Abstract
Inconsistency of reported associations between the Pro919Ser polymorphism in the BRCA1 interacting protein 1 (BRIP1) gene and breast cancer prompted us to undertake a meta-analysis. Although investigated by fewer studies, we have also studied the risk associated with the two additional BRIP1 polymorphisms, C47G and G64A, and breast cancer riskWe conducted searches of the published literature in MEDLINE through PubMed up to October 2012. Individual data on 5,122 cases and 5,735 controls from eight published case–control studies were evaluated for the Pro919Ser polymorphism. Accordingly, C47G and G64A polymorphisms were studied in 1,539 cases and 1,183 controls, and 667 and 782, respectively.In the overall analysis, association was lacking between the Pro919Ser polymorphism and breast cancer risk (odds ratio [OR] 0.98–1.02), materially unchanged when confined to subjects of European ancestry (OR 0.96–1.03) or even in the high-powered studies (OR 0.97–1.03). In the menopausal subgroups, premenopausal women followed the null pattern (OR 0.94–0.98) for the Pro and Ser allele contrasts, but not for the Pro-Ser genotype comparison where significant increased risk was observed (OR 1.39, P = 0.002). The postmenopausal women (>50 years) exhibited a range of pooled effects from protection (OR 0.83, P = 0.11) in the Pro-Ser genotype to slightly increased risk (OR 1.12–1.16, P = 0.28–0.42) in the Pro and Ser allele comparisons. The G64A polymorphism effects were essentially null (OR 0.90–0.98), but C47G was found to confer non-significantly increased risk under all genetic models (OR 1.27–1.40).Upon conclusion, overall summary estimates imply no associations but suggest susceptibility among carriers of the C47G polymorphism and Pro-Ser genotype in premenopausal women. The premenopausal findings and variable outcomes in postmenopausal women require more studies for confirmation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
BRCA1-interacting protein 1 (BRIP1) or BRCA1-associated C-terminal helicase-1 (BACH1), located at chromosome 17q23, belongs to a DEAH helicase family. Also known as FANCJ, BRIP1/BACH1 is a tumor suppressor gene identified through mutations in breast cancer and Fanconi anemia, a childhood cancer [1]. Fanconi anemia is a genetic disease (autosomal recessive or X-linked) characterized by multiple congenital abnormalities, bone marrow failure and cancer susceptibility [2]. BRIP1 mutations were found in Fanconi anemia patients belonging to the complementation group J (FANCJ) [3, 4]. BRIP1 interacts directly with BRCA1, mutations of which accounts for <25 % of excess familial risk for breast cancer [5]. This interaction is mediated through BRCT domains of BRCA1 that is required for establishing the G2 cell-cycle checkpoint response to DNA damage [6], where BRIP1 contributes to BRCA1-associated DNA double strand break repair and homologous recombination repair function [7, 8]. BRIP1 is thought to unwind DNA in the vicinity of the DNA damage and facilitate access of BRCA1 to these sites [9]. It has been suggested that BRIP1 function is required for timely arrival of BRCA1 into DNA damage foci for normal kinetics of double strand break repair. Using BRIP1 deficient cells, Peng et al. [9] provide direct evidence for involvement of BRIP1 in DNA repair as well as for localizing BRCA1.
Evidence suggests the presence of an association of the BRIP1 Pro919Ser polymorphism (dbSNP ID: rs4986764) with breast cancer [10] although other evidence point to its absence [11, 12]. The Ser allele variant in Pro919Ser has recently been associated with an increased breast cancer risk in a kin-cohort study [13]. However, a SNP tagging approach as well as a case–control study [12] found no associations of this polymorphism with this disease [14]. Other approaches such as mutational analyses elicited variable outcomes [12, 15–18]. Two other BRIP1 polymorphisms (C47G and G64A) have received less attention, but availability of genotype data in the literature warranted inclusion of these in our analysis. The G64A polymorphism (rs2048718) might affect gene regulation [17, 19]; however, studies have demonstrated no associations of this polymorphism with breast cancer [19, 20]. On the other hand, the C47G polymorphism (rs4988351) has been demonstrated to have a study-specific increased risk effect for the G variant [21]. The increasing number of reports and discrepancy of the findings for the BRIP1 polymorphisms in breast cancer prompted us to perform a meta-analysis.
Materials and methods
The literature search
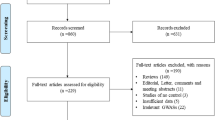
We adopted four search strategies in MEDLINE using PubMed to look for association studies as of October 2012. In all four, we used “breast cancer” in combination with each of the following terms: “BRIP1” “BACH1”, “FANCJ” and “Fanconi anemia.” The resulting four combinations or strategies yielded 79, 54, 42, and 215 citations, respectively. The last three strategies culminated in the same identity of articles as the first one (“BRIP1” and “breast cancer”), which we thus opted to use, outlined in Fig. 1. Of the 79 citations, 16 were retrieved as full text for further evaluation. Studies were eligible if they had genotypic data with a case–control design. Eight studies [10–12, 16–19, 21] were eventually included in this meta-analysis, which focused on the Pro919Ser polymorphism. Of the eight, two other polymorphisms (C47G and G64A) were included in three [16, 17, 21] and two [16, 19] studies, respectively (Table 1).
Data extraction and power calculations
Two investigators independently extracted data and reached consensus on all the items. The following information was obtained from each publication: first author’s name, published year, country of origin, dominant ancestry of the study populations, matching criteria, sample source, genotype data, number of cases and controls. We also calculated frequencies of the variant allele, deviations of controls from the Hardy–Weinberg equilibrium (HWE) as well as statistical power of each study. Assuming odds ratio (OR) of 1.5 at a genotypic risk level of α = 0.05 (two-sided), power was considered adequate at ≥80 %.
Meta-analysis
Risks (ORs) of breast cancer with the Pro919Ser BRIP1 polymorphisms were estimated for each study. Frequency of the Ser allele is minor in six of the eight studies but not in Vahteristo et al. [12] and Frank et al. [19]. Given non-uniformity of the minor allele frequency across the studies, we thus compared the following for Pro919Ser: (i) Ser allele with Ser-Pro/Pro-Pro genotype, (ii) Pro allele with Ser-Pro/Pro-Pro genotype, and (iii) Pro/Ser genotype with homozygous Pro-Pro and Ser-Ser genotypes.
Given uniformity of the minor allele frequencies for G64A and C47G in the included studies, pooled ORs were calculated following the standard genetic models: (i) additive: (AA and GG genotypes compared with the GG and CC, respectively in G64A and C47G), (ii) allelic: (frequency of variant alleles [A and G] assuming the risk could differ across all three genotypes), (iii) recessive (AA vs. AG + GG; CC vs. CG + GG) and (iv) dominant: (AA + AG vs. GG; CC + CG vs. GG).
To compare effects on the same baseline, we used raw data to calculate pooled ORs which were obtained using either the fixed [22] (in the absence of heterogeneity) or random [23] (in its presence) effects models. Heterogeneity between studies was estimated using the χ2-based Q test [24]. Given the low power of this test [25], significance threshold was set at P = 0.10. Heterogeneity between studies was estimated using the χ2-based Q test [24], explored using subgroup analysis [24] with menopausal status as variables, and quantified with the I 2 statistic which measures degree of inconsistency among studies [26]. Data were analyzed using Review Manager 4.3 and SigmaStat 2.03. Significance was set at a P value of ≤0.05 throughout except in heterogeneity estimation. Publication bias was not investigated because of the low sensitivity of the qualitative and quantitative tests when the number of studies is lower than ten [27].
Results
Overall
We undertook a meta-analysis using data from eight studies (5,122 cases/5,735 controls) where we investigated association of three BRIP1 polymorphisms with breast cancer risk. Table 2 shows the Pro919Ser findings where null effects (OR 0.98–1.02) were detected in the overall analysis. Confined to the majority of samples represented by European originated Caucasians (six studies: 4,458 cases/5,041 controls), the null effects remained (OR 0.96–1.03). Confining the studies with >80 % statistical power (five studies: 4,813 cases/5,402 controls) also showed null associations (ORs 0.97–1.03). Consistent with the Pro919Ser findings, G64A effects, from two studies (667 cases/782 controls) were also null (OR 0.90–0.98) as shown in Table 3. Tables 2 and 3 show that all these findings were observed in the absence of heterogeneity (I 2 = 0 %) indicating that the studies are similar enough to be pooled. Only the C47G polymorphism, from three studies (1,539 cases/1,183 controls) differed in pooled effects where non-significant increased risks for the Pro-Ser genotype were observed (OR 1.27–1.40, P = 0.12–0.26) under variable heterogeneous conditions (I 2 = 25–74 %) (Table 3).
Subgroup analysis
Table 2 shows the stratified analysis by age (below and above 50) which corresponds to premenopausal (<50 years) and postmenopausal (≥50 years) status [12]. Results in the premenopausal group (682 cases/843 controls) were essentially null for the Pro (OR 0.94) and Ser alleles (OR 0.98). Pro-Ser genotype associations in this group of women, however, indicated significant increased risk (OR 1.39, P = 0.002). Pooled ORs among postmenopausal women (755 cases/600 controls) spanned a range of non-significantly protective effects (OR 0.83, P = 0.11) for the Pro-Ser genotype to slightly increased risk (OR 1.12–1.16, P = 0.28–0.42) for the Ser and Pro alleles. Except for premenopausal Pro-Ser genotype (I 2 = 60 %), all were obtained in the absence of heterogeneity (I 2 = 0 %).
Assessment of study quality
We examined association of three BRIP1 polymorphisms (Pro919Ser, G64A, and C47G) with breast cancer risk from a population with a European dominated ancestry indicating minimal admixture. Table 1 summarizes features of the included studies. In all three polymorphisms, the eight studies had control frequencies that conformed to the HWE. Of the eight studies, five [10–12, 19, 21] had a total sample size >1,000 which corresponded to high statistical power (93–99.9 %). Controls were either healthy or cancer-free in six (75 %) studies [11, 12, 16–19]. Six studies (75 %) mentioned tissue source samples, and of the six, five (83 %) used blood as source material [10, 12, 16, 18, 19]. Controls for seven studies (88 %) were matched on levels of age, ethnicity, and geography. Of the seven, three (43 %) used the age criterion [10, 11, 21] and two studies each (29 %) used ethnicity [17, 19] as well as geography [12, 18].
Discussion
With a sample size of over 10,800, our meta-analysis has shown overall null association between the Pro and Ser alleles as well as Pro-Ser genotype and breast cancer. This effect was materially unaltered even when confined to subjects of European ancestry, which comprise the majority (75 %) of our study populations. Allowing a Type I error of 5 %, the present meta-analysis has power greater than 80 % to detect an effect size of 1.5 for the overall and European analyses. Confined to high (>80 %) statistical power, the BRIP1 Pro919Ser findings were still null, all without evidence of heterogeneity. These null findings agree with two studies [12, 14] which found no evidence of association between Pro919Ser and breast cancer risk.
Subgrouped by age and thus menopausal status, the Pro919Ser polymorphism outcomes were still null for the Pro and Ser alleles in the premenopausal (<50 years) studies, without evidence of heterogeneity. However, our Pro-Ser genotype analysis yielded significant 1.4-fold increased risk effects which concur with another study that found a 7-fold increased risk among women to the age 50 from a kin-cohort population [13]. In that study, the relative cumulative risk by the age 50 years was 6.9 for Ser homozygotes (P = 0.02). When extended to age 70 years, no significant association was seen (OR 1.3, P = 22) which was similar to our postmenopausal findings of non-significant 1.2-fold increased risk. Although our meta-analysis data suggest that Pro919Ser polymorphism is not a breast cancer predisposition allele, a low risk effects cannot be discounted. Joint effect of the Pro919Ser variant as well as other epidemiological risk factors such as genetic background and environmental influence may be possible.
Null findings of the G64A polymorphism concur with two other studies that observed no associations with breast cancer [13, 14]. On the other hand, C47G had up to 1.4-fold increased risk in the dominant and allelic models with evidence of heterogeneity. Heterogeneity was not evident in the homozygous and recessive models where susceptibility effects were 1.3-fold. In these models, however, the Garcia-Closas et al. [21] study accounted for 91 % weight contribution to the summary effects (data not shown). This indicates that the summary effects of C47G polymorphism is attributed towards this one study. More studies required to achieve more robust conclusions especially confirm the increased risk outcomes in C47G and null results of G64A.
The strength of our meta-analysis includes: (i) large sample sizes in the overall and European analyses, (ii) ethnic homogeneity in three-fourth of the studies, (iii) controls were healthy, (iv) a high proportion (88 %) of the studies were matched to cases; (v) majority (63 %) of the component studies had high statistical power and (vi) consistent no significant associations in the allele/genotype comparisons and across all genetic models under conditions of consistent zero heterogeneity, rendering chance effects less likely. All these features indicate unlikelihood of selection bias as well as non-differential misclassification bias because the issue of different risks in the control groups of developing breast cancer has been modulated.
Conclusion
To our knowledge, this is the first meta-analysis approach that investigates into associations of three BRIP1 polymorphisms with breast cancer risk. Our results demonstrate that variant alleles in two (Pro919Ser and G64A) of the three BRIP1 polymorphisms elicited no associations with breast cancer risk, confirmed in the subgroup analysis for Pro919Ser. Thus, both polymorphisms are not independent risk factors for breast carcinogenesis. Such analysis may shed light on the complexities in the BRCA pathway providing hypotheses for future functional studies. Increased risk effects of the C47G polymorphism are beset with heterogeneity, although consistent for its increased risk effects in all genetic models. However, these results require more studies for confirmation.
References
Cantor SB, Guillemette S (2011) Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future oncol 7(2):253–261
Taniguchi T, D’Andrea AD (2006) Molecular pathogenesis of Fanconi anemia: recent progress. Blood 107(11):4223–4233
Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, Elghalbzouri-Maghrani E, Steltenpool J, Rooimans MA, Pals G et al (2005) The DNA helicase BRIP1 is defective in Fanconi anemia complementation group. Nat Genet 37(9):934–935
Levran O, Attwooll C, Henry RT, Milton KL, Neveling K, Rio P, Batish SD, Kalb R, Velleuer E, Barral S et al (2005) The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat Genet 37(9):931–933
Easton DF (1999) How many more breast cancer predisposition genes are there? Breast Cancer Res 1(1):14–17
Yu X, Chini CC, He M, Mer G, Chen J (2003) The BRCT domain is a phospho-protein binding domain. Science 302(5645):639–642
Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA et al (2001) BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105(1):149–160
Shiozaki EN, Gu L, Yan N, Shi Y (2004) Structure of the BRCT repeats of BRCA1 bound to a BACH1 phosphopeptide: implications for signaling. Mol Cell 14(3):405–412
Peng M, Litman R, Jin Z, Fong G, Cantor SB (2006) BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene 25(15):2245–2253
Seal S, Thompson D, Renwick A, Elliott A, Kelly P, Barfoot R, Chagtai T, Jayatilake H, Ahmed M, Spanova K et al (2006) Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat Genet 38(11):1239–1241
Huo X, Lu C, Huang X, Hu Z, Jin G, Ma H, Wang X, Qin J, Wang X, Shen H et al (2009) Polymorphisms in BRCA1, BRCA1-interacting genes and susceptibility of breast cancer in Chinese women. J Cancer Res Clin Oncol 135(11):1569–1575
Vahteristo P, Yliannala K, Tamminen A, Eerola H, Blomqvist C, Nevanlinna H (2006) BACH1 Ser919Pro variant and breast cancer risk. BMC Cancer 6:19
Sigurdson AJ, Hauptmann M, Chatterjee N, Alexander BH, Doody MM, Rutter JL, Struewing JP (2004) Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer 4:9
Song H, Ramus SJ, Kjaer SK, Hogdall E, Dicioccio RA, Whittemore AS, McGuire V, Hogdall C, Jacobs IJ, Easton DF et al (2007) Tagging single nucleotide polymorphisms in the BRIP1 gene and susceptibility to breast and ovarian cancer. PLoS ONE 2(3):e268
Cao AY, Huang J, Hu Z, Li WF, Ma ZL, Tang LL, Zhang B, Su FX, Zhou J, Di GH et al (2009) Mutation analysis of BRIP1/BACH1 in BRCA1/BRCA2 negative Chinese women with early onset breast cancer or affected relatives. Breast Cancer Res Treat 115(1):51–55
Guenard F, Labrie Y, Ouellette G, Joly BC, Simard J, Durocher F (2008) Mutational analysis of the breast cancer susceptibility gene BRIP1/BACH1/FANCJ in high-risk non-BRCA1/BRCA2 breast cancer families. J Hum Genet 53(7):579–591
Rutter JL, Smith AM, Davila MR, Sigurdson AJ, Giusti RM, Pineda MA, Doody MM, Tucker MA, Greene MH, Zhang J et al (2003) Mutational analysis of the BRCA1-interacting genes ZNF350/ZBRK1 and BRIP1/BACH1 among BRCA1 and BRCA2-negative probands from breast-ovarian cancer families and among early-onset breast cancer cases and reference individuals. Hum Mutat 22(2):121–128
Silvestri V, Rizzolo P, Falchetti M, Zanna I, Masala G, Bianchi S, Palli D, Ottini L (2011) Mutation analysis of BRIP1 in male breast cancer cases: a population-based study in Central Italy. Breast Cancer Res Treat 126(2):539–543
Frank B, Hemminki K, Meindl A, Wappenschmidt B, Sutter C, Kiechle M, Bugert P, Schmutzler RK, Bartram CR, Burwinkel B (2007) BRIP1 (BACH1) variants and familial breast cancer risk: a case-control study. BMC Cancer 7:83
Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA (2007) Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet 3(3):e42
Garcia-Closas M, Egan KM, Newcomb PA, Brinton LA, Titus-Ernstoff L, Chanock S, Welch R, Lissowska J, Peplonska B, Szeszenia-Dabrowska N et al (2006) Polymorphisms in DNA double-strand break repair genes and risk of breast cancer: two population-based studies in USA and Poland, and meta-analyses. Hum Genet 119(4):376–388
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Lau J, Ioannidis JP, Schmid CH (1997) Quantitative synthesis in systematic reviews. Ann Intern Med 127(9):820–826
Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327(7414):557–560
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Ioannidis JP, Trikalinos TA (2007) The appropriateness of asymmetry tests for publication bias in meta-analyses: a large survey. CMAJ 176(8):1091–1096
Acknowledgments
We thank Ofelia Francisco-Pabalan and Hong Li.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Pabalan, N., Jarjanazi, H. & Ozcelik, H. Association between BRIP1 (BACH1) polymorphisms and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 137, 553–558 (2013). https://doi.org/10.1007/s10549-012-2364-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-012-2364-2