Abstract
A major side effect of aromatase inhibitor (AI) therapy is AI-related arthralgia (AIA), which often leads to therapy discontinuation. We aimed to identify genetic variants associated with AIA and therapy discontinuation in the first year of AI treatment. Our prospective cohort study included 343 postmenopausal women with early breast cancer starting AI therapy. Single nucleotide polymorphisms (SNPs) in candidate genes involved in estrogen and vitamin D signaling were selected. Univariate and multivariate linear/logistic regressions were fitted in order to asses the association between studied SNPs and AIA intensity (visual analogic scale score) at 3 and 12 months of follow-up, worsening pain, and therapy discontinuation. We also tested for a priori-defined interactions by introducing multiplicative terms in the regression equations. SNPs in CYP17A1 and VDR genes appeared significantly associated with AIA (P = 0.003, P = 0.012, respectively). One SNP in CYP27B1 gene was related to therapy discontinuation [P = 0.02; OR 0.29 (0.09–0.99)]. We revealed interactions between CYP27B1 and both CYP17A1 (P = 0.01) and VDR SNPs (P = 0.06). Furthermore, an additive effect on pain intensity was shown for unfavorable alleles, with two points higher mean absolute pain increase and up to 5.3-fold higher risk of worsening pain compared to favorable genotypes. SNPs in CYP17A1, VDR, and CYP27B1 genes predict the risk of AIA. Their determination would be useful to trigger the monitoring strategies in women at risk of therapy discontinuation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Third-generation aromatase inhibitors (AIs) are first-line therapies for early-stage breast cancer (EBC) [1, 2]. Unfortunately, 25–30 % of the women treated with AIs experience AI-related arthralgia (AIA) [3], which remains the most common cause of therapy discontinuation [4]. Although, Henry et al. [3] recently described some clinical predictors of arthralgia, these same authors acknowledged their poor predictive ability and stated an urgent need for genetic studies to help us disentangle the pathophysiology of AIA. Some genetic studies have been performed to establish a genetic background of AIA [5, 6], but its total heritability remains unknown.
This study focuses on the investigation of genetic determinants of AIA and on therapy discontinuation. In order to decide a priori candidate genes for our study, two hypotheses were postulated. The first is that estrogens have an antinociceptive role through a direct effect on the opioid pain fibers [7, 8]. In addition, estrogens have a tissue-specific effect on pro-inflammatory cytokines, acting as a protective modulator of inflammation, which could account for enhanced local inflammatory activity related to the estrogen depletion caused by AI therapy [9–11].
Second, some observational data suggest that vitamin D insufficiency, which is very common in postmenopausal women with EBC [12], might also contribute to arthralgia [13].
We performed an association analysis with single nucleotide polymorphisms (SNPs) within genes of estrogen and vitamin D signaling in the B-ABLE cohort.
Methods
Study design and participants
Details on the B-ABLE study methods have been published elsewhere [14] and are outlined below.
Briefly, B-ABLE is a prospective, observational, clinical cohort study, conducted at the Breast Cancer Unit and Bone Metabolism Unit, Hospital del Mar–Parc de Salut Mar, Barcelona (Spain).
Eligible participants were excluded if they had a history of any metabolic or endocrine disease potentially affecting bone and cartilage metabolism, rheumatoid arthritis or concurrent or previous treatment with bisphosphonates, oral corticosteroids, or any other bone-active drug.
Measurements
AI-related arthralgia
A visual analogic scale (VAS) was used to score the intensity of self-reported joint pain at baseline (before starting AI therapy), 3 months and 1 year of follow-up. Score ranged from 0 (no pain) to 10 (maximum pain).
Absolute increase in pain at 3 and 12 months of AI therapy (VAS score at 3 and 12-month follow-up minus baseline, respectively) was the primary outcome for this study.
Assuming that some participants had some degree of pain at baseline, we also defined a binary pain phenotype named “worsening pain” (VAS score at follow-up > baseline VAS) versus “stable/improving” (if VAS score at follow-up ≤ baseline VAS) as a secondary outcome.
AI therapy discontinuation
Patients were asked at 3 and 12 months whether they were still on AI-treatment, and if not, the main reason for discontinuation, out of the following options: arthralgia, cancer-related events, concomitant disease, and personal concerns. Therapy discontinuation due to AIA was defined as a secondary outcome for these analyses.
Other assessments
Information on a large number of clinical variables was recorded at the time of enrollment, including age at recruitment, age at menarche and at menopause, lactation, parity, previous chemotherapy and radiotherapy, adjuvant treatments, weight, height, plasma levels of 25(OH)D, and smoking status.
Interventions
Participants were treated with AIs (letrozole, exemestane, or anastrozole) according to the American Society of Clinical Oncology recommendations [15], starting within 6 weeks after surgery or 1 month after the last cycle of chemotherapy, or alternatively, after 2–3 years of tamoxifen therapy.
All participants were supplemented with calcium and vitamin D tablets (1,000 mg and 800 IU daily), and those with baseline vitamin D deficiency (<30 ng/ml) received an additional dose of 16,000 IU of oral cholecalciferol every 2 weeks.
Selection of candidate genes
Three candidate genes were selected to test our estrogen hypothesis: estrogen receptor 1 (ESR1), aromatase (CYP19A1), and steroid 17-alpha-hydroxylase/17,20 lyase (CYP17A1) (Fig. 1).
Schematic diagram of steroid and vitamin D signaling pathway. Metabolites and selected enzymes involved in the Vitamin D3 signaling [7-dehydrocholesterol reductase (encoded by DHCR7 gene), vitamin D 25-hydroxylase (encoded by CYP2R1 gene), 25-hydroxyvitamin D 1-alpha-hydroxylase (encoded by CYP27B1 gene), 1,25-dihydroxyvitamin D 24-hydroxylase (encoded by CYP24A1 gene), vitamin D-binding protein (DBP, encoded by GC gene), and vitamin D receptor (encoded by VDR gene)], and the estrogen signaling [steroid 17-alpha-monooxygenase (encoded by CYP17A1 gene), aromatase (encoded by CYP19A1 gene), and the estrogen receptor (encoded by ESR1 gene)]
Six genes were selected for the vitamin D hypothesis: 7-dehydrocholesterol reductase (DHCR7), vitamin D-binding protein (GC), vitamin D 25-hydroxylase (CYP2R1), 25-hydroxyvitamin D 1-alpha hydroxylase (CYP27B1), vitamin D3 receptor (VDR), and 1,25-dihydroxyvitamin D 24-hydroxylase (CYP24A1) (Fig. 1).
Selection of SNPs
SNPs were selected on the basis of the following criteria: (1) minor allele frequency (MAF) >0.05; (2) haplotype tagging (tag-SNPs) according to HapMap project in Caucasian population; (3) putative functional polymorphisms; and (4) previous association with other interesting phenotypes: 25(OH)D serum levels in a Genome Wide Study [16] and bone mineral density [17–20].
DNA extraction and polymorphism genotyping
Genomic DNA was obtained from leukocytes by two procedures: Autopure LS, a Robotic workstation for automated purification of genomic DNA using Autopure chemistry (LABS Laboratory Biomedical Support Services in the IMIM), and Wizard Genomic DNA Purification Kit (Promega).
Polymorphism genotyping in 343 individuals was carried out using KASPar v4.0 genotyping systems and Kraken allele calling algorithm at the K-bioscience facilities (KBioscience, Herts, England).
To check for the quality of the genotyping, three SNPs were sequenced in 5 % of the population. The results showed a 98.5 % concordance between the two techniques.
Statistical analyses
Accepting an alpha risk of 0.0014 (corrected P value considering 35 included polymorphisms) and a beta risk of 0.2 in a two-sided test, at least 67 subjects were necessary in each group to recognize as statistically significant a minimum clinically relevant difference [21] of 1.8 U in VAS pain increase. The common deviation is assumed to be 2.5. It has been anticipated a drop-out rate of 5 %. Taking into account that the lowest MAF for the genotyped SNPs was 0.13, the number of individuals carrying the minor allele is approximately 75 in our cohort.
Hardy–Weinberg equilibrium (HWE) was calculated by χ 2. HWE P values for all the SNPs were calculated using the Tufts University website template (http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls).
Univariable and multivariable linear regressions (log-additive, dominant, and recessive models) were used to assess the association between the studied SNPs and absolute increase in VAS at 3 and 12 months follow-up. Multivariable models were adjusted for age, body mass index (BMI), and years since menopause. Further, potential confounding for baseline and follow-up 25(OH)D concentrations was assessed. As a sensitivity analysis, all linear regression models were repeated after excluding participants with baseline 25(OH)D levels ≥30 ng/ml. To minimize false discovery due to multiple testing, we performed the false discovery rate (FDR) correction as proposed by Benjamini and Hochberg [22] accepting all predictions with q value <0.05 as significant.
We also tested for a priori-defined interactions by introducing multiplicative terms in the regression equations: (1) vitamin D levels into the effect of the tested SNPs; and (2) interactions between rs4646536 (CYP27B1), rs11568820 (VDR), and rs6163 (CYP17A1).
The associations between these SNPs and both secondary outcomes (worsening pain and therapy discontinuation) were analyzed using logistic regression.
All analyses were two-tailed. Statistical analyses were performed using SPSS for Windows version 13.0 and R for Windows version 2.13.2 (packages: SNPassoc, foreign, multtest, epicalc, and rms).
Results
Baseline characteristics
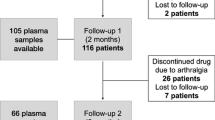
A total of 343 women were recruited from March 2006 to April 2012. Of these, 334 (97.4 %) completed at least 3 months of treatment, and 321 (93.6 %) attended the 1-year follow-up visit (Fig. 2). Baseline characteristics and pain phenotypes at 3 and 12 months follow-up for the study participants are shown in Table 1. Amongst them, 161/334 (48.2 %) and 176/321 (54.8 %) reported worsening pain at 3 and 12 months, respectively. A total of 15 (4.4 %) women discontinued treatment due to severe AIA.
Genetic association with absolute VAS increase
MAF and HWE P values for each SNP in the B-ABLE cohort are reported in Table 2.
Table 3 shows average absolute difference in VAS score for each genotype compared to the reference group (beta coefficients) and P values for SNPs significantly associated with the studied outcome. A number of these SNPs, belonging to two genes, CYP17A1 and VDR, appeared significantly associated with AIA.
Since the majority of the genotyped SNPs in CYP17A1 reached a significant association with the outcome (Table 3), one representative SNP (rs6163), located inside the gene sequence, was chosen for interaction analyses. One SNP (rs4646536) in the CYP27B1 gene showed a P = 0.008 in the association with VAS increment at 3 months post-AI treatment but did not withstand FDR correction. However, this SNP was tested for an interaction with VDR due to their potential relationship (Fig. 1): the interaction term was borderline significant at 3 months (P = 0.06), and consistent with this, a stratified analysis showed that the effect of rs4646536 genotypes on intensity of arthralgia was significant (P = 0.02) in presence of C:C genotype for rs11568820 (VDR), but not among participants with other genotypes. Another significant interaction was observed between rs6163 in CYP17A1 and rs4646536 in CYP27B1 (P = 0.01). The stratified analysis demonstrated different pain intensities among rs4646536 genotypes (P = 0.0006 at 3 months and P = 0.003 at 12 months follow-up) only in presence of C:C genotype for the rs6163, and no such association in A:A or A:C patients. Other interactions tested between SNPs in CYP17A1 and VDR did not yield significance (data not shown). The crude effect of each genotype for the significant SNPs on arthralgia is shown in Fig. 3.
Further adjustment for baseline serum vitamin D, AI, and tamoxifen use
We adjusted the multivariable models for baseline 25(OH)D serum levels and vitamin D increments (3 month-baseline and 12 month-baseline) and tested for an interaction with the SNPs in genes related to vitamin D (VDR and CYP27B1). None of these substantially changed either the beta coefficients or the P values for the associations previously described. A sensitivity analysis looking only at patients with vitamin D deficiency at baseline gave similar results in terms of effect size and direction of the associations previously observed, but higher P values due to reduced power secondary to the exclusion of 25 (7.33 %) participants with optimal levels of 25(OH)D at baseline (data not shown). All these associations were independent of the AI prescribed to the patient (data not shown). In order to evaluate if patients treated previously with tamoxifen could have altered AI-tolerance levels, we compared baseline join pain scores and AIA at 3 and 12 month between patients with or without tamoxifen treatment. In our cohort, no differences were found and therefore, the further adjustment for previous tamoxifen therapy did not affect the association results (data not shown).
Genetic association with worsening pain
The same SNPs associated with pain intensity were also related to worsening pain: adjusted odds ratios (ORs) (95 % CI) at 3 and 12 months for rs6163 were 0.61 (0.43–0.87; P = 0.005) and 0.70 (0.49–0.99; P = 0.04), respectively. OR for worsening pain at 1 year for rs11568820 was 2.14 (1.34–3.43; P = 0.001) in the dominant model. An additional SNP (rs4775936) in the promoter region of CYP19A1 gene (aromatase) was associated with worsening pain at 3 months [multivariable adjusted OR 1.62 (1.16–2.25; P = 0.003)].
Compound genotypes analysis
Table 4 shows the effect of compound genotypes on AIA. An additive effect of the unfavorable alleles for rs4646536 (CYP27B1) and rs6163 (CYP17A1) on pain intensity was observed: participants with four unfavorable alleles (13.54 and 13.59 % at 3 and 12 months, respectively) had an average increase of two points in VAS scale compared to those with only one unfavorable allele (8.61 and 8.97 % at 3 and 12 months, respectively) (P < 0.001). An additive effect was also observed with worsening pain for participants with four unfavorable alleles: OR 3.88 (1.42–11.12) (P < 0.01) at 3 months and OR 5.27 (1.81–16.44) (P < 0.001) at 12 months.
Combination of rs6163 and rs11568820 compound genotypes also appeared associated with both absolute VAS increment and worsening pain at 12 months after therapy initiation in participants with 2 (37.94 %) and 3 (15.43 %) unfavorable alleles compared to the reference group with zero unfavorable alleles (8.36 %). The sample size of this combination was not powered to detect the same association in individuals with four unfavorable alleles (six participants).
Finally, a trend for the combination of rs4646536 and rs11568820 has also been observed.
Therapy discontinuation
Finally, the analyses of the association between these same three SNPs and therapy discontinuation showed only one significant result: 12 patients out of the 185 homozygotes (6.49 %) for the unfavorable genotype of rs4646536 discontinued AI therapy due to severe arthralgia, compared to only three out of the 133 (2.26 %) heterozygotes, and none of the remaining 19 participants [adjusted OR 0.29 (95 %CI 0.09–0.99), P = 0.02].
Discussion
We demonstrate that AIA is genetically determined: SNPs located in genes encoding key factors for the metabolism of estrogens and vitamin D (CYP17A1, VDR, and CYP27B1) are associated with self-reported arthralgia during AI therapy. We report a major interaction between CYP17A1 and CYP27B1 in our study. Accordingly, patients who carry a bad–bad genotype for both genes have the worst clinical response, with two points higher increase in pain at 3 and 12 months follow-up, compared to participants with the most favorable genotype for these same SNPs. Stronger effects were detected when worsening pain was the outcome, with an OR above five for the most unfavorable genotypes. This suggests that both metabolic hormone pathways might be AIA determinants.
CYP17A1 is a key enzyme in the steroidogenic pathway [23]. Assuming that aromatase would maintain some minimum activity during AI therapy, the amount of substrate available might be crucial for this process and would therefore determine the levels of final product. Hence, this estrogen background could contribute to the prevention of AIA.
The other two genes associated with AIA in our data are involved in vitamin D signaling. The potential non-skeletal effects of vitamin D include the synovium and muscle, with putative consequences in the development of arthritis and arthralgia [13]. Consistent with our data, a correlation between musculoskeletal pain intensity and lower vitamin D plasma concentrations has been shown among breast cancer survivors on AI therapy [24].
The SNP rs11568820 is in a cdx-2-binding site in the VDR promoter and can modulate gene expression levels. Arai et al. [25] demonstrated in an in vitro study that rs11568820 alleles have different transcriptional activities. Interestingly, other genotyped SNPs in the VDR gene did not appear associated to the outcome. This would suggest a possible functionality of rs11568820 in the AIA pathogenesis.
CYP27B1 catalyzes the conversion of calcidiol (25(OH)D) to calcitriol (1,25(OH)2D) [26] which binds to VDR. Two tag SNPs were genotyped: rs10877012 in the promoter and rs4646536 in the intronic region. Both SNPs reached a P < 0.05 in the association with AIA at 3 months of treatment, but did not withstand for FDR correction. However, the fact that this CYP acts in a previous step of VDR signaling would suggest that this association is biologically plausible. Supporting this suggestion, we have also shown an association between these SNPs and therapy discontinuation in our cohort, stressing the main role of vitamin D in the occurrence of arthralgia. We hypothesize that genetic variations in CYP27B1 gene reduce the efficiency of the hydroxylation of 25(OH)D to 1,25(OH)2D resulting in suboptimal concentrations of the active ligand.
There is observational [13] and experimental data [27] suggesting a potential effect of vitamin D supplementation in preventing AIA. All the participants in our study were supplemented with vitamin D at the time of AI initiation according to current guidelines [28, 29], with a 71.38 % of patients achieving 25(OH)D levels >30 ng/ml. The mean (SD) was of 44.24 (27.75) ng/ml after 3 months of treatment. Low drop-out rates in our cohort (<7 % after 1 year of follow-up) compared to those described by other AIA studies could be explained by this supplementation with vitamin D amongst other factors.
No significant differences in 25(OH)D serum levels were found among genotypes for any SNP at 3 or 12 months of follow-up. Furthermore, the observed associations between selected genetic variants and AIA remained significant after adjusting for both baseline and increments in vitamin D serum levels. This finding suggests that the effect of these genetic determinants on pain intensity is independent of serum 25(OH)D. Accordingly, the two genes related to vitamin D response, which are associated to AIA in our study, work downstream of 25(OH)D.
We hypothesize that some patients with normal vitamin D (25(OH)D) serum levels, but with a less functional genotype for the described SNPs of CYP27B1 and VDR, within a context of reduced estrogen levels, would be more likely to experience AIA. Thus, vitamin D-supplemented women who still experience pain have a genetic background that could explain this response. Some studies have demonstrated that the conversion of 25(OH)D to 1,25(OH)2D by CYP27B1 depends on the substrate concentration [30]. Hence, higher supplementation of 25(OH)D may help to compensate the lack of function of the enzyme, achieving higher serum concentrations of the active ligand (1,25(OH)2D), which in turn would benefit a less functional VDR, contributing to decrease pain levels. However, this intervention may be also insufficient, and therefore alternative therapies should be implemented.
We also describe a borderline significant interaction between CYP27B1 (rs4646536) and VDR (rs11568820). Our study was not powered to detect this interaction. However, the significant differences among rs4646536 genotypes are detected only in a CC context of the rs11568820, suggesting that this interaction could constitute a true finding. This allows us to speculate on the existence of a complex regulatory mechanism of vitamin D response where all proteins and factors involved in the signal pathway are tightly coupled. Therefore, the addition of small functionality changes in different proteins of the pathway drive to determine the final vitamin D response. However, further research is required in order to clarify the genetic architecture underlying 1,25(OH)2D serum concentrations, and to unravel the mechanisms of action responsible for these associations and interactions.
Finally, the additive effect of the alleles observed suggests that the combination of functional genetic variants in CYP17A1, VDR, and CYP27B1 genes are decisive for the occurrence and/or magnitude of arthralgia.
Moreover, CYP19A1 has been associated to worsening pain in our study, replicating previous work by Mao et al. [6]. Interestingly, both studies defined AIA as a binary outcome.
The main limitation of our study is that the sample size available (343 women) does not allow us to study rare genotypes or to examine the interactions with a low effect size. Hence, our results need further replication in larger studies. Nonetheless, our cohort is the largest source of prospective observational data on women with adjuvant AI treatment to our knowledge. Furthermore, the low drop-out rates can be considered as a potential strength allowing the continual follow-up of the patients with AI-treatment.
Conclusion
Genetic variants in the CYP17A1, VDR, and CYP27B1 genes lead to assess the risk of AIA. The determination of these SNPs could be useful in clinical practice to identify women at high risk of AIA and therapy discontinuation, who could then be targeted for higher dose vitamin D supplementation and/or monitoring strategies to improve quality of life and compliance.
References
Kudachadkar R, O’Regan RM (2005) Aromatase Inhibitors as adjuvant therapy for postmenopausal patients with early stage breast Cancer. CA Cancer J Clin 55:145–163. doi:10.3322/canjclin.55.3.145
Baum M, Buzdar A, Cuzick J, Forbes J, Houghton J, Howell A, Sahmoud T (2003) Anastrozole alone or in combination with tamoxifen versus tamoxifen alone for adjuvant treatment of postmenopausal women with early-stage breast cancer: results of the ATAC (arimidex, tamoxifen alone or in combination) trial efficacy and safety update analyses. Cancer 98:1802–1810. doi:10.1002/cncr.11745
Henry NL, Azzouz F, Desta Z, Li L, Nguyen AT, Lemler S, Hayden J, Tarpinian K, Yakim E, Flockhart DA, Stearns V, Hayes DF, Storniolo AM (2012) Predictors of aromatase inhibitor discontinuation as a result of treatment-emergent symptoms in early-stage breast cancer. J Clin Oncol 30:936–942. doi:10.1200/jco.2011.38.0261
Crew KD, Greenlee H, Capodice J, Raptis G, Brafman L, Fuentes D, Sierra A, Hershman DL (2007) Prevalence of joint symptoms in postmenopausal women taking aromatase inhibitors for early-stage breast cancer. J Clin Oncol 25:3877–3883. doi:10.1200/jco.2007.10.7573
Ingle JN, Schaid DJ, Goss PE, Liu M, Mushiroda T, Chapman J-AW, Kubo M, Jenkins GD, Batzler A, Shepherd L, Pater J, Wang L, Ellis MJ, Stearns V, Rohrer DC, Goetz MP, Pritchard KI, Flockhart DA, Nakamura Y, Weinshilboum RM (2010) Genome-wide associations and functional genomic studies of musculoskeletal adverse events in women receiving aromatase inhibitors. J Clin Oncol 28:4674–4682. doi:10.1200/jco.2010.28.5064
Mao J, Su HI, Feng R, Donelson M, Aplenc R, Rebbeck T, Stanczyk F, DeMichele A (2011) Association of functional polymorphisms in CYP19A1 with aromatase inhibitor associated arthralgia in breast cancer survivors. Breast Cancer Res 13:R8. doi:10.1186/bcr2813
Flores CA, Shughrue P, Petersen SL, Mokha SS (2003) Sex-related differences in the distribution of opioid receptor-like 1 receptor mRNA and colocalization with estrogen receptor mRNA in neurons of the spinal trigeminal nucleus caudalis in the rat. Neuroscience 118:769–778
Blomqvist A (2000) Sex hormones and pain: a new role for brain aromatase? J Comp Neurol 423:549–551. doi:10.1002/1096-9861(20000807
Felson DT, Cummings SR (2005) Aromatase inhibitors and the syndrome of arthralgias with estrogen deprivation. Arthr Rheum 52:2594–2598
Cvoro A, Tatomer D, Tee M-K, Zogovic T, Harris HA, Leitman DC (2008) Selective estrogen receptor-Î2 agonists repress transcription of proinflammatory genes. J Immunol 180:630–636
Morales L, Pans S, Verschueren K, Van Calster B, Paridaens R, Westhovens R, Timmerman D, De Smet L, Vergote I, Christiaens MR, Neven P (2008) Prospective study to assess short-term intra-articular and tenosynovial changes in the aromatase inhibitor-associated arthralgia syndrome. J Clin Oncol 26:3147–3152. doi:10.1200/JCO.2007.15.4005
Nogues X, Servitja S, Peña MJ, Prieto-Alhambra D, Nadal R, Mellibovsky L, Albanell J, Diez-Perez A, Tusquets I (2010) Vitamin D deficiency and bone mineral density in postmenopausal women receiving aromatase inhibitors for early breast cancer. Maturitas 66:291–297. doi:10.1016/j.maturitas.2010.03.012
Prieto-Alhambra D, Javaid M, Servitja S, Arden N, Martinez-GarcÃa M, Diez-Perez A, Albanell J, Tusquets I, Nogues X (2010) Vitamin D threshold to prevent aromatase inhibitor-induced arthralgia: a prospective cohort study. Breast Cancer Res Treat 125:869–878. doi:10.1007/s10549-010-1075-9
Servitja S, Nogués X, Prieto-Alhambra D, Martínez-García M, Garrigós L, Peña MJ, de Ramon M, Díez-Pérez A, Albanell J, Tusquets I (2011) Bone health in a prospective cohort of postmenopausal women receiving aromatase inhibitors for early breast cancer. The Breast 21(1):95–101. doi:10.1016/j.breast.2011.09.001
Winer EP, Hudis C, Burstein HJ, Wolff AC, Pritchard KI, Ingle JN, Chlebowski RT, Gelber R, Edge SB, Gralow J, Cobleigh MA, Mamounas EP, Goldstein LJ, Whelan TJ, Powles TJ, Bryant J, Perkins C, Perotti J, Braun S, Langer AS, Browman GP, Somerfield MR (2005) American Society of clinical oncology technology assessment on the use of aromatase inhibitors as adjuvant therapy for postmenopausal women with hormone receptor-positive breast cancer: status report 2004. J Clin Oncol 23:619–629. doi:10.1200/jco.2005.09.121
Wang TJ, Zhang F, Richards JB, Kestenbaum B, van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, Peltonen L, Cooper JD, O’Reilly PF, Houston DK, Glazer NL, Vandenput L, Peacock M, Shi J, Rivadeneira F, McCarthy MI, Anneli P, de Boer IH, Mangino M, Kato B, Smyth DJ, Booth SL, Jacques PF, Burke GL, Goodarzi M, Cheung C-L, Wolf M, Rice K, Goltzman D, Hidiroglou N, Ladouceur M, Wareham NJ, Hocking LJ, Hart D, Arden NK, Cooper C, Malik S, Fraser WD, Hartikainen A-L, Zhai G, Macdonald HM, Forouhi NG, Loos RJF, Reid DM, Hakim A, Dennison E, Liu Y, Power C, Stevens HE, Jaana L, Vasan RS, Soranzo N, Bojunga J, Psaty BM, Lorentzon M, Foroud T, Harris TB, Hofman A, Jansson J-O, Cauley JA, Uitterlinden AG, Gibson Q, Järvelin M-R, Karasik D, Siscovick DS, Econs MJ, Kritchevsky SB, Florez JC, Todd JA, Dupuis J, Hyppönen E, Spector TD (2010) Common genetic determinants of vitamin D insufficiency: a genome-wide association study. The Lancet 376:180–188. doi:10.1016/S0140-6736(10)60588-0
Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, Orimo H (1996) Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res 11:306–311
Enjuanes A, Garcia-Giralt N, Supervía A, Nogués X, Ruiz-Gaspà S, Bustamante M, Mellibovsky L, Grinberg D, Balcells S, Díez-Pérez A (2006) A new SNP in a negative regulatory region of the CYP19A1 gene is associated with lumbar spine BMD in postmenopausal women. Bone 38:738–743
Zarrabeitia MT, Hernandez JL, Valero C, Zarrabeitia AL, Garcia-Unzueta M, Amado JA, Gonzalez-Macias J, Riancho JA (2004) A common polymorphism in the 5′-untranslated region of the aromatase gene influences bone mass and fracture risk. Eur J Endocrinol 150:699–704. doi:10.1530/eje.0.1500699
Langdahl G, Brixen E (2000) Polymorphisms in the vitamin D receptor gene and bone mass, bone turnover and osteoporotic fractures. Eur J Clin Invest 30:608–617
Todd KH, Funk JP (1996) The minimum clinically important difference in physician-assigned visual analog pain scores. Acad Emerg Med 3:142–146. doi:10.1111/j.1553-2712.1996.tb03402.x
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 57:289–300. doi:10.2307/2346101
Dhir V, Ivison HE, Krone N, Shackleton CHL, Doherty AJ, Stewart PM, Arlt W (2007) Differential inhibition of CYP17A1 and CYP21A2 activities by the P450 oxidoreductase mutant A287P. Mol Endocrinol 21:1958–1968. doi:10.1210/me.2007-0066
Waltman NL, Ott CD, Twiss JJ, Gross GJ, Lindsey AM (2009) Vitamin D insufficiency and musculoskeletal symptoms in breast cancer survivors on aromatase inhibitor therapy. Cancer Nurs 32:143–150. doi:10.1097/01.NCC.0000339262.44560.92
Arai H, Miyamoto K-I, Yoshida M, Yamamoto H, Taketani Y, Morita K, Kubota M, Yoshida S, Ikeda M, Watabe F, Kanemasa Y, Takeda E (2001) The polymorphism in the caudal-related homeodomain protein Cdx-2 binding element in the human vitamin D receptor gene. J Bone Miner Res 16:1256–1264. doi:10.1359/jbmr.2001.16.7.1256
Takeyama K-I, Kitanaka S, Sato T, Kobori M, Yanagisawa J, Kato S (1997) 25-Hydroxyvitamin D3 1α-hydroxylase and vitamin D synthesis. Science 277:1827–1830. doi:10.1126/science.277.5333.1827
Rastelli A, Taylor M, Gao F, Armamento-Villareal R, Jamalabadi-Majidi S, Napoli N, Ellis M (2011) Vitamin D and aromatase inhibitor-induced musculoskeletal symptoms (AIMSS): a phase II, double-blind, placebo-controlled, randomized trial. Breast Cancer Res Treat 129:107–116. doi:10.1007/s10549-011-1644-6
Reid DM, Doughty J, Eastell R, Heys SD, Howell A, McCloskey EV, Powles T, Selby P, Coleman RE (2008) Guidance for the management of breast cancer treatment-induced bone loss: a consensus position statement from a UK expert group. Cancer Treat Rev 34(suppl 1):S3–S18. doi:10.1016/j.ctrv.2008.03.007
Hadji P, Body JJ, Aapro MS, Brufsky A, Coleman RE, Guise T, Lipton A, Tubiana-Hulin M (2008) Practical guidance for the management of aromatase inhibitor-associated bone loss. Ann Oncol 19:1407–1416. doi:10.1093/annonc/mdn164
Rejnmark L, Vestergaard P, Heickendorff L, Mosekilde L (2008) Plasma 1,25(OH)2D levels decrease in postmenopausal women with hypovitaminosis D. Eur J Endocrinol 158:571–576. doi:10.1530/EJE-07-0844
Acknowledgments
This work was supported by Grants from the Generalitat de Catalunya (DIUE 2009 SGR 818) and the Red Temática de Investigación Cooperativa en Envejecimiento y Fragilidad (RETICEF). Grant FIS PI10/01464 (Carlos III Health Institute, Science and Innovation Ministry) is also acknowledged. The authors thank Elaine M. Lilly, Ph.D., for helpful advice and critical reading of the manuscript.
Disclosures
The authors state that they have no conflicts of interest.
Ethics Approval
The protocol of this study was approved by the local ethics committee (CEIC Parc de Salut Mar). Written informed consent was obtained from all participants for their inclusion in the study as well as for DNA extraction.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Garcia-Giralt, N., Rodríguez-Sanz, M., Prieto-Alhambra, D. et al. Genetic determinants of aromatase inhibitor-related arthralgia: the B-ABLE cohort study. Breast Cancer Res Treat 140, 385–395 (2013). https://doi.org/10.1007/s10549-013-2638-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-013-2638-3