LSD1, H3K4me1, H3K4me2, H3K4me3, H3K9me1, H3K9me2, H3K9me3, AcH3K9, H3K27me1, H3K36me2, H3K27me3, AcH4K16, H3, PCNA (Millipore, Billerica, MA), LSD2 (Novus Biologicals, Littleton, CO), β-actin (Abgent, San Diego, CA), DNMT1 (provided by Dr. William Nelson at Johns Hopkins University), PR (Santa Cruz Biotechnologies, Santa Cruz, CA), and PARP (Active Motif, Carlsbad, CA) specific antibodies. Li-Cor (Lincoln, NE) Odyssey Blocking Buffer and secondary antibodies were used. Bands were scanned on the Li-Cor CLx Imager and quantified using Image Studio 2.1 Software (Li-Cor).
Colony formation
Scramble and LSD2-KD MDA-MB-231 cells were plated in 6-well plates (500 cells/well) in complete media. After 14 days, cells were stained with crystal violet (Sigma), dried overnight, and colonies were counted.
Demethylation assays
Nuclear extracts were used in two ELISA-like assays MethylFlash 5-methylcytosine (5-mC) Quantification Kit (colorimetric, sensitivity = 1 nM of 5-mC), and Epigenase LSD1 Demethylase activity Kit (fluorometric, sensitivity = 2 ng purified protein) (Epigentek, Farmingdale, NY) according to manufacturer’s recommendations.
DNA fragmentation
After treatment with DAC (5-aza-2′-deoxycytidine, Decitabine, Cayman Chemical, Ann Arbor, MI) for 96 h, DNA ladder fragments were prepared as described previously [16], and analyzed by agarose (2 %) gel electrophoresis.
Propidium iodide staining
Cells were trypsinized, fixed in 70 % ethanol, centrifuged, and washed. The cell pellet was then resuspended in 50 μg/ml propidium iodide (Sigma) containing 100 μg/ml RNaseI (Roche, Indianapolis, IN). Samples were analyzed on the Accuri C6 (BD Biosciences, San Jose, CA) in the University of Pittsburgh Cancer Institute Cytometry Facility. BD CSampler Software was used to assess cell cycle.
RNA analysis
Total RNA was isolated from cells using the RNeasy Kit (Qiagen, Valencia, CA) according to manufacturer’s specifications and treated with DNaseI (Roche). 3 μg of RNA was reverse transcribed and quantitative real-time PCR was performed using 3 μl of cDNA as previously described using Taqman probes (ABI) (Suppl Table 1).
Crystal violet and drug combination index analysis
To obtain IC50 values for chemotherapeutic drugs, scramble or LSD2-KD cells were plated in 96-well plates and treated with increasing doses of DAC, SAHA (Vorinostat, Cayman Chemical), carboplatin, 4-OH-tamoxifen, lapatinib, doxorubicin, ABT-888, paclitaxel, or TCP (Sigma, St. Louis, MO). To obtain the combination index (CI), parental MDA-MB-231 cells were treated with increasing doses of DAC or TCP for 120 h and IC50 values for each were obtained after 4 experiments (DAC = 5.3 µM, TCP = 582.3 µM, ratio = 1:109.87). Combinatorial doses of 4×, 2×, 1×, 0.75×, 0.5×, and 0.25× were calculated using the IC50 values. Cells were then simultaneously treated with TCP and DAC using the combinatorial doses for 120 h. Cells were stained with crystal violet, dried overnight, crystals were dissolved in 0.1 M sodium citrate, and read at 450 nm. Calcusyn software (Biosoft, Cambridge, UK) was used to calculate IC50 and CI values. The Chou–Talalay median effect/CI model was used to determine synergy, additivity, or antagonism of combination therapy [17].
Statistics
GraphPad Prism 5.0 or Excel software was used to determine the statistical differences between various experimental and control groups through a one-way or two-way analysis of variance or Student’s t test.
Results
Inhibition of LSD2 reduces colony formation of breast cancer cells
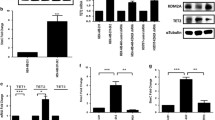
The Oncomine database shows that LSD2 is highly expressed in invasive breast cancer compared to normal tissue in two data sets (www.oncomine.org), and is altered in 11 % of human breast cancers (www.cbioportal.org) (Supp Fig 1) raising the question of whether enhanced LSD2 expression in breast cancer plays a role in tumor cell growth. To address this question, we developed stable LSD2-KD cells using shRNA in the breast cancer cell line, MDA-MB-231. LSD2 targeting shRNA effectively reduced endogenous LSD2 protein expression over 90 % without effecting LSD1 protein levels in two LSD2-KD colonies which arose from single cells (Fig. 1a). The two LSD2-KD clones chosen for use throughout these studies had the lowest levels of LSD2 protein and were created using two different shRNAs with unique sequences. In a 2D colony formation assay loss of LSD2 led to a 25–50 % reduction in colonies formed compared to scramble control MDA-MB-231 cells in both LSD2-KD clones (Fig. 1b, c). These results demonstrate a survival promoting role for LSD2 in breast cancer cells.
Stable LSD2-KD leads to decreased colony formation. a A representative immunoblot of MDA-MB-231 scramble control and LSD2-KD nuclear extracts probed for LSD2, LSD1, and β-actin protein is shown. b LSD2-KD and scramble control cells were allowed to form colonies for 2 weeks and colonies were counted. Bars represent the mean of 3 experiments ± SEM (t test, *** = p < 0.01). c A representative image from 3 colony formation assays for each clone
LSD2 specifically demethylates H3K4me1/2 in breast cancer cells
Our recent study has shown that transient suppression of LSD2 mRNA expression by siRNA in MDA-MB-231 cells increases nuclear levels of H3K4me2, but fails to alter the level of AcH3K9 [12]. In another study using dendritic cells, LSD2 has been shown to demethylate H3K9me2 at specific gene promoters [18]. To investigate the impact of stable LSD2 deficiency on nuclear levels of H3K4me1/2 and other histone marks two approaches were utilized; an ELISA-like assay to determine the demethylase activity of LSD2-KD and scramble nuclear extracts, and a western blot analysis to assess global histone marks. The in vitro ELISA-like assay quantitated H3K4 demethylation by LSD2 using short 21 amino acid H3K4me peptides as substrates. This assay demonstrated that significantly less H3K4 is demethylated by nuclear protein lysates from LSD2-KD cells compared to scramble control counterparts (Fig. 2a). Western blot analysis confirmed this finding showing that LSD-KD led to a significant increase in both H3K4me1 and H3K4me2, without affecting global H3K9me2 levels (Fig. 2b). The increased H3K4 methylation in LSD2-KD cells was accompanied by a 30 % reduction in Acetyl H3K9, a chromatin mark associated with active transcription. H3K27me2, H3K36me1 & 3 proteins were undetectable, while levels of H3K4me3, H3K9me1, H3K9me2, H3K9me3, H3K27me1, H3K27me3, AcH4K16, and H3K36me2 remain unchanged, suggesting that LSD2 acts as a bona fide demethylase specific to H3K4me1/2 in breast cancer cells (Fig. 2b, c).
Stable LSD2-KD is associated with H3K4 methylation and has little effect on the Jmj-C histone demethylases or HDAC activity. a MDA-MB-231 scramble or LSD2-KD nuclear extracts were used as input for an ELISA-like H3K4 demethylation assay (Epigentek). Bars represent the mean of 3 experiments ± SEM (t test, *** = p < 0.01). b Nuclear extracts from scramble or LSD2-KD MDA-MB-231 cells were probed using the indicated antibodies by western blot. Images depicted are representative of 3 independent experiments. c Image Studio software was used to quantitate western blot band intensity. Bars represent the mean of 3 experiments ±SEM (t test, * = p < 0.05). d Jmj mRNA expression was assessed by quantitative PCR using specific Taqman probes in scramble and LSD2-KD MDA-MB-231 cells. Bars represent the mean of 3 experiments ±SEM (t test). e Scramble and LSD2-KD MDA-MB-231 cells were treated with increasing doses of SAHA for 72 h and a crystal violet assay was performed. Each point represents the mean of 3 experiments ±SEM
We also examined the effect of LSD2 deficiency on mRNA expression of JmjC-domain-containing histone demethylases which catalyze Fe and α-ketoglutarate-dependent histone demethylation. JARID1B (KDM5B) is an H3K4 demethylase specifically targeting H3K4me3/2 [19], while JMJD2B (KDM4B) antagonizes H3K9me3/2 and H3K36me3/2 [20, 21]. Loss of LSD2 has no effect on mRNA expression of these enzymes (Fig. 2d).
Recently, we demonstrated that LSD1 interacts with HDACs in breast cancer cells and stable knockdown of LSD1 repressed mRNA expression of most HDAC isozymes. Additionally, HDAC inhibition led to significant growth inhibition and apoptotic death in LSD1-KD MDA-MB-231 cells [15]. To understand whether LSD2, in concert with LSD1, interacts with HDACs in human breast cancer cells, we examined the effect of LSD2-KD on cellular sensitivity in response to the HDAC inhibitor SAHA, finding similar sensitivity in scramble and LSD2-KD cells (Fig. 2e). These data indicate that LSD2 may not function in association with HDAC activity in the way LSD1 does in breast cancer cells.
Inhibition of LSD2 reduces global DNA methylation
In cancer cells, DNA hypermethylation frequently acts in collaboration with abnormal histone modifications culminating in decreased chromatin activating marks. However, the impact of histone demethylases on DNA methylation in cancer cells has not been fully investigated. We assessed global DNA methylation in LSD2-KD cells using an ELISA-like assay to detect 5-mC levels in genomic DNA. Global DNA methylation was significantly decreased in both LSD2-KD clones to a similar extent as DAC treatment (Fig. 3a). Quantitative RT-PCR results showed that LSD2-KD increased the mRNA expression of DNMT1 and DNMT3L without altering mRNA levels of DNMT3a and DNMT3c or TET1 (Fig. 3b). DNMT3L is a DNMT-like protein which binds directly to DNMT3a and DNMT3b. However, neither DNMT1 nor DNMT3L protein expression was markedly changed by LSD2-KD as evidenced by western blot (Fig. 3c). These data suggest that loss of LSD2 reduces DNA methylation likely through blockade of DNMT activity rather than downregulation of the protein expression of DNMTs.
LSD2-KD reduces global DNA methylation in breast cancer cells without changing DNMT1 protein levels. a 5-Methylcytosine (5-mC) levels in genomic DNA from scramble control, or LSD2-KD MDA-MB-231 cells were measured by an ELISA-like assay (Epigentek). Bars represent the mean of 3 experiments ±SEM (t test, * = p < 0.05). b DNMT or TET1 mRNA levels were analysed by quantitative PCR using Taqman gene expression assays. Bars represent the mean of 3 experiments ± SEM (t test, * = p < 0.05). c A representative western blot image of 3 experiments of DNMT1 and DNMT3L protein expression in nuclear lysate is shown. In all panels t tests were used (n = 3, * = p < 0.05)
Loss of LSD2 enhances DNMT inhibitor-induced reexpression of aberrantly silenced genes in breast cancer cells
In cancer cells, the occupancy of H3K4me2 is typically found to be at a low level in the promoters of epigenetically silenced genes that are frequently associated with DNA hypermethylation [22, 23]. The combination of LSD2-KD with DNMT inhibition leads to enhanced reexpression of several epigenetically silenced candidate genes in breast cancer cells including PR, RARβ, SFRP1, SFRP2, ERα, and CDH1 (Fig. 4a). Reexpression of PR, SFRP1, and SFRP2 reached statistical significance in both LSD2-KD clones, while reexpression of RARβ was significant only in clone 2 and CDH1 reexpression was greatly enhanced only in LSD2-KD clone 2. ERα gene reexpression was greatly enhanced in both clones but did not reach statistical significance. PR was the most significantly reexpressed gene by combined inhibition of LSD2 and DNMT. PR plays an important role in breast cancer biology and has been shown to be silenced in breast cancer due to DNA methylation [24], and this silencing is known to be associated with worse prognosis [25]. Next, we investigated further if the reexpression of PR mRNA translated to enhanced protein expression. Treatment with 1 µM DAC for 72 h led to robust increase of PRA protein reexpression in both LSD2-KD clones, while PRB levels were unaffected (Fig. 4b). Quantitation in Fig. 4c shows that DAC treatment leads to an increase in PRA of about 2-fold in scramble 1 compared to untreated with no change in scramble 2, and an approximate 7-fold increase in protein in each LSD2-KD clone. These data indicate that combined inhibition of LSD2 and DNMT leads to reexpression of the epigenetically silenced PRA gene which translates to increased protein expression.
Combined LSD2-KD and DNMT inhibition greatly enhances reexpression of epigenetically silenced candidate genes. a Scramble or LSD2-KD MDA-MB-231 cells were treated with DAC (1 µM) or vehicle for 48 h and RNA was collected. Relative fold change expression is presented in both scramble and LSD2-KD clones 1 and 2 for the genes indicated. Two-way ANOVA was implemented to determine statistically significant differences. Bars represent the mean of 3 experiments ± SEM (t test, * = p < 0.05, ** = p < 0.01, *** = p < 0.001). b A representative immunoblot of MDA-MB-231 scramble, or LSD2-KD clones 1 & 2 ± DAC (1 µM) or vehicle for 72 h and probed for PR and β actin protein is shown. c PRA bands were quantitated and the mean ± SE of 3 experiments is presented
LSD2-KD renders breast cancer cells more susceptible to apoptosis in response to DNMT inhibition
The apparent synergy between the LSD2-KD and a DNMT inhibitor for gene re-expression raised the important question of whether such an effect might also translate into therapeutic efficacy in breast cancer. To address this issue, MDA-MB-231 LSD2-KD cells were treated with DAC for 120 h, and cell number was assessed by crystal violet staining. MDA-MB-231 cells stably expressing LSD2 shRNA were more sensitive to DAC-induced growth inhibition as evidenced by significantly decreased IC50 values (Fig. 5a). A similar result was observed in MCF-7 LSD2-KD cells, suggesting that loss of LSD2 significantly sensitizes breast cancer cells to DAC-induced growth inhibition and exerts a similar effect in different subtypes of breast cancer cells (Fig. 5b).
LSD2-KD cells display increased sensitivity to DAC. a MDA-MB-231 and b MCF7 scramble and stable LSD2-KD cells were treated with increasing doses of DAC, and a crystal violet assay was performed after 120 h. Each point represents the mean of 3 experiments ±SEM. Calcusyn software was used to calculate IC50 values. c Parental MDA-MB-231 cells were treated simultaneously with increasing doses of DAC and TCP at 4×, 2×, 1×, 0.75×, 0.5×, and 0.25× of each IC50 (DAC = 21.2, 10.6, 5.3, 4.0, 2.7, and 1.3 µM; TCP = 2 329.2, 1 164.6, 582.3, 436.7, 291.2, and 145.6 µM, DAC = 5.3 µM, TCP = 582.3 µM, ratio = 1:109.87) for 120 h and stained with crystal violet. Mean ± SEM of combination index values from 3 experiments is presented. Values <1 are defined as synergism and values >1 as antagonism
Next, we investigated the combinatorial effect of tranylcypromine, an identified LSD2 inhibitor, and DAC on cell growth using the median effect/CI model as described in “Materials and methods” section. Using a concomitant 120 h treatment schedule of agents, significant synergistic growth inhibition (CI < 1) was observed at low, median, or higher dose combination (ED 50, 75 and 90) (Fig. 5c). This result clearly suggests that combination therapy targeting LSD2 and DNMT exhibits great synergy in inhibiting growth of breast cancer cells.
To determine if DAC-induced growth inhibition was due to cell death by apoptosis, PARP cleavage and cell cycle parameters were analyzed. PARP cleavage was induced by DAC treatment in both scramble and LSD2-KD cells and a quantitative analysis showed DAC induced approximately 2-fold more PARP cleavage in LSD2-KD cells compared to scramble counterparts (Fig. 6a). LSD2-KD in combination with DAC treatment led to a significant induction of DNA fragmentation, a typical feature of apoptotic cell death, in both LSD2-KD clones (Fig. 6b). Furthermore, flow cytometry analysis indicated that the sub G0/G1 population containing apoptotic cells was significantly enhanced in the LSD2-KD cells treated with DAC compared to DAC-treated scramble cells (p < 0.05) (Fig. 6c). As shown in Fig. 6d, LSD2-KD cells had 20 % less cells in G0/G1 population than scramble cells which is replicated in DAC-treated cells (p < 0.01). LSD2-KD cells treated with DAC accumulated in S phase (p < 0.01), with only a slight increase of cell numbers in G2/M phase. Representative images of scramble and LSD2-KD cells ±1 μM DAC for 96 h clearly indicate that inhibition of LSD2 sensitizes MDA-MB-231 cells to DAC-induced apoptosis (Fig. 6e).
Increased DAC sensitivity in LSD2-KD cells is due to induction of apoptosis. MDA-MB-231 LSD2-KD and scramble cells were treated with 1 µM DAC for 96 h. a A representative western blot of cleaved PARP with the relative density below each band is shown. b A representative image of 3 experiments of fragmented DNA indicative of apoptosis was analysed by electrophoresis. c Propidium iodide stained cells analyzed by flow cytometry. Percentage of cell cycle distribution was quantitated (n = 4, two-way ANOVA, *** = p < 0.001, ** = p < 0.01, * = p < 0.05). d Representative images of MDA-MB-231 scramble and LSD2-KD cells in the presence or absence of 1 μM DAC after 96 h
To determine if decreased growth in LSD2-KD cells in response to DAC reflects a general deficiency in survival after an insult, or if it is specific to DAC, we assessed sensitivity of scramble and LSD2-KD cells to a panel of clinically used breast cancer therapeutic reagents. LSD2-KD cells were not more sensitive to any other breast cancer therapeutics tested including 4-OH-tamoxifen, paclitaxel, doxorubicin, carboplatin, or ABT-888 (PARP inhibitor). Thus, the increased sensitivity to growth inhibition in LSD2-KD cells is specific to the DNMT inhibitor, DAC (Table 1 and Supp Fig. 2).
Discussion
DAC is an efficacious therapy for leukemia and myelodysplastic syndrome and a number of studies suggest that epigenetic agents, like DAC, exert their effect in part through the reexpression of epigenetically silenced genes. However, the success of DAC and other epigenetic modifiers as a treatment for solid tumors including breast cancer has been more limited. To improve the potential of DNMT inhibitors to act as effective antitumor agents in breast cancer, it is necessary to better understand the epigenetic mechanisms by which DNMT activity is regulated. The potential of developing novel and effective combination strategies to improve the efficacy of current epigenetic agents in breast cancer treatment is also worthy of study.
Our studies showed that LSD2-KD sensitizes breast cancer cells to the DNMT inhibitor, DAC, through induction of S phase cell cycle arrest and apoptosis as evidenced by enhanced PARP cleavage, DNA fragmentation, and percent of cells in the apoptotic peak by flow cytometry. We suggest a novel strategy to overcome the refractoriness of solid tumors to DAC through combination therapy with LSD2 inhibition. Inhibiting LSD2 and DNMT by TCP and DAC resulted in synergistic growth inhibition, demonstrating a similar effect of pharmacological inhibition and shRNA-mediated LSD2-KD. The mechanisms underlying LSD2’s effects on the efficacy of DNMTi are not clear. A likely explanation is that H3K4 methylation by LSD2 inhibition makes genomic DNA more accessible to the DNMT inhibitor. Ooi et al. [26] demonstrated that H3K4 methylation strongly inhibits the binding of DNMT3L to H3, suggesting an important role for H3K4 methylation in DNMT activity. However, increased H3K4 methylation is clearly not the only chromatin alteration contributing to reduced DNMT activity. Interestingly, increased sensitivity was not observed in any clinically used anti-neoplastic agents with different mechanisms of action assessed, implying a unique role for LSD2 in mediation of antitumor efficacy of DNMTi in breast cancer cells. Further studies are needed to understand the precise mechanisms underlying the distinct roles of histone demethylase family members in regulation of DNMT activity in breast cancer cells.
In this study, we explored abnormal histone methylation in breast cancer, and the possibilities for reversing these alterations as novel therapeutic targets. We demonstrated for the first time that inhibition of a novel FAD-dependent histone demethylase, LSD2, by shRNA significantly reduces colony formation in breast cancer cells. Although the precise role of LSD2 in breast tumorigenesis remains unclear, these results provide evidence for a growth promoting role for LSD2 in breast cancer and point to a potential utility of LSD2 inhibition as an effective therapeutic approach for this disease.
We have demonstrated that LSD2 is a bona fide histone demethylase specific for H3K4me1/2. Recent studies have suggested a role for LSD2 in H3K9 methylation [5, 18, 27], but we observed no change in H3K9 methylation in LSD2-KD breast cancer cells. Unlike LSD1, the increased H3K4 methylation by LSD2 depletion was not accompanied by increased acetyl-H3K9, suggesting that LSD2 may not be functionally associated with HDACs. This is further supported by our findings that LSD2-KD failed to change expression of the majority of HDAC isozymes (data not shown) and did not alter the cellular sensitivity to the HDAC inhibitor, SAHA. We also found that the level of global DNA methylation is significantly attenuated in LSD2-KD. The loss of LSD1 demethylase activity was reported to directly result in reduced levels of DNMT1 and global DNA methylation during mouse embryogenesis [28]. Loss of LSD2 in oocytes resulted in a decreased ability to methylate DNA during oogenesis [7]. This suggests that LSD2 might be required for the stabilization or activity of DNMTs.
The precise mechanisms by which inhibition of LSD2 globally decreases DNA methylation remain elusive. H3K4me2 is a critical histone mark that is associated with open chromatin and active gene transcription. H3K4 methylation is reduced in the promoters of a number of epigenetically silenced genes such as tumor suppressor genes that may lead to tumorigenesis [29]. Enhanced expression of LSD2 in breast cancer cells depresses levels of H3K4me2 providing a suitable chromatin environment for DNMT recruitment. Oocytes from LSD2-deficient mice display significantly increased H3K4 methylation and failed to recruit DNMTs at important imprinted genes, suggesting a critical role of LSD2 in establishing DNA methylation at imprinted gene loci during oogenesis [7]. Combined inhibition of LSD2 and DNMT led to a robust reactivation of epigenetically silenced genes in breast cancer cells, including several nuclear receptors (PR, ERα, and RARβ) and tumor suppressor genes (SFRP1, SFRP2 and CDH1). Aberrant silencing of these genes has been implicated in breast tumor development and resistance to targeted therapies [4, 11]. In triple negative breast cancer (TNBC), the absence of ER and PR may contribute to resistance to hormonal therapy. Induction of these nuclear factors by combined inhibition of LSD2 and DNMT suggests a novel epigenetic mechanism contributing to aberrant loss of these genes as well as a potential therapeutic intervention in the treatment of TNBC through restoration of sensitivity to hormonal therapy.
In sum, our studies provide solid evidence that LSD2 is a key regulator of DNA methylation and antitumor efficacy of DNMTi in breast cancer cells. Inhibition of LSD2 promotes the apoptotic response of breast cancer cells to DNMTi. Based on these findings, we conclude that combined inhibition of LSD2 and DNMT represents a novel and effective therapeutic target for breast cancer therapy. It is anticipated that these data will advance a novel approach of targeting multiple epigenetic pathways in breast cancer and lead to the development of novel inhibitors of histone lysine demethylases, which will be more effective than current strategies in breast cancer therapy.
Abbreviations
- LSD:
-
Lysine-specific demethylase
- KD:
-
Knockdown
- HDAC:
-
Histone deacetylase
- DAC:
-
Decitabine
- TCP:
-
Tranylcypromine
- DNMT:
-
DNA methyltransferase
- PGR (PR):
-
Progesterone receptor
- ESR1 (ER):
-
Estrogen receptor
- SFRP:
-
Secreted frizzled related protein
- RAR:
-
Retinoic acid receptor
- CDH1:
-
E-cadherin
References
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119(7):941–953
Lee MG, Wynder C, Cooch N, Shiekhattar R (2005) An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437(7057):432–435
Karytinos A, Forneris F, Profumo A, Ciossani G, Battaglioli E, Binda C, Mattevi A (2009) A novel mammalian flavin-dependent histone demethylase. J Biol Chem 284(26):17775–17782. doi:10.1074/jbc.M109.003087
Huang Y, Nayak S, Jankowitz R, Davidson NE, Oesterreich S (2011) Epigenetics in breast cancer: what’s new? Breast Cancer Res 13(6):225. doi:10.1186/bcr2925
Fang R, Barbera AJ, Xu Y, Rutenberg M, Leonor T, Bi Q, Lan F, Mei P, Yuan GC, Lian C, Peng J, Cheng D, Sui G, Kaiser UB, Shi Y, Shi YG (2010) Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell 39(2):222–233. doi:10.1016/j.molcel.2010.07.008
Yang Z, Jiang J, Stewart DM, Qi S, Yamane K, Li J, Zhang Y, Wong J (2010) AOF1 is a histone H3K4 demethylase possessing demethylase activity-independent repression function. Cell Res 20(3):276–287. doi:10.1038/cr.2010.12
Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T (2009) KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461(7262):415–418. doi:10.1038/nature08315
Mersin H, Yildirim E, Berberoglu U, Gulben K (2008) The prognostic importance of triple negative breast carcinoma. Breast 17(4):341–346. doi:10.1016/j.breast.2007.11.031
Munster PN, Thurn KT, Thomas S, Raha P, Lacevic M, Miller A, Melisko M, Ismail-Khan R, Rugo H, Moasser M, Minton SE (2011) A phase II study of the histone deacetylase inhibitor vorinostat combined with tamoxifen for the treatment of patients with hormone therapy-resistant breast cancer. Br J Cancer 104(12):1828–1835. doi:10.1038/bjc.2011.156
Pathiraja TN, Stearns V, Oesterreich S (2010) Epigenetic regulation in estrogen receptor positive breast cancer—role in treatment response. J Mammary Gland Biol Neoplasia 15(1):35–47. doi:10.1007/s10911-010-9166-0
Stearns V, Zhou Q, Davidson NE (2007) Epigenetic regulation as a new target for breast cancer therapy. Cancer Invest 25(8):659–665. doi:10.1080/07357900701719234
Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE (2012) Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat 131(3):777–789. doi:10.1007/s10549-011-1480-8
Lim S, Janzer A, Becker A, Zimmer A, Schule R, Buettner R, Kirfel J (2009) Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive tumor biology. Carcinogenesis 31(3):512–520. doi:10.1093/carcin/bgp324
Zhu Q, Huang Y, Marton LJ, Woster PM, Davidson NE, Casero RA Jr (2012) Polyamine analogs modulate gene expression by inhibiting lysine-specific demethylase 1 (LSD1) and altering chromatin structure in human breast cancer cells. Amino Acids 42(2–3):887–898. doi:10.1007/s00726-011-1004-1
Vasilatos SN, Katz TA, Oesterreich S, Wan Y, Davidson NE, Huang Y (2013) Crosstalk between lysine specific demethylase 1 (LSD1) and histone demethylases mediates antineoplastic efficacy of HDAC inhibitors in breast cancer cells. Carcinogenesis 34 (6):1196–1207. doi:10.1093/carcin/bgt033
Huang Y, Hager ER, Phillips DL, Dunn VR, Hacker A, Frydman B, Kink JA, Valasinas AL, Reddy VK, Marton LJ, Casero RA Jr, Davidson NE (2003) A novel polyamine analog inhibits growth and induces apoptosis in human breast cancer cells. Clin Cancer Res 9(7):2769–2777
Chou TC, Talalay P (1984) Quantitative analysis of dose–effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55
van Essen D, Zhu Y, Saccani S (2010) A feed-forward circuit controlling inducible NF-kappaB target gene activation by promoter histone demethylation. Mol Cell 39(5):750–760. doi:10.1016/j.molcel.2010.08.010
Seward DJ, Cubberley G, Kim S, Schonewald M, Zhang L, Tripet B, Bentley DL (2007) Demethylation of trimethylated histone H3 Lys4 in vivo by JARID1 JmjC proteins. Nat Struct Mol Biol 14(3):240–242. doi:10.1038/nsmb1200
Katoh Y, Katoh M (2007) Comparative integromics on JMJD2A, JMJD2B and JMJD2C: preferential expression of JMJD2C in undifferentiated ES cells. Int J Mol Med 20(2):269–273
Fodor BD, Kubicek S, Yonezawa M, O’Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, Jenuwein T (2006) Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev 20(12):1557–1562. doi:10.1101/gad.388206
Baylin SB, Ohm JE (2006) Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer 6(2):107–116. doi:10.1038/nrc1799
McGarvey KM, Van Neste L, Cope L, Ohm JE, Herman JG, Van Criekinge W, Schuebel KE, Baylin SB (2008) Defining a chromatin pattern that characterizes DNA-hypermethylated genes in colon cancer cells. Cancer Res 68(14):5753–5759. doi:10.1158/0008-5472.CAN-08-0700
Lapidus RG, Ferguson AT, Ottaviano YL, Parl FF, Smith HS, Weitzman SA, Baylin SB, Issa JP, Davidson NE (1996) Methylation of estrogen and progesterone receptor gene 5′ CpG islands correlates with lack of estrogen and progesterone receptor gene expression in breast tumors. Clin Cancer Res 2(5):805–810
Pathiraja TN, Shetty PB, Jelinek J, He R, Hartmaier R, Margossian AL, Hilsenbeck SG, Issa JP, Oesterreich S (2011) Progesterone receptor isoform-specific promoter methylation: association of PRA promoter methylation with worse outcome in breast cancer patients. Clin Cancer Res 17(12):4177–4186. doi:10.1158/1078-0432.CCR-10-2950
Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, Cheng X, Bestor TH (2007) DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448(7154):714–717. doi:10.1038/nature05987
Holmes A, Roseaulin L, Schurra C, Waxin H, Lambert S, Zaratiegui M, Martienssen RA, Arcangioli B (2012) LSD1 and LSD2 control programmed replication fork pauses and imprinting in fission yeast. Cell Rep 2(6):1513–1520. doi:10.1016/j.celrep.2012.10.011
Wang J, Hevi S, Kurash JK, Lei H, Gay F, Bajko J, Su H, Sun W, Chang H, Xu G, Gaudet F, Li E, Chen T (2009) The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet 41(1):125–129. doi:10.1038/ng.268
McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB (2006) Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res 66(7):3541–3549. doi:10.1158/0008-5472.CAN-05-2481
Acknowledgments
Partially supported by Breast Cancer Research Foundation, Samuel Winters Foundation, and Competitive Medical Research Fund of UPMC. These studies used the UPCI Genomics Core Facility supported by P30CA047904.
Conflicts of interest
The authors declare no competing interests.
Ethical standard
All experiments comply with the current laws of the United States of America.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Fig. 1
LSD2 gene expression is increased in breast cancer. Three data sets from the oncomine.org database are presented, as well as LSD2 OncoPrint data in cBioPortal.org. (PPTX 8122 kb)
Supplemental Fig. 2
Increased sensitivity in LSD2-KD cells is specific for DAC. Dose responses for 4-OH-tamoxifen, carboplatin, and lapatinib after 120 h and for paclitaxel, ABT-888, and doxorubicin after 96 h were assessed by crystal violet assay. Each point represents the mean of 3 experiments ± SEM (t test, * = p < 0.05) (PPTX 209 kb)
Rights and permissions
About this article
Cite this article
Katz, T.A., Vasilatos, S.N., Harrington, E. et al. Inhibition of histone demethylase, LSD2 (KDM1B), attenuates DNA methylation and increases sensitivity to DNMT inhibitor-induced apoptosis in breast cancer cells. Breast Cancer Res Treat 146, 99–108 (2014). https://doi.org/10.1007/s10549-014-3012-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-3012-9