Abstract
Insulin-like growth factor-1 receptor (IGF-1R) is expressed in normal and malignant breast tissue and has been implicated in cell survival and resistance to cytotoxic therapies. We sought to assess the prognostic impact of IGF-1R expression among patients with early breast cancer and among breast cancer subtypes. Patients with stages I–III breast cancer with archival tumor tissue were included. Paraffin tissue blocks were used to construct a tissue microarray that was stained for ER, PR, Ki-67, HER2, EGFR, and cytokeratins 5/6 to classify the breast subgroups and for expression of IGF-1R, p27, and Bcl2 by immunohistochemistry. Kaplan–Meier plots were created by subtypes. Associations between IGF-1R and prognostic variables were examined in multivariate analysis. Among 2,871 eligible women the prognostic cut point for IGF-1R expression for breast-cancer-specific survival (BCSS) was Allred score <7 versus ≥7. IGF-1R was ≥7 in 52% (LuminalA), 57.5% (LuminalB), 44.8% (LuminalHER2), 9.7% HER2-enriched, and 22.5% (Basal-like), P = 1.3 × 10−52. IGF-1R+ was associated with age ≥50, lower histopathology grade, ER+, HER2 negativity (−), high p27 and high Bcl2 score. IGF-1R ≥7 was associated with better BCSS among LuminalB patients, hazard ratio = 0.64 (0.49–0.84); P = 1.2 × 10−3, and worse outcome in the HER2-enriched subtype, hazard ratio = 2.37 (1.21–4.64); P = 0.012. IGF-1R correlates with good prognostic markers among patients with early breast cancer and is differentially expressed with variable prognostic impact among breast cancer subtypes. Results may have relevance to the development of therapeutics targeting IGF-1R.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Insulin-like growth factor-1 receptor (IGF-1R) is a homodimeric receptor tyrosine kinase activated by IGF I/II ligand binding which results in tumor growth and apoptosis blockade [1–6]. This receptor is present in breast cancer as well as other malignancies [7]. Recently, Law et al. [8] have shown that activated IGF-1R may be expressed in all breast cancer subtypes, regardless of estrogen receptor (ER) or HER2 status.
The prognostic and predictive role of IGF-1R is not clearly defined in the literature. Although some investigators have found no correlation with IGF-1R and outcome [9], a recent study of 438 patients with early breast carcinoma reported inferior breast-cancer-specific survival (BCSS) for cases with high levels of phosphorylated IGF-1R [8]. In another study with 126 breast cancer patients, patients with ER negative (−), IGF-1R positive (+) tumors had a worse prognosis [10]. This is in contrast to other reports of IGF-1R as a favorable prognostic factor [11–13]. Differing techniques for assessing the marker and defining its expression may have contributed to the prognostic difference.
Targeting the IGF-1R pathway is an active area of research and a number of agents are in various stages of development. These include antibodies directed at IGF-1R, antisense agents, and small molecules [14–25]. Further understanding of the pattern of IGF-1R expression in breast cancer subtypes and its impact on prognosis may be useful as these agents are being developed.
To define IGF-1R expression and its prognostic relevance in early breast cancer we set out to:
-
(1)
Determine an optimal prognostic cut-point for IGF-1R expression by immunohistochemistry.
-
(2)
Compare IGF-1R expression in benign versus malignant breast tissues.
-
(3)
Describe IGF-1R expression and determine its prognostic impact in breast cancer subtypes in multivariable analysis using conventional prognostic markers.
-
(4)
Examine associations between IGF-1R and p27, a cell cycle inhibitor [26, 27], and Bcl2, an anti-apoptotic marker [28, 29].
Patients and methods
Patients with AJCC stage I–III breast cancer [30, 31] referred to the BC Cancer Agency between 1986 and 1992 with archival tissue were included (n = 4,046). Benign breast tissue from 120 patients without a breast cancer diagnosis was obtained from the Vancouver Coastal Health Pathology Department. Benign histological diagnosis included sclerosing adenosis, radiation scar, and papillomas. The full list is in Table 1.
Among patients with invasive breast cancer, tissue cores were extracted from archival blocks of the primary breast tumor and used to construct tissue microarrays as previously described [32, 33]. Patient and tumor characteristics were reported and included age, tumor size (T), nodal status (N), estrogen receptor (ER), progesterone receptor (PR), and grade. IGF-1R staining was performed using Santa Cruz rabbit polyclonal antibody Cat# sc-713, lot C3005.
Staining of all cases was done as a single run on the automated Ventana immunostainer. Positive controls were applied and the marker was scored according to Allred scoring system from 0 to 8 [7, 34–37]. IGF-1R stained tissue microarrays were digitally scanned. Stains can be seen on the web site: http://www.gpecimage.ubc.ca username: igf1r password: abc123.
Immunohistochemical staining for the biomarkers ER, PR, HER2, Ki-67, EGFR, and CK 5/6 on each of the tissue microarray slides used the standard streptavidin–biotin complex method with DAB chromogen. Staining and interpretation of ER, PR, HER2, Ki-67, EGFR, and CK 5/6 have been previously described [38, 39]. ER and PR positivity were defined as any positive nuclear staining (i.e., ≥1%) and HER2 positive cases were defined as IHC 3+ or if IHC 2+, FISH with amplification ratio ≥2.0. Samples with fewer than 50 tumor cells in the TMA cores were considered uninterpretable and were excluded from analysis. Pathologists scoring the tissue microarrays were blinded to the clinico-pathological characteristics and outcome of each case.
For Bcl2 staining the Dako, mouse clone 124, cat# M0887, 1:200 was used and the staining was performed on an automated Bond-MAX platform (Leica Microsystems), with Tris–EDTA pH 9.0 for 20 min using Bond polymer Refine (F protocol) detection kit. The p27 staining was done with mouse monoclonal antibody (1:100 dilution, clone 57, cat#610241, BD Transduction), and the staining was performed on a semi-automated Ventana Discovery XT System using pre-diluted Ventana Universal Secondary Antibody and DAB MAP detection system. Antigen retrieval included a mild Cell Conditioner 1. For the analysis a binarized score was used. Cases with more than 50% of the tumor nuclei staining for p27 were considered positive for p27. Bcl2 was scored by staining intensity (0 no staining, 1 weak staining, 2 moderate staining, 3 strong staining) and the percentage of positive cells (1–100). Positive cases were defined as those with 1 or higher staining intensity in more than 10% of the cells.
Breast cancer molecular subtypes were classified according to a gene expression profile-validated immunohistochemical surrogate panel [38–40]. LuminalA (ER+ and/or PR+, and Her2− and Ki-67 < 14%), LuminalB (ER+ and/or PR+ and Her2− and Ki-67 ≥ 14%), Luminal/HER2 (ER+ and/or PR+ and Her2+, regardless of Ki67 status), HER2-enriched (ER− and PR− and Her2+), and Basal-like (ER− and PR− and Her2− and (EGFR+ and/or CK 5/6+)).
Cases were excluded due to uninterpretable IGF-1R staining (n = 717 in breast cancer cases and 10 among benign breast cases) or undetermined subtype (n = 548). Tumors staining negative for ER, PR, HER2, and negative for either CK5/6 or EGFR (non-Basal triple negative) were excluded as well (n = 304). Three cases were excluded due to unknown cause of death (Fig. 1).
Statistical analysis
The malignant breast cancer study cohort was divided into a training (n = 1,433) and validation (n = 1,438) set for the purpose of a split-sample validation analysis approach. Exploratory analyses were performed on the training set and repeated on the validation cohort. The details and rationale behind this approach are described in a previous publication [41]. Statistical analyses were performed using SPSS 17.0 (SPSS, Inc.) and R 2.9.1 (http://www.r-project.org/). A two-sided alpha level of 0.05 was used for all statistical tests. Survival analyses were performed using Kaplan–Meier plots and Cox proportional hazards regression models. Generalized Wilcoxon test using the Breslow method was used to compare survival curves on the Kaplan–Meier plots. Survival end points includes breast-cancer-specific (BCSS), overall (OS), and relapse-free (RFS) survival. Local, regional, distant relapse, and breast cancer death events were included in the RFS endpoint. Proportional hazard assumption of the Cox regression models were tested by examining scaled Schoenfeld residual plots. Kendall’s tau-b and Mann–Whitney U tests were used to measure the correlation of IGF-1R expression to clinical parameters and other biomarkers. To assess the prognostic effect of IGF-1R Allred score as a dichotomized variable, the X-tile version 3.6.1 was used. The X-tile program split the cohort randomly into matched training and validation set as a method for selecting optimal cut-points. It is a graphical method that shows the robustness of the relationship between a biomarker and outcome [42]. The optimal cut-off point for IGF-1R was determined by applying this program on the training set (n = 1,432 with 1 case with unknown cause of death excluded) using BCSS as the end point. In X-tile analysis the Allred score which maximized differences in BCSS (based on Log-rank statistics) was chosen. The cut-off point analysis was performed on the training set only to avoid biased results due to “over-fitting.” Further analyses were done using IGF-1R Allred score as a continuous variable on breast-cancer-specific survival using various smoothing methods [43].
Results of the exploratory analyses of IGF-1R continuous Allred score using smoothing methods, Schoenfeld residual plots on selected Cox regression models, and cohort characteristics as well as IGF-1R correlation with selected clinicopathological variables on the training/validation set are presented as Supplemental material.
The study was approved by the University of British Columbia Research Ethics Board and methodology is consistent with REMARK criteria [44, 45].
Results
A total of 2,871 eligible patients with early breast cancer and complete scoring data for IGF-1R and an intrinsic subtype assignment based on immunohistochemistry were included. Median follow-up was 10 years. Twenty-six percent of the patients had received adjuvant chemotherapy, 41% received adjuvant hormonal treatment, and 7.1% had received both modalities.
In X-tile analysis on the training set with BCSS as the end point, the prognostic value of IGF-1R was maximized when IGF-1R Allred score was dichotomized as <7 versus ≥7 defined (Fig. 2a–c). Log-rank test on the training set with Miller–Siegmund correction for multiple comparisons indicates that IGF-1R is significantly associated with BCSS (P = 0.0059). When X-tile analysis was repeated on the validation and the entire cohort the optimal cut-point remained <7 versus ≥7. Using this cut-off point, a total of 1,335 patients, 46% of the entire cohort, were scored as IGF-1R positive (+).
The prognostic value of IGF-1R was further assessed using continuous Allred score. Four smoothing methods were applied to explore any non-linear relationship between IGF-1R Allred score and outcome. Three of the four models showed that the hazard changes are minimal for Allred score between 0 and 6, and Allred score greater or equal to 7 corresponds to lower hazard (superior survival), agreeing with the single-cut point approach (data is shown in Supplemental material).
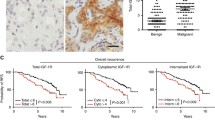
IGF-1R expression was compared between the 2,871 malignant and 110 benign cases. Mean and the median scores were 5.5 and 6.0 in benign tissue and 6.25 and 6 in malignant tissue. Malignant cases were more frequently IGF-1R+ (46%) compared to the benign cohort (15%). Kendall’s rank correlation tau = −0.118, P = 1.4 × 10−10. Detailed scoring for all the study cohort is presented in Table 2.
IGF-1R stratified by prognostic variables
Table 3 provides a correlative analysis between IGF-1R expression and select clinical and pathologic variables. IGF-1R+ status was associated with ER positivity, HER2 negative status, and high Bcl2 score. IGF-1R positivity was weakly correlated with age ≥50, lower histopathology grade and high p27 score. No significant correlation between p27 and IGF-1R was observed on the validation set (P = 0.3). No correlation was found with tumor size and LVI. A low Ki-67 score was correlated with IGF-1R on the validation set but not in the training set. Pertained Kendall’s rank correlation tau and P values are presented in Table 3.
IGF-1R and breast cancer subtypes
The study cohort was subsequently divided into five breast cancer subtypes (Table 4) as defined by immunohistochemical markers and described above: LuminalA (1,356), LuminalB (779), Luminal/HER2 (203), HER2-enriched (226), and Basal-like (307). Median age was generally similar with exception of the Basal-like subtype with a median of 10 years younger. LuminalA cases were more likely to present with T1 stage disease (P < 2×10−16). Negative axillary nodal status was found in more than half of the LuminalA and core Basal-type cases. IGF-1R high expression (Allred score ≥ 7) was seen in a significant proportion of LuminalA (52%), LuminalB (57.5%) and Luminal/HER2+ (44.8%) patients, whereas 90.3% with HER2-enriched and 77.5% with Basal-like were IGF-1R negative (Allred score < 7).
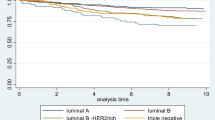
Survival estimates according to IGF-1R positivity were generated for the whole cohort and among breast cancer subtypes. IGF-1R positivity was associated with superior relapse-free survival, breast-cancer-specific survival, and overall survival (OS). Ten-year BCSS was 77.5% for IGF-1R positive versus 69.6% for IGF-1R negative, P = 2.9 × 10−8 (Table 5). IGF-1R had variable effects on survival among individual breast cancer subtypes (Fig. 3a–e, Table 6). Among LuminalB patients IGF-1R+ conferred an improved BCSS (P = 1.9 × 10−4). A trend for superior outcomes was also observed among Luminal/HER2 tumors, P = 0.076. The opposite effect was seen in patients with HER2-enriched subtype where IGF-1R positivity conferred a trend of inferior outcomes (P = 0.069), Fig. 3d.
The prognostic effect of IGFR-1R expression was further evaluated in multivariate models among patients with LuminalB and in HER2-enriched tumors. IGF-1R positivity was associated with improved BCSS among patients with LuminalB, HR = 0.64 (95% CI 0.49, 0.84), P = 1.2 × 10−3. In multivariate analysis of HER2-enriched tumors, IGF-1R was associated with an inferior prognosis, HR = 2.37 (95% CI 1.21. 4.64), P = 0.012. Multivariate analyses included age, grade, LVI, number of positive nodes, tumor size, chemotherapy, hormonal treatment, and IGF-1R expression (Tables 7, 8). Analyses were repeated according to adjuvant systemic treatment as was delivered during the study era. The women were divided into one of the three groups: no adjuvant systemic therapy, tamoxifen only, chemotherapy ± tamoxifen. Cox model was adjusted to age (<50 vs. ≥50 years), T size (≤2 vs. >2 cm), number of positive lymph nodes (0 vs. >0), grade, LVI and IGF-1R (0–6 vs. 7, 8). These additional analyses supported our results: IGF-1R was a good prognostic marker for LuminaB subtype and a bad prognostic marker for HER2-enriched patients (though for HER2 enrich subtype P value was not statistically significant) (Table 9).
Discussion
In this large study, we set out to define a prognostic cut-off point for IGF-1R expression and define its prognostic impact in uni- and multi-variate analysis among breast cancer subtypes. IGF-1R was highly expressed in half of the cohort and was associated with improved outcomes. To our knowledge this is the largest report to study the association and the prognostic value of IGF-1R among the different breast subtypes. The observation that IGF-1R expression has a differential prognostic impact in LuminalB and HER2-enriched tumors is novel.
An accepted cut-point for IGF-1R positivity has not been described in the literature. Various cut-points such as an Allred score 0–2 versus 3–8 [7, 36] or score 0 versus 2–4 versus 5–6 versus 7–8 [34] have been reported. Others investigators avoided any dichotomy and only reported the 0–8 Allred score values [35]. There is no consensus on the cut-off points or mode of reporting IGF-1R which precludes comparisons between studies. A standard methodology and scoring system as well as accepted cut-off points may facilitate assessment of this marker and comparison of results from different laboratories. In this study we have defined a prognostic cut-point for IGF-1R in a training and validation large cohort of patients and optimized this cut-off by X-tile analysis.
Previous studies have shown that IGF-1R overexpression or constitutive activation is sufficient to induce mammary tumor development in vivo [46]. Our analysis of IGF-1R expression in benign versus malignant breast tissue indicates a significantly higher level of IGF-1R positivity as defined by an Allred score of ≥7. Benign tissue included in this study included a number of different entities with different proliferative rates which may affect IGF-1R expression.
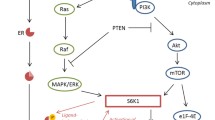
The observation that IGF-1R has a varied prognostic role in the different subtypes might be related to cross-talk between signaling pathways. The favorable outcome for the LuminalB in correlation with high expression of the receptor is in contrast to a previous study suggesting that LuminalB tumors have hyperactive GFR/PI3K signaling and are associated with poor prognosis [47]. An earlier study by the same group showed that tumors that manifested IGF-I signature had a poor outcome event, however, it should be noted that this group examined the expression patterns of the genes which are induced or repressed by IGF-I, whereas this study measured the IGF-1R expression [48].
Univariate analysis revealed that high IGF-1R expression was associated with other good prognostic factors such as older age, ER positivity, lower grade, HER2 negativity, and higher p27 levels. p27 is known to be a tumor suppressor gene and protein levels are reduced in most human cancers [49]. Esparis-Ogando et al. have recently reported that breast cancer cells treated with an IGF-1R antagonist show decreased pAkt and increased levels of p27 [50] which may further point to a connection between these pathways.
We also looked at the correlation of Bcl2 and IGF-1R. Bcl2 has been shown to be a good prognostic factor in early breast cancer and associated with hormone receptor positive tumors [51–53]. This is consistent with our results that overexpression of Bcl2 is correlated with high level of IGF-1R breast cancers.
IGF-1R antagonists have been shown to interact with both ER and HER2 pathways. Recent data indicate that the signaling cross-talk between IGF-1R and the HER2 family is bidirectional and can occur through the various members of the HER receptor family [24, 54]. This cross talk between ER and HER2 suggests that IGF-1R may be an attractive treatment target especially for the “Luminals” and HER2 positive breast cancers. This is supported by in vitro experiments showing a synergistic effect when co-targeting the IGF-1R receptor along with antiestrogen agent [55]. Moreover, growth of tamoxifen resistant MCF-7 cells declines when anti-IGF-1R antibody is added to the cells [56].
With respect to HER2 positive breast cancers there is preclinical evidence that IGF-IR signaling may provide a mechanism of resistance against therapies that target members of the EGF receptor family, including HER2/neu [57–61].
In a randomized phase II study, no benefit was seen among patients with ER positive metastatic breast cancer treated with AMG 479, a fully humanized monoclonal antibody targeting IGF-1R, in combination with endocrine therapy [62]. Our results indicate that the IGF-1R pathway may have particular relevance in the LuminalB and HER2 breast tumors. A recent study on colorectal cell lines demonstrated that IGF-1 is only one part in the endocrine signaling system and that IGF-1R alone is most probably not sufficient to predict drug sensitivity [63, 64]. However, it may be relevant to evaluate benefit among the individual subtypes in the early stages of clinical development. An understanding of the expression levels and their predictive value in the different breast subtypes may provide a rational approach to the development of new agents directed to IGF-1R including stratification or enrollment criteria based on its expression.
References
Kasuya J, Paz IB, Maddux BA et al (1993) Characterization of human placental insulin-like growth factor-I/insulin hybrid receptors by protein microsequencing and purification. Biochemistry 32:13531–13536
Pandini G, Vigneri R, Costantino A et al (1999) Insulin and insulin-like growth factor-I (IGF-I) receptor overexpression in breast cancers leads to insulin/IGF-I hybrid receptor overexpression: evidence for a second mechanism of IGF-I signaling. Clin Cancer Res 5:1935–1944
Adams TE, Epa VC, Garrett TP et al (2000) Structure and function of the type 1 insulin-like growth factor receptor. Cell Mol Life Sci 57:1050–1093
Shang Y, Mao Y, Batson J et al (2008) Antixenograft tumor activity of a humanized anti-insulin-like growth factor-I receptor monoclonal antibody is associated with decreased AKT activation and glucose uptake. Mol Cancer Ther 7:2599–2608
Fagan DH, Yee D (2008) Crosstalk between IGF1R and estrogen receptor signaling in breast cancer. J Mammary Gland Biol Neoplasia 13:423–429
Werner H, Maor S (2006) The insulin-like growth factor-I receptor gene: a downstream target for oncogene and tumor suppressor action. Trends Endocrinol Metab 17:236–242
Ouban A, Muraca P, Yeatman T et al (2003) Expression and distribution of insulin-like growth factor-1 receptor in human carcinomas. Hum Pathol 34:803–808
Law JH, Habibi G, Hu K et al (2008) Phosphorylated insulin-like growth factor-i/insulin receptor is present in all breast cancer subtypes and is related to poor survival. Cancer Res 68:10238–10246
Shimizu C, Hasegawa T, Tani Y et al (2004) Expression of insulin-like growth factor 1 receptor in primary breast cancer: immunohistochemical analysis. Hum Pathol 35:1537–1542
Railo MJ, von Smitten K, Pekonen F (1994) The prognostic value of insulin-like growth factor-I in breast cancer patients. Results of a follow-up study on 126 patients. Eur J Cancer 30A:307–311
Papa V, Gliozzo B, Clark GM et al (1993) Insulin-like growth factor-I receptors are overexpressed and predict a low risk in human breast cancer. Cancer Res 53:3736–3740
Shin A, Ren Z, Shu XO et al (2007) Expression patterns of insulin-like growth factor 1 (IGF-I) and its receptor in mammary tissues and their associations with breast cancer survival. Breast Cancer Res Treat 105:55–61
Bonneterre J, Peyrat JP, Beuscart R et al (1990) Prognostic significance of insulin-like growth factor 1 receptors in human breast cancer. Cancer Res 50:6931–6935
Gualberto A, Pollak M (2009) Emerging role of insulin-like growth factor receptor inhibitors in oncology: early clinical trial results and future directions. Oncogene 28:3009–3021
Blum G, Gazit A, Levitzki A (2000) Substrate competitive inhibitors of IGF-1 receptor kinase. Biochemistry 39:15705–15712
Garcia-Echeverria C, Pearson MA, Marti A et al (2004) In vivo antitumor activity of NVP-AEW541-A novel, potent, and selective inhibitor of the IGF-IR kinase. Cancer Cell 5:231–239
Mitsiades CS, Mitsiades NS, McMullan CJ et al (2004) Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell 5:221–230
Yuen JS, Macaulay VM (2008) Targeting the type 1 insulin-like growth factor receptor as a treatment for cancer. Expert Opin Ther Targets 12:589–603
Stephen RL, Shaw LE, Larsen C et al (2001) Insulin-like growth factor receptor levels are regulated by cell density and by long term estrogen deprivation in MCF7 human breast cancer cells. J Biol Chem 276:40080–40086
Sachdev D, Singh R, Fujita-Yamaguchi Y et al (2006) Down-regulation of insulin receptor by antibodies against the type I insulin-like growth factor receptor: implications for anti-insulin-like growth factor therapy in breast cancer. Cancer Res 66:2391–2402
Burtrum D, Zhu Z, Lu D et al (2003) A fully human monoclonal antibody to the insulin-like growth factor I receptor blocks ligand-dependent signaling and inhibits human tumor growth in vivo. Cancer Res 63:8912–8921
Pandini G, Wurch T, Akla B et al (2007) Functional responses and in vivo anti-tumour activity of h7C10: A humanised monoclonal antibody with neutralising activity against the insulin-like growth factor-1 (IGF-1) receptor and insulin/IGF-1 hybrid receptors. Eur J Cancer 43:1318–1327
Sachdev D, Drug evaluation (2007) CP-751871, a human antibody against type I insulin-like growth factor receptor for the potential treatment of cancer. Curr Opin Mol Ther 9:299–304
Jin Q, Esteva FJ (2008) Cross-talk between the ErbB/HER family and the type I insulin-like growth factor receptor signaling pathway in breast cancer. J Mammary Gland Biol Neoplasia 13:485–498
Schillaci R, Salatino M, Cassataro J et al (2006) Immunization with murine breast cancer cells treated with antisense oligodeoxynucleotides to type I insulin-like growth factor receptor induced an antitumoral effect mediated by a CD8+ response involving Fas/Fas ligand cytotoxic pathway. J Immunol 176:3426–3437
Capodanno A, Camerini A, Orlandini C et al (2009) Dysregulated PI3 K/Akt/PTEN pathway is a marker of a short disease-free survival in node-negative breast carcinoma. Hum Pathol 40:1408–1417
Charpin C, Giusiano S, Secq V et al (2009) Quantitative immunocytochemical profile to predict early outcome of disease in triple-negative breast carcinomas. Int J Oncol 34:983–993
Vendrell JA, Robertson KE, Ravel P et al (2008) A candidate molecular signature associated with tamoxifen failure in primary breast cancer. Breast Cancer Res 10:R88
Bremer TM, Jacquemier J, Charafe-Jauffret E et al (2009) Prognostic marker profile to assess risk in stage I-III hormone receptor-positive breast cancer patients. Int J Cancer 124:896–904
Fleming ID, Cooper JS, Henson DE et al (1997) AJCC Cancer Staging Manual, 5th edn. Lippincott-Raven, Philadelphia
Bearhs OH, Henson DE, Hutter RVP et al (1992) AJCC Cancer Staging Manual, 5th edn. Lippincott-Raven, Philadelphia
Cheang MC, Treaba DO, Speers CH et al (2006) Immunohistochemical detection using the new rabbit monoclonal antibody SP1 of estrogen receptor in breast cancer is superior to mouse monoclonal antibody 1D5 in predicting survival. J Clin Oncol 24:5637–5644
Voduc D, Cheang M, Nielsen T (2008) GATA-3 expression in breast cancer has a strong association with estrogen receptor but lacks independent prognostic value. Cancer Epidemiol Biomarkers Prev 17:365–373
Mulligan AM, O’Malley FP, Ennis M et al (2007) Insulin receptor is an independent predictor of a favorable outcome in early stage breast cancer. Breast Cancer Res Treat 106:39–47
Hakam A, Fang Q, Karl R et al (2003) Coexpression of IGF-1R and c-src proteins in human pancreatic ductal adenocarcinoma. Dig Dis Sci 48:1972–1978
Power KA, Chen JM, Saarinen NM et al (2008) Changes in biomarkers of estrogen receptor and growth factor signaling pathways in MCF-7 tumors after short- and long-term treatment with soy and flaxseed. J Steroid Biochem Mol Biol 112:13–19
Harvey JM, Clark GM, Osborne CK et al (1999) Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer. J Clin Oncol 17:1474–1481
Cheang MC, Chia SK, Voduc D et al (2009) Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst 101:736–750
Cheang MC, Voduc D, Bajdik C et al (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14:1368–1376
Nielson TO, Hsu FD, Jensen K et al (2004) Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res 10:5367–5374
Rajput AB, Turbin DA, Cheang MC et al (2008) Stromal mast cells in invasive breast cancer are a marker of favourable prognosis: a study of 4, 444 cases. Breast Cancer Res Treat 107:249–257
Camp RL, Dolled-Filhart M, Rimm DL (2004) X-tile: a new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 10:7252–7259
Govindarajulu US, Malloy EJ, Gangali B et al. (2009) The comparison of alternative smoothing methods for fitting non-linear exposure-response relationships with cox models in a simulation study. Int J Biostat 5. doi: 10.2202/1557-4679.1104
McShane LM, Altman DG, Sauerbei W et al (2005) Reporting recommendations for tumor marker prognostic studies. J Clin Oncol 23:9067–9072
Hayes DF, Ethier S, Lippman ME (2006) New guidelines for reporting of tumor marker studies in breast cancer research and treatment: REMARK. Breast Cancer Res Treat 100:237–238
Robert A, Toupance B, Tremblay M et al (2009) Impact of inbreeding on fertility in a pre-industrial population. Eur J Hum Genet 17:673–681
Creighton CJ, Fu X, Hennessy BT et al (2010) Proteomic and transcriptomic profiling reveals a link between the PI3K pathway and lower estrogen receptor (ER) levels and activity in ER+ breast cancer. Breast Cancer Res 12:R40. doi:101186/bcr2594
Creighton CJ, Casa A, Lazard Z et al (2008) Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol 26:4078–4085
Chu IM, Hengst L, Slingerland JM (2008) The cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer 8:253–267
Esparis-Ogando A, Ocana A, Rodriguez-Barrueco R et al (2008) Synergic antitumoral effect of an IGF-IR inhibitor and trastuzumab on HER2-overexpressing breast cancer cells. Ann Oncol 19:1860–1869
Binder C, Marx D, Overhoff R et al (1995) Bcl-2 protein expression in breast cancer in relation to established prognostic factors and other clinicopathological variables. Ann Oncol 6:1005–1010
Joensuu H, Pylkkanen L, Toikkanen S (1994) Bcl-2 protein expression and long-term survival in breast cancer. Am J Pathol 145:1191–1198
Lee KH, Im SA, Oh DY et al (2007) Prognostic significance of bcl-2 expression in stage III breast cancer patients who had received doxorubicin and cyclophosphamide followed by paclitaxel as adjuvant chemotherapy. BMC Cancer 7:63
Haluska P, Carboni JM, TenEyck C et al (2008) HER receptor signaling confers resistance to the insulin-like growth factor-I receptor inhibitor, BMS-536924. Mol Cancer Ther 7:2589–2598
Chakraborty AK, Welsh A, Digiovanna MP (2010) Co-targeting the insulin-like growth factor I receptor enhances growth-inhibitory and pro-apoptotic effects of anti-estrogens in human breast cancer cell lines. Breast Cancer Res Treat 120:327–335
Weroha SJ, Haluska P (2008) IGF-1 receptor inhibitors in clinical trials—early lessons. J Mammary Gland Biol Neoplasia 13:471–483
Guix M, Faber AC, Wang SE et al (2008) Acquired resistance to EGFR tyrosine kinase inhibitors in cancer cells is mediated by loss of IGF-binding proteins. J Clin Invest 118:2609–2619
Camirand A, Lu Y, Pollak M (2002) Co-targeting HER2/ErbB2 and insulin-like growth factor-1 receptors causes synergistic inhibition of growth in HER2-overexpressing breast cancer cells. Med Sci Monit 8:BR521-6
Jones HE, Goddard L, Gee JM et al (2004) Insulin-like growth factor-I receptor signalling and acquired resistance to gefitinib (ZD1839; iressa) in human breast and prostate cancer cells. Endocr Relat Cancer 11:793–814
Nahta R, Yuan LX, Zhang B et al (2005) Insulin-like growth factor-I receptor/human epidermal growth factor receptor 2 heterodimerization contributes to trastuzumab resistance of breast cancer cells. Cancer Res 65:11118–11128
Lu Y, Zi X, Zhao Y et al (2001) Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (herceptin). J Natl Cancer Inst 93:1852–1857
Kaufman PA, Ferrero JM, Bourgeois H et al (2010) A randomized, double-blind, placebo-controlled, phase 2 study of AMG-479 with Exemestane (E) or Fulvestrant (F) in postmenopausal women with hormone receptor positive (HR+) metastatic (M) or locally advanced (LA) breast cancer. In: San Antonio breast cancer symposium, CTRC-AACR, San-Antonio, Texas, December 2010
Pitts TM, Tan AK, Kulikowski GN et al (2010) Development of an integrated genomic classifier for novel agent in colorectal cancer: approach to individualized therapy in early development. Clin Cancer Res 16:3193–3204
Yee D (2010) How to train your biomarker. Clin Cancer Res 16:3091–3093
Acknowledgments
Thanks to Prof. Torsten Nielsen’s (MD/PHD) helpful comments. GPEC lab is supported through unrestricted educational funds from Sanofi Aventis Canada.
Conflict of interest
No conflict of interest; declared by all authors.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yerushalmi, R., Gelmon, K.A., Leung, S. et al. Insulin-like growth factor receptor (IGF-1R) in breast cancer subtypes. Breast Cancer Res Treat 132, 131–142 (2012). https://doi.org/10.1007/s10549-011-1529-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-011-1529-8