Abstract
BRCA1 promoter methylation reportedly plays an important part in the pathogenesis of human breast cancer. In the present study, we investigated whether or not BRCA1 promoter methylation in peripheral blood cells (PBCs) can serve as a risk factor for developing breast cancer. The association of BRCA1 promoter methylation in PBCs with breast cancer risk was examined in a case–control study (200 breast cancer patients and 200 controls). BRCA1 promoter methylation in PBCs and breast tumors was determined with a methylation-specific quantitative PCR assay. BRCA1 promoter methylation in PBCs was seen in 43 (21.5%) of the breast cancer patients and in 27 (13.5%) of the controls. The odds ratio for breast cancer adjusted for other epidemiological risk factors was 1.73 (1.01–2.96) and was statistically significant (P = 0.045). When breast tumors were classified into those with and without BRCA1 promoter methylation, the odds ratio was 0.84 (0.43–1.64) (P = 0.61) for BRCA1 promoter methylation-negative and 17.78 (6.71–47.13) (P < 0.001) for BRCA1 promoter methylation-positive breast tumors. BRCA1 promoter methylation in PBCs is significantly associated with risk of breast cancer with BRCA1 promoter methylation. This seems to indicate that BRCA1 promoter methylation in PBCs may constitute a novel risk factor for breast cancer with BRCA1 promoter methylation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
BRCA1 is a well-established breast cancer susceptibility gene [1], and its germline mutations are found in 40–50% of hereditary breast cancers [2, 3]. It is a typical tumor suppressor gene, and loss of a wild allele has been reported in almost all breast tumors originating in BRCA1 germline mutation carriers [4–6]. Although its function is still not fully understood, it has been shown that BRCA1 is implicated in repair of double strand DNA breaks [7, 8], various transcriptional pathways [9], and regulation of the cell cycle [10, 11]. Moreover, recent studies have shown that most BRCA1 functions can be explained by its E3 ubiquitin ligase activity in cooperation with BARD1 [12, 13].
Somatic mutations of BRCA1 are very rare [14, 15], but its promoter methylation, which leads to silencing of the gene, is reportedly observed in 10–30% of sporadic breast tumors [16–18], suggesting that this gene is involved in the pathogenesis of a significant proportion of sporadic breast cancers. Interestingly, phenotypes of breast tumors with BRCA1 promoter methylation, which lack expression of the BRCA1 protein due to BRCA1 gene silencing [19, 20], are reported to be similar to those arising in patients with germline BRCA1 mutations in that they are more likely to be ER, PR [21–23], and HER2 negative tumors [24] as well as histologically high grade tumors [17]. These results seem to indicate that BRCA1 promoter methylation in sporadic breast tumors can be considered to occur relatively early in their pathogenesis. Such tumors thus appear to share similar phenotypes with hereditary breast tumors with BRCA1 germline mutations, which, according to the two-hit theory, are thought to lose their BRCA1 function from the beginning of pathogenesis.
Snell et al. [25] recently studied BRCA1 promoter methylation in peripheral blood cells (PBCs) from seven familial breast cancer patients whose tumors were pathologically similar to BRCA1-mutated tumors but lacked BRCA1 and BRCA2 germline mutations, and were able to demonstrate BRCA1 promoter methylation in PBCs of three of them. They also reported that BRCA1 promoter methylation was observed in all breast tumors in these three patients, which suggests that BRCA1 promoter methylation in PBCs is associated with risk of developing breast cancer, especially breast cancer with BRCA1 promoter methylation. Moreover, Flanagan et al. [26] recently demonstrated that gene body hypermethylation of the ATM gene in PBCs is significantly associated with breast cancer risk [26], indicating a possible association of tumor suppressor gene methylation in PBCs with breast cancer risk.
These results taken together led us to hypothesize that BRCA1 promoter methylation in PBCs may indicate the propensity of a woman’s normal breast tissue to have BRCA1 promoter methylation and that such would become susceptible to breast cancer pathogenesis. This would mean that BRCA1 promoter methylation in PBCs could constitute a risk factor for breast cancer. In the case–control study presented here, we, therefore, investigated whether BRCA1 promoter methylation in PBCs is associated with risk of breast cancer, especially breast cancer with BRCA1 promoter methylation.
Materials and methods
Patients
Cases were breast cancer patients who underwent breast conserving surgery or mastectomy at Osaka University Hospital between October 2000 and November 2008, and controls were women who received breast cancer screening by physical examination and mammography and/or ultrasonography at the screening institutes in Osaka and were found to be free of breast cancer during the period of June 2001 to September 2005. In order to minimize the influence of age, cases were composed of 50 patients in every 10 years-stratum (30–39, 40–49, 50–59, and 60–69) and controls were also composed of 50 patients in every 10 years-stratum (30–39, 40–49, 50–59, and 60–69) as shown in Table 1. Cases were selected in order of date of surgery, and controls were selected in order of date of breast cancer screening in each stratum. Peripheral blood was collected from breast cancer patients before surgery, and the patients who received neoadjuvant chemotherapy and/or hormonal therapy were excluded. Informed consent was obtained from each patient or control before blood collection.
DNA extraction and sodium bisulfite treatment
DNA was extracted from whole peripheral blood cells (PBCs) of 200 cases and 200 controls and from 162 breast tumor tissues as previously described [27–29] and 1 μg of the DNA was subjected to sodium bisulfite treatment using the EpiTect Bisulfite Kit (48) (QIAGEN, Inc. Valencia, CA) according to the manufacturer’s protocol.
Fresh tumor tissues were obtainable from 162 tumors and kept at −80°C until use. They were not obtainable in the remaining 38 patients mostly due to their small size for sampling. The backgrounds of breast tumors with fresh tumors available were not significantly different from those with fresh tumor tissues unavailable in terms of age, tumor size, histological type, lymph node status, histological grade, and positivity of ER, PR, and HER2, except for tumor size, i.e., the former tumors included Tis (n = 0), T1 (n = 105), T2 (n = 57), and T3 (n = 0), and the latter tumors included Tis (n = 3), T1 (n = 27), T2 (n = 7), and T3 (n = 1)(P < 0.001).
Analysis of BRCA1 promoter methylation
Methylation of the BRCA1 promoter was assessed with real-time methylation-specific polymerase chain reaction (MSP). The methylated primer set and probe for BRCA1 were used as previously reported [30]. The forward primer was 5′-TTTCGTGGTAACGGAAAAGCG-3′, the backward primer was 5′-CCGTCCAAAAAATCTCAACGAA-3′, and the probe was FAM-5′-CTCACGCCGCGCAATCGCAATTT-3′-DDQ1. The unmethylated primer set for BRCA1 was modified very slightly from the one previously reported [17] with forward primer 5′-TGGTTTTTGTGGTAATGGAAAAGTGTG-3′, backward primer 5′-CCCATCCAAAAAATCTCAACAAA-3′, and probe FAM-5′-CTCACACCACACAATCACAATTTTAAT-3′-DDQ1. For a quantification of the methylated and unmethylated BRCA1 promoter, methylated standard oligoDNA, 5′-TTTCGTGGTAACGGAAAAGCGCGGGAATTATAGATAAATTAAAATTGCGATTGCGCGGCGTGAGTTCGTTGAGATTTTTTGGACGG-3′ and unmethylated standard oligoDNA, 5′-TGGTTTTTGTGGTAATGGAAAAGTGTGGGAATTATAGATAAATTAAAATTGTGATTGTGTGGTGTGAGTTTGTTGAGATTTTTTGGATGGG-3′, were synthesized so as to include the aforementioned forward primer, backward primer, and probe sequences. EpiTect® Control DNA (human) (QIAGEN) was used as both the positive and negative control for methylated alleles.
For PCR amplification, 2 μl of bisulfite-modified DNA was added in a final volume of 20 μl PCR mix containing 10 μl Fast Start Universal Probe Master (Rox; Roche Applied Science, Mannheim, Germany), 8 μl distilled water, probe, and primers. PCR amplifications were performed using the Light Cycler® 480 Real-Time PCR System (Roche Applied Science) under the following conditions: 1 cycle at 95°C for 10 min followed by 45 cycles at 95°C for 30 s, at 58°C for 30 s, and at 72°C for 30 s. The standard curve was raised for each run for quantification of the methylated BRCA1 promoter by using standard oligo DNA from 120 to 120,000 copies (Fig. 1). Each sample was analyzed in triplicate assays, and those which showed 120 or more copies in all samples were considered positive. Finally, PCR products were also confirmed by means of 2% agarose gel electrophoresis, staining with ethidium bromide and visualization under UV illumination. The percentage of methylation was calculated as M% = M/(M + UM). Finally, all data were corrected by quantification of the positive control.
MSP assay standard curve. a Amplification plot Samples containing five different copies (120000, 12000, 1200, 120, and 12) of standard oligo DNA were subjected to MSP assay. Cycle numbers are plotted against changes in normalized reporter signals. b Standard curve plot Log starting copy number was plotted against C t. Dots represent data obtained from standard curve point samples
Detection sensitivity of MSP assay for methylated against unmethylated BRCA1 promoter
Human genome DNA completely methylated by Sss1 methylase treatment (EpiScope™Methylated HeLa Genomic DNA, Takara Bio, Shiga, Japan) was diluted against unmethylated human genome DNA obtained from the healthy controls at various ratios [methylated DNA/(methylated DNA + unmethylated DNA) = 1, 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6]. These mixtures (total DNA content: 1 μg) were then treated with sodium bisulfite treatment and subjected to the MSP assay as described above.
Antibodies
The antibodies used were: BRCA1 (Ab-1, monoclonal, IgG isotype, 1:70; Calbiochem, San Diego, CA), estrogen receptor (ER) (polyclonal, IgG isotype, 1:100; Santa Cruz Biotechnology, Santa Cruz, CA), progesterone receptor (PR) (clone 636, monoclonal, IgG isotype, 1:800; Dako, Kyoto, Japan), and c-erbB2 (HER2) (polyclonal, IgG isotype, 1:100; Nichirei, Kyoto, Japan),
Immunohistochemical assay
Paraffin sections (3 μm) prepared from the formalin-fixed paraffin-embedded tumor specimens were immunohistochemically stained with the avidin–biotin-peroxidase method. The paraffin sections were deparaffinized and then rehydrated in graded alcohols. In brief, antigens for ER and PR were retrieved by heating the samples in a target retrieval solution (Dako) at 98°C for 40 min while antigen BRCA1 was retrieved with microwave oven treatment at 500 W for 15 min. Sections to be stained for HER2 were not pretreated. After quenching of endogenous peroxidase with 3% H2O2 in methanol for 20 min, non-specific binding was blocked by incubating the slides with Block Ace (Dainippon Sumitomo Pharma, Osaka, Japan) for 30 min, followed by incubation of the sections with the primary antibody at 4°C overnight. Next, the sections were treated with a biotin-conjugated secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA). Finally, after treatment with the avidin–biotin-peroxidase complex system (VECTASTAIN® Elite ABC kit, Vector Laboratories, Burlingame, CA), the sections were visualized with 3,3′-diaminobenzidine tetrahydrochloride (Merck, Darmstadt, Germany). The sections were then counter stained with hematoxylin.
Nuclear-stained tumor cells in three non-overlapping fields were identified with a 20× lens and counted. When tumor cells positive for nuclear BRCA1, ER, or PR staining were observed in more than 10% of all tumor cells, the tumor was considered positive for the corresponding protein. Staining of HER2 was scored with four grades (0, 1, 2, and 3) according to a previously described method [31], and tumors with scores of 2+ and 3+ were considered to be HER2 (+).
Statistics
BRCA1 promoter methylation status in PBCs of cases and controls was assessed and compared by using a logistic regression method to obtain the odds ratios (OR) and 95% confidence intervals (CI) after adjustment for other epidemiological risk factors such as age, family history, age at menarche, parity, menopausal status, and body mass index (BMI). These factors were categorized as (1) family history of breast cancer or ovarian cancer in a first-degree relative (yes or no); (2) age at menarche (12 years or younger, 13–14 years, and 15 years or older); (3) parity (first live birth at 25 years or younger, 26–29 years, 30 years or older, or nulliparity); (4) menopausal status (post-menopausal at less than 50 years, 50 years or older, premenopausal); (5) BMI (<20 kg/m2, 20 to <23 kg/m2, and 23.0 kg/m2 or more). The association between BRCA1 promoter methylation status and clinicopathological characteristics of breast tumors was assessed with a chi-square test. A P value of <0.05 was considered significant. SPSS software (SPSS Inc., Chicago, IL) was used for all statistical analyses.
Results
BRCA1 promoter methylation in PBCs from breast cancer patients and controls
DNA samples extracted from PBCs from 200 breast cancer patients and 200 healthy controls were subjected to MSP assay for BRCA1 promoter methylation. Demographics of breast cancer patients and controls are shown in Table 1. BRCA1 promoter methylation in PBCs could be detected in 43 (21.5%) breast cancer patients and in 27 (13.5%) controls. BRCA1 promoter methylation was not significantly associated with age, body mass index, age at menarche, age at menopause, or family history of either breast cancer patients or controls. The average percentages of methylated against total BRCA1 genes were 1.82% (S.E. 0.49) for BRCA1 promoter methylation-positive breast cancer patients and 1.58% (S.E. 0.72) for BRCA1 promoter methylation-positive controls.
BRCA1 promoter methylation in breast tumors
DNA samples available from 162 tumor tissues were subjected to MSP assay for BRCA1 methylation and 31 (19.1%) were found to have BRCA1 promoter methylation. BRCA1 promoter methylation-positive tumors were more likely to be ER-negative (P = 0.068) and PR-negative (P = 0.006) (Table 2). Furthermore, when breast tumors were classified into four intrinsic subtypes based on the immunohistopathological results of ER, PR, and HER2 [32], the BRCA1 promoter methylation-positive tumors were significantly more likely to be triple-negative tumors (P = 0.025) (Table 2). The average percentage of methylated against total BRCA1 genes was 19.0% (S.E. 5.12) for the BRCA1 promoter methylation-positive breast tumors.
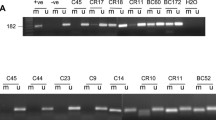
The relationship between BRCA1 promoter methylation in tumor tissues and BRCA1 protein expression was also examined immunohistochemically in 50 representative breast tumors (Fig. 2). Of the 40 tumors without BRCA1 promoter methylation in tumor tissues, 33 were immunohistochemically positive for BRCA1, but only one of the ten tumors with BRCA1 promoter methylation in tumor tissues showed such positivity (P < 0.001).
Association between BRCA1 promoter methylation in PBCs and breast cancer risk
Association of BRCA1 promoter methylation with breast cancer risk was evaluated by means of a case–control study (Table 3), which demonstrated that the odds ratio for breast cancer adjusted for other epidemiological risk factors was 1.73 (1.01–2.96) and had statistical significance of P = 0.045.
The breast tumors were then classified into those with and without BRCA1 promoter methylation, and odds ratios for each class of breast tumors were calculated (Table 3). The odds ratios were 0.84 (0.43–1.64) (P = 0.61) for BRCA1 promoter methylation-negative and 17.78 (6.71–47.13) (P = 0.001) for BRCA1 promoter methylation-positive breast tumors, indicating a significant association of BRCA1 promoter methylation in PBCs with a risk of developing breast tumors with BRCA1 promoter methylation but not of tumors without such methylation.
No interference with MSP assay by circulating tumor cells
PBCs may be contaminated by circulating tumor cells with BRCA1 promoter methylation so that the MSP assay may detect BRCA1 promoter methylation of such tumor cells even though the normal PBCs do not show methylation. To rule out this possibility, we carried out a study to assess the detection sensitivity of the MSP assay using the method described in “Materials and methods” section (Fig. 3). The detection sensitivity of the MSP assay was found to be 10−3, which was far higher than the reported ratios (10−6–10−5) of circulating tumor cells to PBCs [33]. It is thus unlikely that our MSP assay was interfered with by contamination from circulating tumor cells.
Detection sensitivity of MSP assay for methylated against unmethylated BRCA1 promoter. Human genome DNA completely methylated by Sss1 methylase treatment was diluted against unmethylated human genome DNA obtained from healthy controls at various ratios (methylated DNA/(methylated DNA + unmethylated DNA) = 10−1, 10−2, 10−3, 10−4, 10−5, and 10−6). These mixtures (total DNA content = 1 μg) were subjected to sodium bisulfite treatment and MSP assay and detection sensitivity of the assay was established as 10−3. Amplification plots of the MSP assay for these mixtures are shown
Discussion
We detected BRCA1 promoter methylation in PBCs in 21.5% of breast cancer patients and 13.5% of controls, while women with BRCA1 promoter methylation showed a significantly higher risk (odds ratio = 1.73) of breast cancer as compared with those without it. We next examined the association between the presence of BRCA1 promoter methylation in PBCs and risk of breast cancer with BRCA1 promoter methylation. We first analyzed BRCA1 promoter methylation in 162 breast tumors and found BRCA1 promoter methylation in 31 of them. These tumors were more likely to be ER- and PR-negative [21–23] and lacked BRCA1 protein expression [23, 34–36], which is consistent with the findings of previous studies. Of special interest is that we could establish a significant association (odds ratio = 17.78; P < 0.001) between BRCA1 promoter methylation in PBCs and risk of breast cancer with BRCA1 promoter methylation, but not with breast cancer without such methylation (odds ratio = 0.84, P = 0.61).
Our results are consistent with those reported by Snell et al. [25], who showed that BRCA1 promoter methylation in PBCs could be detected in three of seven familial breast cancer patients and that all breast tumors arising in such patients carried BRCA1 promoter methylation. However, Chen et al. [37] reported that BRCA1 promoter methylation could not be detected in PBCs of any of 41 familial breast cancer patients. The reason for this discrepancy is explained, at least in part, by the fact that Snell et al. [25] examined only those tumors which were pathologically similar to BRCA1-mutated tumors, whereas Chen et al. [37] did not select breast tumors in this manner.
It is well established that the aging process is associated with a global hypomethylation of the genome but an increase in methylation of specific gene promoters is also observed in conjunction with aging [38, 39]. Interestingly, a significant number of genes that are hypermethylated during aging are also hypermethylated during carcinogenesis [16, 40], indicating that hypermethylation of such genes in normal tissues may be associated with heightened cancer susceptibility. The association between DNA methylation and aging seems to imply that DNA methylation can be a somatic and acquired change which can accrue under the influence of various environmental factors later in life. However, no association of frequency of BRCA1 promoter methylation in PBCs with age was observed in our study nor in a study by Kontorovich et al. [41]. These observations seem to suggest that BRCA1 promoter methylation may represent a germ line change (either trans-generational or single-generational) or a somatic change occurring in the early developmental stages and that it may not be influenced very much by environmental factors later in life. Since the presence of a heritable germline epimutation (MSH2 methylation) has been reported in a family with hereditary non-polyposis colon cancer [42], it is possible that BRCA1 promoter methylation is also a heritable germline epimutation. This possibility could not be investigated in our study, however, because DNA samples were not available from the family members of the breast cancer patients with BRCA1 promoter methylation in their PBCs. Lack of a significant association of BRCA1 promoter methylation in PBCs with a family history of breast cancer may indicate that BRCA1 promoter methylation, even if it is a heritable germline epimutation, is unlikely to be a major cause of hereditary breast cancer.
Our present study, though still preliminary, has suggested that analysis of BRCA1 promoter methylation in PBCs might be clinically useful in the identification of women at high risk for developing breast tumors with BRCA1 promoter methylation. It is well established that breast tumors with BRCA1 methylation are more likely to be triple-negative (basal-like) tumors [43, 44]. Since tamoxifen or raloxifene has been shown to be efficacious in the prevention of ER-positive tumors but not ER-negative tumors [45, 46], women with BRCA1 promoter methylation are expected to gain no benefits from these agents. Recently, it has been reported that poly-ADP ribose polymerase (PARP) inhibitors are very efficacious for triple-negative tumors [47, 48], suggesting a possibility that PARP inhibitors would be useful in the prevention of breast tumors with BRCA1 methylated tumors. Breast tumors are essentially heterogeneous, and recent development of gene expression profiling-based analysis has classified breast tumors into the several intrinsic subtypes [49–51]. Since it is speculated that causes of carcinogenesis might be different among these intrinsic subtype, it would be infeasible to identify a marker(s) associated with an increased risk for every type of breast tumors. More feasible approach would be detecting a marker(s) associated with a specific type of breast tumors. Such a tailored approach seems to be important for the development of chemoprevention of breast tumors in future.
Recently, it has been found that circulating tumor cells (CTCs) can occur in a significant proportion of breast cancer patients, even in those without metastases. Since we extracted DNA from whole PBCs, it is possible that the PBCs were contaminated by CTCs with BRCA1 promoter methylation and that the results of our MSP assay might be flawed. However, and as already mentioned under “Results” section, we analyzed the detection sensitivity of our MSP assay and found that it was around 10−3, which is far higher than the reported ratios (10−6–10−5) of circulating tumor cells to PBCs. It is, therefore, very unlikely that the results of our MSP assay have been affected by CTCs, and can thus be considered to represent the actual BRCA1 promoter methylation status of PBCs.
In conclusion, we detected BRCA1 promoter methylation in PBCs in 21.5% of breast cancer patients and in 13.5% of controls, indicating that BRCA1 promoter methylation in PBCs is significantly associated with a risk of developing breast cancer with BRCA1 promoter methylation. These results suggest that BRCA1 promoter methylation may constitute a novel risk factor for breast cancer with BRCA1 promoter methylation.
References
Miki Y, Swensen J, Shattuck-Eidens D et al (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266(5182):66–71
Easton DF, Bishop DT, Ford D et al (1993) Genetic linkage analysis in familial breast and ovarian cancer: results from 214 families. The Breast Cancer Linkage Consortium. Am J Hum Genet 52(4):678–701
Ford D, Easton DF, Bishop DT et al (1994) Risks of cancer in BRCA1-mutation carriers. Breast Cancer Linkage Consortium. Lancet 343(8899):692–695
Merajver SD, Frank TS, Xu J et al (1995) Germline BRCA1 mutations and loss of the wild-type allele in tumors from families with early onset breast and ovarian cancer. Clin Cancer Res 1(5):539–544
Cornelis RS, Neuhausen SL, Johansson O et al (1995) High allele loss rates at 17q12-q21 in breast and ovarian tumors from BRCAl-linked families. The Breast Cancer Linkage Consortium. Genes Chromosom Cancer 13(3):203–210
Takano M, Aida H, Tsuneki I et al (1997) Mutational analysis of BRCA1 gene in ovarian and breast-ovarian cancer families in Japan. Jpn J Cancer Res 88(4):407–413
Zhang J, Powell SN et al (2005) The role of the BRCA1 tumor suppressor in DNA double-strand break repair. Mol Cancer Res 3(10):531–539
Ting NS, Lee WH et al (2004) The DNA double-strand break response pathway: becoming more BRCAish than ever. DNA Repair (Amst) 3(8–9):935–944
Yoshida K, Miki Y et al (2004) Role of BRCA1 and BRCA2 as regulators of DNA repair, transcription, and cell cycle in response to DNA damage. Cancer Sci 95(11):866–871
Mullan PB, Quinn JE, Harkin DP et al (2006) The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene 25(43):5854–5863
Somasundaram K et al (2003) Breast cancer gene 1 (BRCA1): role in cell cycle regulation and DNA repair–perhaps through transcription. J Cell Biochem 88(6):1084–1091
Xia Y, Pao GM, Chen HW et al (2003) Enhancement of BRCA1 E3 ubiquitin ligase activity through direct interaction with the BARD1 protein. J Biol Chem 278(7):5255–5263
Hashizume R, Fukuda M, Maeda I et al (2001) The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem 276(18):14537–14540
Futreal PA, Liu Q, Shattuck-Eidens D et al (1994) BRCA1 mutations in primary breast and ovarian carcinomas. Science 266(5182):120–122
Xu CF, Solomon E et al (1996) Mutations of the BRCA1 gene in human cancer. Semin Cancer Biol 7(1):33–40
Yang Q, Yoshimura G, Nakamura M et al (2002) BRCA1 in non-inherited breast carcinomas (Review). Oncol Rep 9(6):1329–1333
Matros E, Wang ZC, Lodeiro G et al (2005) BRCA1 promoter methylation in sporadic breast tumors: relationship to gene expression profiles. Breast Cancer Res Treat 91(2):179–186
Wei M, Grushko TA, Dignam J et al (2005) BRCA1 promoter methylation in sporadic breast cancer is associated with reduced BRCA1 copy number and chromosome 17 aneusomy. Cancer Res 65(23):10692–10699
Birgisdottir V, Stefansson OA, Bodvarsdottir SK et al (2006) Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res 8(4):R38
Catteau A, Morris JR et al (2002) BRCA1 methylation: a significant role in tumour development? Semin Cancer Biol 12(5):359–371
Catteau A, Harris WH, Xu CF et al (1999) Methylation of the BRCA1 promoter region in sporadic breast and ovarian cancer: correlation with disease characteristics. Oncogene 18(11):1957–1965
Bianco T, Chenevix-Trench G, Walsh DC et al (2000) Tumour-specific distribution of BRCA1 promoter region methylation supports a pathogenetic role in breast and ovarian cancer. Carcinogenesis 21(2):147–151
Tapia T, Smalley SV, Kohen P et al (2008) Promoter hypermethylation of BRCA1 correlates with absence of expression in hereditary breast cancer tumors. Epigenetics 3(3):157–163
Miyoshi Y, Murase K, Oh K et al (2008) Basal-like subtype and BRCA1 dysfunction in breast cancers. Int J Clin Oncol 13(5):395–400
Snell C, Krypuy M, Wong EM et al (2008) RCA1 promoter methylation in peripheral blood DNA of mutation negative familial breast cancer patients with a BRCA1 tumour phenotype. Breast Cancer Res 10(1):R12
Flanagan JM, Munoz-Alegre M, Henderson S et al (2009) Gene-body hypermethylation of ATM in peripheral blood DNA of bilateral breast cancer patients. Hum Mol Genet 18(7):1332–1342
Arai T, Miyoshi Y, Kim SJ et al (2006) Association of GSTP1 CpG islands hypermethylation with poor prognosis in human breast cancers. Breast Cancer Res Treat 100(2):169–176
Hasegawa S, Miyoshi Y, Egawa C et al (2002) Mutational analysis of the class I beta-tubulin gene in human breast cancer. Int J Cancer 101(1):46–51
Miyoshi Y, Ando A, Hasegawa S et al (2003) Association of genetic polymorphisms in CYP19 and CYP1A1 with the oestrogen receptor-positive breast cancer risk. Eur J Cancer 39(17):2531–2537
Fackler MJ, Malone K, Zhang Z et al (2006) Quantitative multiplex methylation-specific PCR analysis doubles detection of tumor cells in breast ductal fluid. Clin Cancer Res 12(11 Pt 1):3306–3310
Tsuda H, Sasano H, Akiyama F et al (2002) Evaluation of interobserver agreement in scoring immunohistochemical results of HER-2/neu (c-erbB-2) expression detected by HercepTest, Nichirei polyclonal antibody, CB11 and TAB 250 in breast carcinoma. Pathol Int 52(2):126–134
Shibuta K, Ueo H, Furusawa H et al (2010) The relevance of intrinsic subtype to clinicopathological features and prognosis in 4,266 Japanese women with breast cancer. Breast Cancer. doi:10.1007/s12282-010-0209-6
Cristofanilli M, Budd GT, Ellis MJ et al (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351(8):781–791
Niwa Y, Oyama T, Nakajima T et al (2000) BRCA1 expression status in relation to DNA methylation of the BRCA1 promoter region in sporadic breast cancers. Jpn J Cancer Res 91(5):519–526
Miyamoto K, Fukutomi T, Asada K et al (2002) Promoter hypermethylation and post-transcriptional mechanisms for reduced BRCA1 immunoreactivity in sporadic human breast cancers. Jpn J Clin Oncol 32(3):79–84
Mirza S, Sharma G, Prasad CP et al (2007) Promoter hypermethylation of TMS1, BRCA1, ERalpha and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sci 81(4):280–287
Chen Y, Toland AE, McLennan J et al (2006) Lack of germ-line promoter methylation in BRCA1-negative families with familial breast cancer. Genet Test 10(4):281–284
Issa JP, Ottaviano YL, Celano P et al (1994) Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 7(4):536–540
Suijkerbuijk KP, Fackler MJ, Sukumar S et al (2008) Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Ann Oncol 19(11):1870–1874
Gonzalo S et al (2009) Epigenetic alterations in aging. J Appl Physiol 109(2):589–597
Kontorovich T, Cohen Y, Nir U et al (2009) Promoter methylation patterns of ATM, ATR, BRCA1, BRCA2 and P53 as putative cancer risk modifiers in Jewish BRCA1/BRCA2 mutation carriers. Breast Cancer Res Treat 116(1):195–200
Chan TL, Yuen ST, Kong CK et al (2006) Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat Genet 38(10):1178–1183
Galizia E, Giorgetti G, Piccinini G et al (2010) BRCA1 expression in triple negative sporadic breast cancers. Anal Quant Cytol Histol 32(1):24–29
Lee JS, Fackler MJ, Lee JH et al. (2010) Basal-like breast cancer displays distinct patterns of promoter methylation. Cancer Biol Ther 9(12):1017–1024
Smith RE, Good BC et al (2003) Chemoprevention of breast cancer and the trials of the National Surgical Adjuvant Breast and Bowel Project and others. Endocr Relat Cancer 10(3):347–357
Visvanathan K, Chlebowski RT, Hurley P et al (2009) American society of clinical oncology clinical practice guideline update on the use of pharmacologic interventions including tamoxifen, raloxifene, and aromatase inhibition for breast cancer risk reduction. J Clin Oncol 27(19):3235–3258
Rottenberg S, Jaspers JE, Kersbergen A et al (2008) High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci USA 105(44):17079–17084
Fong PC, Boss DS, Yap TA et al (2009) Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 361(2):123–134
Perou CM, Sorlie T, Eisen MB et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Sorlie T, Tibshirani R, Parker J et al (2003) Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA 100(14):8418–8423
Carey LA, Perou CM, Livasy CA et al (2006) Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295(21):2492–2502
Acknowledgments
Grant support: (1) Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science and Technology, Japan, and (2) Promotion of Cancer Research (Japan) for the 3rd Term Comprehensive 10-Year Strategy for Cancer Control.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Iwamoto, T., Yamamoto, N., Taguchi, T. et al. BRCA1 promoter methylation in peripheral blood cells is associated with increased risk of breast cancer with BRCA1 promoter methylation. Breast Cancer Res Treat 129, 69–77 (2011). https://doi.org/10.1007/s10549-010-1188-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-1188-1