Abstract
Small Ubiquitin-like Modifier proteins (or SUMO) modify the function of protein substrates involved in various cellular processes including DNA damage response (DDR). It is becoming apparent that dysregulated SUMO contribute to carcinogenesis by affecting post-transcriptional modification of key proteins. It is hypothesised that SUMO contributes to the aggressive nature of breast cancer particularly those associated with features similar to breast carcinoma arising in patients with BRCA1 germline mutations. This study aims to assess the clinical and biological significance of three members of SUMO in a well-characterised annotated series of BC with emphasis on DDR. The study cohort comprised primary operable invasive BC including tumours from patients with known BRCA1 germline mutations. SUMO proteins PIAS1, PIAS4 and UBC9 were assessed using immunohistochemistry utilising tissue microarray technology. Additionally, their expression was assessed using reverse phase protein microarray utilising different cell lines. PIAS1 and UBC9 showed cytoplasmic and/or nuclear expression while PIAS4 was detected only in the nuclei. There was a correlation between subcellular localisation and expression of the nuclear transport protein KPNA2. Tumours showing positive nuclear/negative cytoplasmic expression of SUMO featured good prognostic characteristics including lower histologic grade and had a good outcome. Strong correlation with DDR-related proteins including BRCA1, Rad51, ATM, CHK1, DNA-PK and KU70/KU80 was observed. Correlation with ER and BRCA1 was confirmed using RPPA on cell lines. SUMO proteins seem to play important role in BC. Not only expression but also subcellular location is associated with BC phenotype.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Small Ubiquitin-like Modifier proteins (or SUMO) are a family of small proteins that covalently attach to amino acid residues of target proteins to modify their functions [1–3]. Through post-translational modification of proteins (PTM), SUMO are involved in various cellular processes including protein stability, response to stress, DNA damage response, nuclear-cytoplasmic transport, transcriptional regulation, cell growth, survival and apoptosis [1, 4, 5]. SUMOylation follows the same enzyme structural design as ubiquitin modification, requiring an E1-activating enzyme (i.e. SAE), E2-conjugating enzyme (i.e. UBC9) and E3-ligating enzymes (i.e. PIAS1-4) [6]. SUMO proteins bind the activating enzyme E1 in an ATP-dependent manner and are transferred to the conjugating enzyme UBC9, which is the only E2 dedicated to SUMO conjugation. It is also reported that UBC9 is able to recognise and transfer SUMO to targets in the absence of a co-adjuvating E3. PIAS (Protein Inhibitor of Activated STAT [Signal Transducer and Activator of Transcription]) act as adaptor proteins that enhance the interaction between UBC9 and the substrate proteins [2].
Genomic instability is a hallmark of cancer and a major contributing factor to tumour development and progression. Central to the maintenance of genome stability is the repair of DNA damage, and it is therefore not surprising that reversible PTM with SUMO have been identified as key contributors to the maintenance of the genome. With regard to the role of SUMO in cancer, previous studies have reported increased expression of UBC9 in carcinomas of the ovary, colon and melanoma, but it was found to be down-regulated in metastatic breast and lung carcinomas [7–9]. Aberrant regulation of PIAS in different tumour types has also been reported [10] including breast cancer (BC) in African women [11].
The study aims to investigate the role of key SUMO proteins including PIAS1, PIAS4 and UBC9 in a well-characterised clinically and molecularly annotated series of BC using immunohistochemistry (IHC) and tissue microarray (TMA) in order to establish the relationship between the SUMO markers, clinico-pathological features, immunoprofile and clinical outcome. To further understand their role in DNA damage response (DDR) pathways and to correlate and confirm their expression in the different molecular classes of BC, the expression level of SUMO markers has been evaluated by reverse phase protein microarray (RPPA) in different cell lines.
Materials and methods
Study cohort
The study cohort was derived from the well-characterised Nottingham Tenovus primary breast carcinoma series. It comprised 1,249 unselected primary operable invasive tumours from female patients presenting between 1989 and 1998. In addition, a further 245 cases unselected primary operable oestrogen receptor (ER) negative BC, from patients presenting between 1998 and 2003 and a cohort of BRCA1 germline mutation carrier (24 cases) were included. Patients’ clinicopathologic features were obtained including age, menopause status, primary tumour size, tumour type, histological grade, nodal status, lymphovascular invasion and Nottingham Prognostic Index (NPI) [12, 13]. Survival data were collected in a prospective way including development of loco-regional and distant recurrences and mortality. BC specific survival (BCSS) is defined as the interval from the date of primary surgery to the time of death because of BC. Death due to other causes is considered as a censored event. Disease-free interval (DFI) is defined as the interval from the date of primary surgery to the time of first loco-regional recurrence or distant metastasis. Both of these parameters were measured in months.
Tumour characteristics have been considered for patient’s managements by selecting NPI and ER status [13]. Patients with NPI excellent prognostic group (score ≤3.4) received no adjuvant therapy, but those patients with NPI > 3.4 received Tamoxifen if ER-positive (± Zoladex in case the patients were pre-menopausal). On the other hand, classical cyclophosphamide, methotrexate and 5-fluorouracil (CMF) were used if the patients were ER-negative and fit to receive chemotherapy.
Data on the following biomarkers were available: ER, progesterone receptor (PgR), HER2, DNA damage respose proteins (Rad51, KU70/KU80, DNA-PK, BRCA1, BARD1, CHK1, MTA1, and ATM), nuclear transport protein importin subunit alpha-2 (KPNA2), basal markers (cytokeratins [CK5, CK14, and CK17] and the proliferation and cell-cycle associated proteins (Ki67, and P53). The immunoreactivity, scoring and categorisation of these markers were defined in this study as previously described [12–16]. In this series, HER-2 was assessed using IHC (DAKO) and dual-colour chromogenic in situ hybridisation (CISH) as previously published [16]. Ki67 labelling index (Ki67LI) was assessed in whole tumour tissue sections and was expressed as the percentage of MIB1 positive cells among a total number of 1,000 malignant cells at high-power magnification (×400) [16]. All other markers were assessed using IHC and TMA prepared sections.
This study was approved by Nottingham Research Ethics Committee 2.
Immunohistochemistry
Immunohistochemistry was carried out using the Novolink Kit-polymer detection system (Leica, Newcastle, UK). Primary antibodies used were PIAS1 (clone Ab32219, Abcam Ltd., Cambridge, UK) and PIAS4 (clone NBP1-31215, Novus Biologicals, Cambridge, UK) with a dilution of 1:425 and 1:250, respectively, and UBC9 (clone Ep2938Y, Novus Biologicals, Cambridge UK) with a dilution of 1:225 and 60-min incubation for all. 3-3′Diam-inobenzidine tetrahydrochloride (Novolink DAB substrate buffer plus) was freshly prepared and used as a chromogen. The TMA sections were counter stained with haematoxylin for 6 min [15].
The conditions of other proteins used in this study are as follow: (1) DDR markers: BARD1; Novus Biologicals, 1:50, BRCA1 Ab-1 (MS110) Calbiochem, 1:150, Rad51; Abcam, 1:70, KU70/KU80; Abcam,1:2500, DNA-PK; Cell signalling, 1:28, BRCA1 down-regulator marker MTA1; Abcam,1:200, DNA damage signal transducer: CHK1 (Phospho S345); Abcam, 1:150, DNA damage sensor: ATM; Abcam, 1:100, (2) cell proliferation marker: Ki-67; Dako-Cytomation, 1:100, and (3) Nucleocytoplasmic transport marker: KPNA2; Abcam, 1:400. All were incubated for 1 h except ATM for overnight.
Immunohistochemical scoring
Two TMA cores (peripheral or central) were evaluated from each tumour. Only immunostaining of invasive cancer cells within the tissue cores was considered. Each core was scored individually; if one core was uninformative, the overall score applied was that of the remaining core. There was immunoreactivity of each target protein in the TMA (nuclear, cytoplasmic, or both). High-resolution digital images (Nanozoomer; Hamamatsu Photonics, Welwyn Garden City, UK) scanned at x20 magnification were used to facilitate the manual scoring of the TMA cores via web-based interface (Distiller; Slidepath, Ltd., Dublin, Ireland).
The three biomarkers (UBC9, PIAS1 and PIAS4) were categorised based on the frequency histogram distributions and X-Tile software (version 3.6.1, Yale University). The cut-off points used were as follows: nuclear PIAS1 (PIAS1n; negative/low <35 and positive ≥35 H-score), cytoplasmic PIAS1 (PIAS1c; negative/low <95 and positive ≥95 H-score), nuclear PIAS4 (negative/low <160 and positive ≥160 H-score) and nuclear UBC9 (negative/low <160 and positive ≥160 H-score) and cytoplasmic UBC9 (negative/low <200 and positive ≥200 H-score).
Reverse phase protein microarray (RPPA)
For the purpose of this study, 4 different cell lines were used. (A) BRCA1 deficient (HeLa SilenciX®) as well as their control BRCA1 proficient (Tebu-Bio) cell lines. SilenciX cells were grown in DMEM medium (with l-Glutamine 580 mg/L, 4,500 mg/L D19 Glucose, with 110 mg/L Sodium Pyruvate) supplemented with 10 % FBS, 1 % penicillin/streptomycin and 125 μg/mL Hygromycin B. (B) MDA-MB-436 cell lines (characterised by negative expression of ER and BRCA1 deficient) were purchased from CLS and were grown in DMEM (Sigma, UK), luminal phenotype MCF-7 cell lines (characterised by positive expression of ER and BRCA1) were purchased from ATCC and were grown in RPMI1640 (Sigma, UK). Lysate extraction and western blotting were carried out by lysing cells in RIPA buffer (20 mM Tris, 150 mM Nacl, 1 % Nonidet p-40, 0. 5 % sodium deoxycholate, 1mMEDTA, 0. 1 % SDS) containing protease inhibitor (Sigma) and phosphatase inhibitor cocktail 2 and 3 (Sigma). An overall total of 50 μg protein was used from each cell line. Cell lysate was resolved on SDS 4-12 % precast gel (Expedeon, UK) after that blotted onto nitrocellulose membrane of Protran BA 85 (Whatman GmbH, Germany). PBS Tween-20 containing 5 % (w/v) non-fat dried milk was applied for the purpose of blocking. The membranes have been incubated for 1 h at room temperature in 1 % (w/v) non-fat dried milk in PBS-T that contains primary antibody (PIAS1, PIAS4 or UBC9) for 1 h at room temperature, developed using GE Enhanced Chemiluminescence substrate (GE Healthcare Life Sciences, Buckinghamshire, UK). After stripping the membrane, β-actin was identified by incubating the membrane for 1 h at room temperature in 1 % (w/v) non-fat dried milk in PBST that contains HRP-conjugated anti-β-actin (Abcam Ltd., Cambridge, UK) and developed using GE Enhanced Chemiluminescence substrate (GE Healthcare Life Sciences, Buckinghamshire, UK).
Reverse phase protein microarray (RPPA)
Cell line lysates were solubilised in 4× Sodium dodecyl sulphate (SDS) sample buffer in a ratio of 1:3, respectively, and boiled for 5 min at 95 °C. Samples were loaded onto a 384-well plate (Genetix, UK), where each sample was serially diluted 5 times in 1× SDS buffer. Samples were robotically spotted in duplicates onto nitrocellulose-coated glass slide (Grace Bio-labs), a microarraying robot (MicroGrid 610, Digilab, Marlborough, MA, USA). Slides were incubated overnight in blocking solution (0.2 % I-block (Tropix, Bedford, MA, USA), 0.1 % Tween-20 in PBS) at 4 °C with shaking. After washing three times 5 min each, the slide was incubated overnight at 4 °C with shaking with the primary antibodies diluted in antibody diluent with reducing background (DAKO). In addition, GAPDH (BioLegend), diluted 1:250 in the same diluent, was used as a house- keeping protein to control protein loading.
Following washing, the slides were incubated with diluted infrared (1:5,000 in washing buffer) secondary antibodies (800 CW anti-rabbit antibody and 700 CW anti-mouse antibodies) for 30 min at room temperature in the dark with shaking. The slide was washed as before, dried by centrifugation at 500×g for 5 min and scanned with a Licor Odyssey scanner (LI-COR, Biosciences) at 21 μm resolution at 800 nm (green) and 700 nm (red). The resultant TIFF images were processed with Axon Genepix Pro-6 Microarray Image Analysis software (Molecular Services Inc.) to obtain fluorescence data for each feature and generate gpr files. Protein signals were finally determined with background subtraction and normalisation to the internal housekeeping targets using RPPanalyzer, a module within the R statistical language on the CRAN (http://cran.r-project.org/).
Statistical analysis
All statistical analyses were performed using SPSS 21.0 IBM statistical software. Analyses of categorical variables were carried out with Chi-Squared test (χ 2). One way ANOVA was used to find out which of different BC classes (by IHC or cell lines) were significantly different from each other (post hoc test; Tukey). Associations with outcome were calculated using Kaplan–Meier curves and log-rank test. Cox-regression was applied for multivariate analyses. A two-sided P value of <0.01 was considered statistically significant.
Results
Expression of SUMO markers in invasive breast cancer
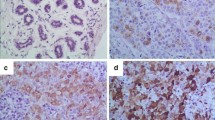
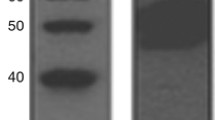
The specificity of SUMO antibodies used in this study was validated using Western blotting as evident by a single band at the correct protein size (Fig. 1). PIAS4 showed nuclear staining, which ranged from negative/weak to strong with no cytoplasmic or membranous staining observed in invasive tumours. PIAS1 and UBC9 showed both nuclear and cytoplasmic staining. Figure 2 shows staining pattern of SUMO markers in BC while Table 1 summarises the frequencies of these markers in BC. The associations between SUMO markers are summarised in Table 2. Positive correlations were identified between nuclear expressions of the SUMO markers apart from PIAS4. PIAS4 was correlated with cytoplasmic UBC9 (UBC9c) and cytoplasmic PIAS1 (PIAS1c). There was a positive correlation between PIAS1c and UBC9c. No correlation was found between UBC9n and PIAS1c or between UBC9c and PIAS1n. Association between PIAS1 and UBC9 and clinico-pathological and molecular features and outcome was carried out after considering a combination of expression and subcellular location of each marker (Tables 3, 4).
Correlation of SUMO proteins with clinico-pathological features
The correlation between SUMO markers expression and the clinico-pathological features indicates that tumours with poor prognostic features were mainly associated with loss of nuclear expression of PIAS1 (PIAS1n−) and UBC9 (UBC9n−), cytoplasmic expression of both markers (PIAS1c+ and UBC9c+) and with the expression of PIAS4 (Table 3). No correlation was identified with patient age (P > 0.01).
Correlation of SUMO markers with other tumour biomarkers
The correlation between SUMO proteins with relevant tumour biomarkers is shown in Table 4. In brief, there was a positive correlation between PIAS4 and DDR markers (Rad51, CHK1, KU70/KU80, BARD1 and DNA-PK). Positive correlations were identified between BARD1 and KU70/KU80 and PIAS1 regardless of its subcellular localisation and between Ki67 and cytoplasmic location of PIAS1 and UBC9. Importantly, there was a correlation between the nuclear transport protein KPNA2 and cytoplasmic location of UBC9 and PIAS1.
The expression of SUMO was assessed in the different molecular classes of BC. Association with HER2-positive and triple negative classes is shown in Table 4. UBC9 expression was correlated with basal-like breast carcinoma (BLBC) in which UBC9c+/n+, UBC9c+/n− and UBC9c−/n− were associated with BLBC compared with UBC9c−/n+. In addition, molecular classification corresponding to DDR status and the phenotype of the cell lines used was performed based on the expression of ER and BRCA1. Class 1; sporadic BRCA1− and ER−, class 2; sporadic BRCA1+ and ER+, class 3; tumours from patients with known BRCA1 germline mutations (hereditary) showing ER− and ER+. The highest level of expression of PIAS1n and UBC9n was seen in sporadic BRCA1+/ER+ BC in comparison to hereditary and sporadic BRCA1−/ER− BC (P < 0.0001) (Fig. 3). No association was observed between molecular classes and PIAS4.
SUMO protein levels detected by IHC in breast cancer showing either hereditary or sporadic BRCA1 deficiencies in addition to ER status. n nuclear and c cytoplasmic expression. Cases were classified based on the expression of BRCA1 and ER. Error bars represent mean (SD) and were created on H-score (ranges 0–300). A = sporadic cases [ER− & BRCA1−] versus sporadic cases [ER+ & BRCA1+], B = sporadic cases [ER− & BRCA1−] versus Hereditary cases [ER−], C = sporadic cases [ER− & BRCA1−] versus Hereditary cases [ER+], D = sporadic cases [ER+ & BRCA1+] versus Hereditary cases [ER−], E = sporadic cases [ER+ & BRCA1+] versus Hereditary cases [ER+] and F = Hereditary cases [ER−] versus Hereditary cases [ER+]. ANOVA test was used for each marker within the classes. The long bars are expected, the mean of H-score does not describe repeated observation, it presents different cases distribution share status of ER and BRCA1 yet with various other variables including grade, stage and size of the tumour, which have some influence on the expression of the protein
SUMO markers expression and patient’s outcome
Univariate survival analysis of the whole series showed a positive association between expression of PIAS1n and better outcome in terms of longer BCSS (χ 2 = 8.95, P = 0.003) and DFI (χ 2 = 8.06, P = 0.005). Cytoplasmic expression of PIAS1 (PIAS1c) showed an association of borderline significance with shorter BCSS (χ 2 = 6.39, P = 0.011). Co-expression of nuclear and cytoplasmic PIAS1 demonstrated that PIAS1n−/c+ is associated with shorter BCSS in comparison with other phenotypes with PIAS1n+/c− associated with the best outcome (χ 2 = 12.62, P = 0.006). There was an association between nuclear expression of UBC9 and longer BCSS (χ 2 = 7.29, P = 0.007) and of borderline significance with DFI (χ 2 = 6.08, P = 0.014). Regarding PIAS4, an association of borderline significance was detected between its expression and longer BCSS (χ 2 = 3.75, P = 0.053).
However, when the cohort was stratified according to chemotherapy treatment, association between PIAS4 and PIAS1n expression and longer BCSS was found in the subgroup of patients who did not receive chemotherapy (χ 2 = 13.60, P < 0.001, χ 2 = 7.53, P = 0.006, respectively) but not in the subgroup who received chemotherapy (P > 0.05). Multivariate analyses including tumour stage, grade, size, lymphovascular invasion and chemotherapy treatment showed that PIAS4 is independent prognostic markers for breast cancer (Table 5). However, PIAS1 and UBC9 were not independent predictor of survival (Fig. 4).
Expression of SUMO markers in breast cancer cell lines by reverse phase protein array
RPPA was used to evaluate the expression level of SUMO markers in the four cell lines; BRCA1 deficient HeLa SilenciX® cells and control HeLa cells (proficient BRCA1), MCF-7 and MDA-MB-436 cells. Although RPPA measures protein expression regardless of its subcellular localisation, there was a correlation between RPPA and IHC results, particularly with regards to nuclear IHC expression. Higher levels of expression of UBC9 in BRCA1 proficient HeLa cell lines and MCF-7 were observed compared to other cell lines (Fig. 5). However, PIAS1 did not show significant difference between different cell lines. Each cell line was compared to a selective cohort in IHC (see Fig. 3), where HeLa control and MCF7 represent sporadic BRCA1+ and ER+; BRCA1 deficient HeLa and MDA-MB-436 represent sporadic BRCA1− and ER−.
The SUMO levels detected by reverse phase protein microarray in different cell lines (BRCA1 deficient HeLa SilenciX® cells and its control [BRCA1 and BRCA1.c, respectively], MCF-7 and MDA-MB-436 cells). For image of nitrocellulose slide spotted with different cell lysates, the red square represents the 700 channel for detection of mouse antibody while green square the 800 channel for rabbit antibody. Images of scanned nitrocellulose slides printed with extracted protein from cell lines and probed with the antibodies against the target proteins. Five twofold dilutions of each sample were printed in duplicate. Background was subtracted and the intensity of each spot was normalised to its corresponding GAPDH level. Each (R) represents different passage of each sample, therefore, three different passages of each sample were used. Error bars represent mean (SD). Hela BRCA1; between passage21 and 30, Hela BRCA1 control; between passage 44 and 50, MCF-7; between passage 25 and 32, and MDA-MB-436; between passage 12 and 20. A = BRCA1 versus BRCA1.C, B = BRCA1 versus MDA-MB-436, C = BRCA1 versus MCF-7, D = BRCA1.C versus MDA-MB-436, E = BRCA1.C versus MCF-7, and F = MDA-MB-436 versus MCF-7. ANOVA test was used
Discussion
SUMOylation is involved in different cellular processes including DDR through post-translational modification of protein [17–21]. Few studies have addressed the role of individual SUMO proteins in small series of breast cancer (BC) [21–24]. In the present study, we have assessed the expression of 3 key SUMO markers (PIAS1, PIAS4 and UBC9) in a large well-characterised series of BC to evaluate their biological and clinical significance with particular interest on their role in DDR. In this study, a large number of the tumours expressed PIAS4, UBC9 and PIAS1c, whereas the frequency of expression of PIAS1n was remarkably low. This is in line with Chen et al. [22] who reported a higher level of UBC9 in BC; Wei et al. who have demonstrated an increased PIAS4 expression in gastric tumours [25] and with Coppola et al. [26] who revealed a significant decrease in PIAS1 protein level in colon cancer.
Our results demonstrate that SUMO proteins expression can be localised exclusively in the nucleus (PIAS4) or expressed in both nucleus and cytoplasm (PIAS1 and UBC9). This observation is consistent with some previous studies [26, 27]. Importantly, the association between the nuclear transport protein KPNA2 and the subcellular localisation of UBC9 and PIAS1 and the distinct functions of cytoplasmic SUMO compared to nuclear SUMO indicate the complexity of the process of controlling the function of these proteins. In the present study, expressions of any of UBC9n+/c−, PIAS1n+/c− and low PIAS4 were related to less aggressive phenotype of BC such as lower histologic grade, ER and BRCA1 positivity [19, 22]. Consistent with that outcome analysis showed an association between nuclear expression of PIAS1 and UBC9 and longer survival compared to cytoplasmic expression.
It is reported that trafficking in and out of the nucleus has role in signal transduction, gene expression, progression of cell-cycle and apoptosis. For this reason, markers of SUMO as nuclear localised proteins, the unpredicted expression in the cytoplasm of the cancer cell could possibly have significant role in tumorigenesis especially they showed distinct roles or features than nuclear expression. It can suggest that when SUMO markers are transferred to the cytoplasm maybe they are unable to function properly. SUMO proteins in the cytoplasm of cancer cells they may be degraded entirely or retained. In this study, a marker that has role in nucleocytoplasmic transport has been investigated (KPNA2) which showed a significant association with both PIAS1n−/c+ and UBC9n−/c+. This result may show the KPNA2 role as nuclear export markers (they bind to cargoproteins in the cytoplasm, following interaction with the nuclear pore complex and passing through its channel) [28].
The relationship between PIAS with ER has been previously discussed [19]. Mutations that normally prevented SUMO modification can damage activated ERα transcription with no need of affecting ERα cellular localisation. Aside from identifying PIAS1 as E3 ligase for ERα, a study by Sentis et al. [19] showed that PIAS1, plus UBC9, modulated ERα-dependent transcription independently from their conjugation activity of SUMO-1. Supporting this observation, all SUMO markers in the present study showed significant associations with ERα.
In this study, the SUMO biomarkers expression in sporadic cases and familial BRCA1-associated tumours were investigated. The findings here suggest that SUMO proteins expression is aberrant and reduced more frequently among BRCA1 familial tumours than in sporadic tumours. In addition, the sporadic IHC BRCA1+ tumours with decreased SUMO proteins were more frequently ER negative. This was also confirmed on cell lines. The low expression of PIAS1n in this study is an additional confirmation that the majority of SUMO proteins could possibly indicate a further characteristic shared by BRCA1 known mutation cancers by showing lack ER, considering that, the expression of PIAS1 is influenced by the presence of ER. As a result, these findings support the hypothesis that SUMOylation as a process modulating ERα-dependent cellular response and provide a relationship somewhere between the SUMO and pathways of ER.
Both UBC9 and PIAS4 have previously been discussed to down regulate BRCA1 expression [20, 29], that is in agreement with the present study where a significant number of tumours that expressed UBC9 or PIAS4 had a positive association with BRCA1 down-regulator proteins such as MTA1. It is documented that repair of DMA breaks is achieved by one or more alternative DDR pathways and they are influenced by each other [30]. This study demonstrated strong association between SUMO proteins and DDR-related proteins including those involved in homologous recombination (BRCA1, ATM, CHK1 and Rad51) and non-homologous end joining (DNA-PK and KU70/KU80).
In breast cancer, Ki-67, p53, CHK1 and ATM have been shown being good predictors of BRCA1 dysfunction [31]. Ki-67 expression is associated with abnormal cell proliferation with poor outcome [32]. Both of UBC9 and PIAS4 could possibly have a role in the cell cycle regulation. SUMOylation of P53 has been discussed previously [33]. Park and his group have identified UBC9 involvement in the cell cycle regulation, where UBC9 negatively controls BRCA1 through several promoters including P21 and P27 [34]. The UBC9 association and PIAS4 with abnormal expression of P53 in breast cancers proposed that these types of tumour could have experienced disorganised control of the cell cycle and as a consequence caused rapid division of any abnormal cell, which in turn a hallmark of tumour aggressiveness. In breast tumours, PIAS1 is probably engaging in a function of tumour suppressor, for the reason that lack of this gene is linked to abnormal cell proliferation. Additional study is necessary on the PIAS1 to determine its potential function as tumour suppressor gene. ATM functions upstream of for example BRCA1 in the same pathway, considering the fact that BRCA1 is directly phosphorylated by ATM kinase on serine residues S1423 and S1524, consequently modulating the function of BRCA1 [35]. In the present study, markers of SUMO were significantly associated with markers involved in cell cycle process such as CHK1, Ki-67 and P53. Considering the idea of the DNA damage response to be an anti-cancer barrier, the results in this study are supported the scenario in which the primary cancer-predisposing defect of for instance BRCA1 may possibly weaken or even damage the control of genome integrity in addition to the aberrantly increased outcomes of unrepaired DSBs could possibly result in an increasingly effective activation of the initially wild-type ATM. This might trigger the ATM-regulated cell cycle checkpoints along with cell death pathways which could determine for ATM inactivation if perhaps these types of lesions will develop in malignancy direction. As a result, the SUMO of BRCA1 defect sooner or later led to enhanced inactivation frequency of ATM.
Previous studies have reported association between expression and activity of PIAS1 and chemoresistance [25, 36]. In this study, PIAS4 showed different association with outcome when cases were stratified based on adjuvant chemotherapy treatment. The association between PIAS4 and better outcome in the group of patients who did not receive chemotherapy and the lack of such association may suggest that PIAS4 negative tumours respond better to chemotherapy than PIAS4 positive tumours. Multivariable analysis showed that PIAS4 is a predictor of outcome independent of therapy, stage size or tumour grade. Although PIAS1 and UBC9 showed some associations with outcome, these associations were not independent of other variable.
In this study, SUMO protein levels were assessed using the high-throughput proteomic technique RPPA in different cell lines representing different phenotypes based on BRCA1 and ER status. Although the results of RPPA were comparable to IHC and demonstrated the relationship between SUMO and BRCA1 and ER status in cells lines, the results of RPPA should be interpreted with caution: (A) Cell lines were used without determining the phases of the cell cycle, although it was not a functional study; each phase of cell cycle could possibly have an impact on the expression of the proteins. (B) RPPA gives quantifiable data for the differential expression levels of proteins, yet the subcellular locational which was evident using IHC; the activation status or the biological triggers of proteins cannot be concluded. However, the findings of the present study combine the power of IHC staining with the parallel analytic capability of protein microarray RPPA.
To conclude, the findings of this study confirm the results of previous studies of the biological function of SUMO and provide evidence that SUMO play important role in BC particularly in DDR and related to hormone receptor. Not only expression but also subcellular location of SUMO may be related to their function. The potential for targeting these markers for therapeutic use needs to be exploited.
References
Pinder JB, Attwood KM, Dellaire G (2013) Reading, writing, and repair: the role of ubiquitin and the ubiquitin-like proteins in DNA damage signaling and repair. Front Genet 4:45
Bologna S, Ferrari S (2013) It takes two to tango: ubiquitin and SUMO in the DNA damage response. Front Genet 4:106
Al-Hakim A et al (2010) The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair (Amst) 9(12):1229–1240
Matunis MJ, Coutavas E, Blobel G (1996) A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol 135(6 Pt 1):1457–1470
Mahajan R et al (1997) A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88(1):97–107
Morris JR (2010) More modifiers move on DNA damage. Cancer Res 70(10):3861–3863
Mo YY et al (2005) A role for Ubc9 in tumorigenesis. Oncogene 24(16):2677–2683
Moschos SJ, Mo YY (2006) Role of SUMO/Ubc9 in DNA damage repair and tumorigenesis. J Mol Histol 37(5–7):309–319
Moschos SJ et al (2010) Expression analysis of Ubc9, the single small ubiquitin-like modifier (SUMO) E2 conjugating enzyme, in normal and malignant tissues. Hum Pathol 41(9):1286–1298
Hoefer J et al (2012) PIAS1 is increased in human prostate cancer and enhances proliferation through inhibition of p21. Am J Pathol 180(5):2097–2107
Agboola A et al (2013) PIASgamma expression in relation to clinicopathological, tumour factors and survival in indigenous black breast cancer women. J Clin Pathol. doi:10.1136/jclinpath-2013-201658
Alshareeda AT et al (2013) Clinicopathological significance of KU70/KU80, a key DNA damage repair protein in breast cancer. Breast Cancer Res Treat 139(2):301–310
Rakha EA et al (2006) Morphological and immunophenotypic analysis of breast carcinomas with basal and myoepithelial differentiation. J Pathol 208(4):495–506
Rakha EA et al (2009) Triple-negative breast cancer: distinguishing between basal and nonbasal subtypes. Clin Cancer Res 15(7):2302–2310
Alshareeda AT et al (2012) Fatty acid binding protein 7 expression and its sub-cellular localization in breast cancer. Breast Cancer Res Treat 134(2):519–529
Aleskandarany MA et al (2012) Prognostic value of proliferation assay in the luminal, HER2-positive, and triple-negative biologic classes of breast cancer. Breast Cancer Res 14(1):R3
Bergink S, Jentsch S (2009) Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458(7237):461–467
Sachdev S et al (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15(23):3088–3103
Sentis S et al (2005) Sumoylation of the estrogen receptor alpha hinge region regulates its transcriptional activity. Mol Endocrinol 19(11):2671–2684
Morris JR et al (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462(7275):886–890
Qin Y et al (2011) Ubc9 mediates nuclear localization and growth suppression of BRCA1 and BRCA1a proteins. J Cell Physiol 226(12):3355–3367
Chen SF et al (2011) Ubc9 expression predicts chemoresistance in breast cancer. Chin J Cancer 30(9):638–644
Agboola AO et al (2014) Clinicopathological and molecular significance of Sumolyation marker (ubiquitin conjugating enzyme 9 (UBC9)) expression in breast cancer of black women. Pathol Res Pract 210(1):10–17
Clevenger CV (2004) Roles and regulation of stat family transcription factors in human breast cancer. Am J Pathol 165(5):1449–1460
Wei J et al (2011) mRNA expression of BRCA1, PIAS1, and PIAS4 and survival after second-line docetaxel in advanced gastric cancer. J Natl Cancer Inst 103(20):1552–1556
Coppola D et al (2009) Substantially reduced expression of PIAS1 is associated with colon cancer development. J Cancer Res Clin Oncol 135(9):1287–1291
Baek SH (2006) A novel link between SUMO modification and cancer metastasis. Cell Cycle 5(14):1492–1495
Chook Y, Blobel G (2001) Karyopherins and nuclear import. Curr Opin Struct Biol 11(6):703–715
Xu J et al (2009) A novel mechanism whereby BRCA1/1a/1b fine tunes the dynamic complex interplay between SUMO-dependent/independent activities of Ubc9 on E2-induced ERalpha activation/repression and degradation in breast cancer cells. Int J Oncol 34(4):939–949
Martin RW et al (2007) RAD51 up-regulation bypasses BRCA1 function and is a common feature of BRCA1-deficient breast tumors. Cancer Res 67(20):9658–9665
Abdel-Fatah TM et al (2010) The biological, clinical and prognostic implications of p53 transcriptional pathways in breast cancers. J Pathol 220(4):419–434
Aleskandarany MA et al (2011) MIB1/Ki-67 labelling index can classify grade 2 breast cancer into two clinically distinct subgroups. Breast Cancer Res Treat 127(3):591–599
Kahyo T, Nishida T, Yasuda H (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell 8(3):713–718
Park MA et al (2008) SUMO1 negatively regulates BRCA1-mediated transcription, via modulation of promoter occupancy. Nucleic Acids Res 36(1):263–283
Cortez D et al (1999) Requirement of ATM-dependent phosphorylation of brca1 in the DNA damage response to double-strand breaks. Science 286(5442):1162–1166
Mohanty S et al (2014) ROS-PIASgamma cross talk channelizes ATM signaling from resistance to apoptosis during chemosensitization of resistant tumors. Cell Death Dis 5:e1021
Acknowledgments
Alaa Alshareeda is sponsored by the higher education of kingdom of Saudi Arabia.
Conflict of interest
None.
Funding
No financial relationship with the organisation that sponsored the research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ola H. Negm is first joint author for this study.
Rights and permissions
About this article
Cite this article
Alshareeda, A.T., Negm, O.H., Green, A.R. et al. SUMOylation proteins in breast cancer. Breast Cancer Res Treat 144, 519–530 (2014). https://doi.org/10.1007/s10549-014-2897-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-014-2897-7