Abstract
There is a growing body of evidence that HER2 status can change during disease recurrence or progression in breast cancer patients. In this context, re-evaluation of HER2 status by assessment of HER2 expression on circulating tumor cells (CTCs) is a strategy with potential clinical application. The aim of this trial was to determine the HER2 status of CTCs in metastatic breast cancer patients comparing two CTC assays. A total of 254 patients with metastatic breast cancer from nine German university breast cancer centers were enrolled in this prospective study. HER2 status of CTCs was assessed using both the FDA-approved CellSearch® assay and AdnaTest BreastCancer™. Using the CellSearch assay, 122 of 245 (50%) patients had ≥5 CTCs, and HER2-positive CTCs were observed in 50 (41%) of these patients. Ninety of 229 (39%) patients were CTC positive using AdnaTest BreastCancer, and HER2 positivity rate was 47% (42 of 90). The rate of breast cancer patients with HER2-negative primary tumors but HER2-positive CTCs was 32% (25 of 78) and 49% (28 of 57) using the CellSearch assay and AdnaTest BreastCancer, respectively. Considering only those patients who had CTCs on both tests (n = 62), concordant results regarding HER2 positivity were obtained in 50% of the patients (31/62) (P = 0.96, κ = −0.006). HER2-positive CTCs can be detected in a relevant number of patients with HER2 negative primary tumors. Therefore, it will be mandatory to correlate the assay-dependent HER2 status of CTCs to the clinical response on HER2-targeted therapies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human epidermal growth factor receptor 2 (HER2) is a transmembrane tyrosine kinase receptor encoded by a proto-oncogene located on chromosome 17q21. The HER2 proto-oncogene is amplified or overexpressed in approximately 20% of invasive primary breast cancers [1–3]. A positive HER2 status has been linked with aggressive tumor behavior and resistance to cytotoxic and endocrine therapy [3–5]. Patients with HER2 amplification and/or overexpression are eligible for HER2-targeted treatment [6–8]. As a consequence, strategies for response prediction and monitoring are of high clinical relevance.
The methods used to select patients that are suitable for HER2-targeted therapy are based on the immunohistochemical (IHC) detection of HER2 protein overexpression and the demonstration of HER2 gene amplification by fluorescence in situ hybridization (FISH) in the primary tumor [9]. In recent years, there has been a growing body of evidence that the HER2 status of the primary tumor may be different from metastatic disease and changes might occur during treatment. A discrepancy between the primary tumor and distant metastases has been observed in 7–26% of cases [10–19].
In many patients with metastatic breast cancer, the re-evaluation of HER2 status by a tissue biopsy of the metastatic lesion is not feasible due to location of the metastatic site. In addition, HER2 status may vary between different metastatic sites and also change during treatment. Therefore, determination of the HER2 status of circulating tumor cells (CTCs) might be a strategy with potential clinical application. So far, no large prospective studies have been reported comparing different methods for CTC detection and characterization.
The aim of this prospective multicenter trial was to compare the HER2 status of CTCs in 254 patients with metastatic breast cancer at the time of first diagnosis or disease progression obtained by the antibody-based CellSearch® System and the reverse transcriptase polymerase chain reaction (RT-PCR) approach, AdnaTest BreastCancer™, and to assess the concordance rate between these two techniques. The CellSearch assay is an automated, standardized, and FDA-approved method for detecting and enumerating tumor cells [20–22]. AdnaTest BreastCancer is based on detection of three tumor-associated transcripts by RT-PCR after immunomagnetic enrichment of tumor cells [23, 24]. Both assays have been frequently used in studies evaluating CTC detection [19–26]. However, comparison between the two methods, notably for HER2 expression, has not been performed in a prospective, large, multicenter cohort to date.
Methods
Patients
A total of 254 patients with metastatic breast cancer from nine German university breast cancer centers [Düsseldorf (n = 4), Erlangen (n = 30), Essen (n = 46), Freiburg (n = 9), Hamburg (n = 79), Heidelberg (n = 18), Munich (n = 16), Regensburg (n = 2), and Tübingen (n = 50)] were enrolled in this prospective, open-label, non-randomized study. Inclusion criteria were epithelial invasive carcinoma of the breast with distant metastatic disease (M1), age ≥18 years, the availability of primary tumor tissue results for ER, PR, and HER2 and first diagnosis of metastatic disease or disease progression (before start of new treatment regimen). Blood was drawn before the start of a new line of therapy. This included any therapy including hormonal therapy and any line of treatment. All patients gave their informed consent for the use of their blood samples. A web-based databank was designed for data management and on-line documentation (www.detect-study.de). By the use of this interface, clinical investigators were blinded for results of CTC testing and the investigators performing CTC testing were blinded for the clinical data of the patients and the result of the CTC test results from the other centers. The study was approved by local institutional review boards (2007/B01). The trial was registered in the Current Controlled Trials Registry at http://www.controlled-trials.com (no. ISRCTN59722891).
Enumeration and characterization of CTCs
Detection of CTCs and assessment of HER2 status of CTCs was performed using both AdnaTest BreastCancer (Adnagen AG, Langenhagen, Germany) and the CellSearch assay (Veridex LLC, Raritan, NJ, USA) according to the manufacturers’ instructions without modifications. Table 1 summarizes the technical features of each assay [20, 25, 26]. Sample preparation and analysis by AdnaTest BreastCancer were performed by either of two centers (Departments of Gynecology and Obstetrics, Essen (SK-B) or Tübingen (TF); 173 and 81 tests, respectively). CTC analysis by the CellSearch assay was performed by either of two centers (Institute of Tumor Biology, University Medical Center, Hamburg-Eppendorf (KP) or Department of Gynecology, Munich (BR); 173 and 81 tests, respectively). These centers have previously conducted a validation study and demonstrated that samples could be stored and transported (up to 72 h) as well as examining the high inter- and intra-assay concordance of the results in a multicenter setting [20].
Before the study was started, each breast cancer center was assigned to send its samples only to the designated laboratory for the CellSearch assay and AdnaTest BreastCancer, respectively. Blood samples for AdnaTest BreastCancer were shipped in cooled boxes at 4°C whereas samples for the CellSearch assay were sent at room temperature based on the manufacturer’s recommendation. All blood samples were processed within 48 h for AdnaTest BreastCancer and 96 h for the CellSearch assay, or otherwise discarded. Both AdnaTest BreastCancer and the CellSearch assay were performed independently and the investigators were blinded to the results obtained by the other method.
AdnaTest BreastCancer
Two 5-ml EDTA blood samples were collected for CTC isolation using the AdnaCollect™ blood collection tubes (Adnagen AG) and stored at 4°C until further analysis. Establishment and validation of AdnaTest BreastCancer has been described in detail elsewhere [23, 24, 26]. In brief, blood samples were incubated with a ready-to-use antibody mixture (against GA 73.3 and MUC1) commercialized as AdnaTest BreastCancerSelect™ (Adnagen AG) according to the manufacturer’s instructions. The labeled cells were extracted by a magnetic particle concentrator. Subsequently, mRNA isolation from lysed, enriched cells was performed with a Dynabeads mRNA DIRECT™ Micro Kit (Dynal Biotech GmbH, Hamburg, Germany). Sensiscript™ Reverse Transcriptase (QIAGEN GmbH, Hilden, Germany) was used for the reverse transcription in combination with oligo(dT) coupled Dynabeads® of the mRNA DIRECT™ Micro Kit (Dynal Biotech GmbH) [26]. The analysis of tumor-associated mRNA isolated from CTC tumor cells was performed in a multiplex polymerase chain reaction (PCR) for three tumor-associated transcripts (HER2, MUC1, and GA733-2) and the housekeeping gene β-actin. GA 73.3 refers to the EpCAM epitope and the GA733-2 transcript refers to EpCAM mRNA. The primers generate fragments of the following sizes: GA733-2, 395 base pairs (bp); MUC1, 293 bp; and HER2, actin, 114 bp. Visualization of the PCR fragments was carried out with a 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA, USA) using DNA 1000 LabChips and the Expert Software Package (version B.02.03.SI307). The test was considered CTC positive if a PCR fragment of at least one tumor-associated transcript (MUC-1, GA 773-2, or HER2) and a fragment of the control gene β-actin (internal PCR control) was clearly detected (peak concentration of >15 ng/μl) in both blood samples. CTCs were considered HER2 positive if a PCR fragment of the HER2 transcript (peak concentration of >15 ng/μl) was present.
CellSearch assay
Two 7.5-ml blood samples were collected into CellSave tubes (Veridex Inc.). The CellSearch Epithelial Cell Test (Veridex Inc.) was applied for CTC enrichment and enumeration. The method has been described in detail elsewhere [20]. In brief, CTCs are captured from peripheral blood by anti-epithelial cell adhesion molecule (EpCAM)-antibody-bearing ferrofluid and subsequently identified by cytokeratin-positivity/negativity for the leukocyte common antigen CD45 and 4′,6-diamidino-2-phenylindole (DAPI) staining to ensure the integrity of the nucleus. A blood sample was positive when at least five CTCs were present based on the prognostically relevant cut-off as previously published [21, 22]. HER2 expression of CTCs was characterized within the CellSearch assay by addition of a fluorescein isothiocyanate (FITC)-labeled anti-HER2 antibody (CellSearch tumor phenotyping reagent HER2, Veridex Inc.), as described previously [25, 27, 28]. To evaluate the intensity of HER2 immunostaining, approximately 500 breast cancer cells from cell lines with known HER2 status (MCF-7: no HER2 gene amplification, MDA-MB-453: 2- to 3-fold HER2 gene amplification, SK-BR-3 and BT474: 5- to 7-fold HER2 gene amplification) were spiked into 7.5 ml blood from healthy donors and were processed under identical conditions with the CellSearch assay. The intensity of the HER2-specific immunofluorescence was categorized into negative (0), weak (1+), moderate (2+), and strong (3+). CTCs were considered HER2 positive if at least one CTC had strong HER2 staining (3+) based on the cut-off level published by Riethdorf et al. [25].
Determination of the HER2 status in the primary tumor
The HER2 status of the primary tumor was obtained from the patient’s chart. In all participating centers, the HER2 status had been determined by the HercepTest™ (Dako, Glostrup, Denmark) and/or the Pathvysion® Kit (Vysis, Downers Grove, IL, USA). All pathology laboratories had participated in ring experiments and were certified laboratories for HER2 detection. A central review of the HER2 status of the primary tumor was therefore not performed. The cut-off level for HER2 positivity were based on the ASCO/CAP guidelines [9].
Statistical analysis
Primary endpoint of the study was the rate of HER2-positive CTCs with each method. Secondary endpoints were the concordance between the two methods in HER2-positive CTC detection. The study was performed in accordance with REMARK criteria [29, 30]. Relationships between categorical variables were investigated using contingency tables. In case of independent data, Fisher’s exact test was used to evaluate the relationship, whereby P values <0.05 indicate statistically significance. When paired data were considered in terms of assessing the reliability of test results of the methods, agreement and consistency were regarded via Cohen’s kappa (κ) and McNemar-test, respectively. Statistical analysis was performed using SAS version 9.2.
Results
Patient characteristics
A total of 254 patients with metastatic disease were enrolled between December 2007 and May 2009. The clinical characteristics of the patients are summarized in Table 2 for the total cohort and the sub-groups that were CTC positive by CellSearch assay and AdnaTest BreastCancer, respectively.
Detection of CTCs and determination of HER2 expression with the CellSearch assay
A total of 245 of 254 blood samples (two 7.5-ml blood tubes) could be analyzed for the presence of CTCs by the CellSearch assay. Nine samples had to be excluded due to technical issues: test failure (n = 6), hemolysis of blood sample (n = 2), and insufficient blood volume (n = 1). At least one CTC was detected in 180 of 245 patients (73%) (Table 3). The average number of tumor cells was 177 cells per 7.5 ml (range, 1–6389; median, 4). Using the established cut-off level of five cells, 122 of 245 (50%) metastatic patients were considered CTC positive at the time of first diagnosis or disease progression. Presence of CTCs was only associated with extent of metastatic disease (P < 0.05) (Table 2).
All CTCs were further characterized for HER2 expression within the CellSearch system by addition of an FITC-labeled anti-HER2 antibody. Cases were categorized as HER2-positive CTC if at least five CTCs were detected and at least one CTC showed strong immunostaining (3+) for HER2. Based on this definition, 72 (59%) of the 122 CTC-positive patients, were classified as HER2 negative and 50 (41%) as HER2 positive by immunofluorescence. Table 4 summarizes the distribution of combinations regarding HER2 immunostaining of CTCs in individual patients.
The mean ratio between number of HER2-positive CTCs and total number of CTCs was 0.31, ranging from 0.1 to 1 in HER2-positive blood samples. The percentage of HER2-positive CTCs was >10% in 64% of patients (32/50), >30% in 40% of patients (20/50), and >50% in 26% patients (13/50).
Correlation of HER2 status between CTCs and corresponding primary tumor was determined. Of those patients with detectable CTCs, primary tumors were HER2 negative in 78 patients and HER2 positive in 31 patients. HER2 status was unknown or inconclusive due to missing FISH analysis in 15 cases. HER2-positive CTCs in HER2-negative primary tumors were seen in 25 of the 76 patients (33%). Discordant HER2 expression was also found in patients with HER2-positive primary tumors, where 13 of 31 (42%) patients had exclusively HER2-negative CTCs. The correlation between HER2 status of CTC and corresponding primary tumor was fair (P = 0.02, κ = 0.23). Results are summarized in Table 5.
Detection of CTCs and determination of HER2 expression with AdnaTest BreastCancer
A total of 229 blood samples (two 5-ml blood tubes) could be analyzed for presence of CTCs by AdnaTest BreastCancer. Twenty-five blood samples had to be excluded, in most cases, due to insufficient blood volume (n = 12) followed by failure of the assay to pass quality control (n = 7), and time until processing >48 h (n = 6). The overall detection rate for CTCs was 39% (90 of 229 patients). No correlations were observed between CTC positivity and any clinicopathological characteristic (Table 2).
A blood sample was considered as HER2-positive CTC, if the blood sample was considered CTC positive based on the cut-off level described above and the transcript for HER2 was present. HER2-positive CTCs were detected in 42 of 90 CTC-positive patients (47%). Nine of these patients had a HER2-positive primary tumor, whereas 28 patients were initially HER2 negative based on the expression profile of the primary tumor (Table 5). There was no correlation between HER2 expression of CTCs and the corresponding primary tumor (P = 0.51, κ = −0.09).
Comparison between CellSearch assay and AdnaTest BreastCancer
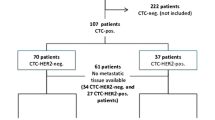
Both methods could be performed successfully in 221 cases. A significant difference in CTC-positivity rates was observed. The CTC-positivity rate was 53% for the CellSearch assay compared to 40% for AdnaTest BreastCancer (P < 0.05). Concordance between the two assays was 64%. The correlation between the two assays was fair (P < 0.01, κ = 0.28). Sixty-two patients (28%) were CTC positive by both methods, whereas either the CellSearch assay or AdnaTest BreastCancer were exclusively positive in 24% (54/221) and 12% (26/221), respectively (Table 6).
Concordance between the HER2 status revealed by either the CellSearch assay or AdnaTest BreastCancer could be only evaluated in the 62 patients who were CTC positive with both assays. Concordant results were obtained in 50% of the patients (31/62) (P = 0.96, κ = −0.006). Thirteen patients had HER2-positive CTCs by both assays. HER2-positive CTCs were only detected by the CellSearch assay in 15 patients (24%) and only by the AdnaTest BreastCancer in 16 patients (26%) (Table 6). Data are presented for different cut-off levels for the CellSearch assay including 1 and 2 cells, respectively, in Table 3.
Discussion
HER2 is the most prominent target for novel therapeutic approaches in breast cancer [6–8]. Currently, HER2 status is determined at the time of initial disease diagnosis by analyzing primary tumor tissue. However, several studies have demonstrated that HER2 status may change during disease progression. CTCs may be an ideal tool for reassessment of predictive markers since the phenotype of CTCs reflect the phenotype of the metastatic load at different sites [27–30].
In this study, we used the CellSearch assay and AdnaTest BreastCancer because they are suitable for use in a multicenter setting due to a highly standardized procedure for CTC enrichment and detection [20, 23, 25, 26].
The FDA-approved CellSearch assay is currently the most frequently used approach, particularly, in on-going clinical trials. CTCs are isolated by immunomagnetic beads coated with antibodies against EpCAM and identified by cytokeratin positivity, positive nuclear staining, and CD45 negativity [20]. Using only EpCAM to enrich CTCs from blood is considered as one of the major limitations of this assay. EpCAM might be heterogeneously expressed by CTCs and be down-regulated as a consequence of the metastatic process [31]. In addition, experimental data not confirmed in the clinical setting suggest that normal-like breast cancer cells are less efficiently captured by an EpCAM-based approach, which may reduce the sensitivity of this assay [32].
Most prognostic studies have used the cut-off level for CTC positivity of five cells (in 7.5 ml blood) in metastatic breast cancer to distinguish patients with statistically significant clinical outcome [20–22]. Based on this cut-off level, 50% of our patients had at least five CTCs at the time of first diagnosis or disease progression, which is in concordance with the reported positivity rates from 36 to 61% in other large studies [19–22, 33, 34]. The determination of HER2 status of CTCs has been based on an immunofluorescence staining score established by Meng et al. [28] and Riethdorf et al. [25]. FISH would be the optimal method for this task but the relocation of CTCs for FISH analysis may be difficult and does not currently seem feasible in a large clinical trial. A high concordance between immunofluorescence and FISH analysis has been demonstrated by Meng et al. [28], Riethdorf et al. [25], and Pestrin et al. [35], suggesting that the immunofluorescence approach is accurate. However, in the study of Pestrin et al. the correlation between HER2 status of CTCs and primary tumor was higher using the FISH technique. An appropriate cut-off level for HER2 positivity of CTC-positive blood samples has not yet been established. Pestrin et al. [35] defined CTC samples as HER2 positive if at least 50% of CTCs demonstrated HER2 expression. Meng et al. [28] suggested that at least 10 CTCs are necessary for optimal HER2 evaluation but did not exclude the possibility that fewer CTCs may be sufficient to indicate that a patient is a candidate for HER2-targeted therapy. The cut-off level used in that study was also 50%. Riethdorf et al. [25] investigated the percentage of HER2-positive CTCs in a neoadjuvant setting using the threshold of one HER2-positive CTC; however, the number of CTCs in non-metastatic patients is much lower than in the metastatic setting.
In our trial, a case was defined as HER2-positive CTC when at least one CTC had a HER2 immunostaining score of 3+ and ≥5 CTCs were present (criterion for CTC positivity). Based on these definitions, 72 (59%) of the 122 CTC-positive patients were classified as HER2 negative and 50 (41%) as HER2 positive by immunofluorescence staining. HER2-positive CTCs in HER2-negative primary tumors were seen in 25 of the 78 patients (33%), which is in the range of conversion rates of 29–38% for initially HER2-negative patients reported in other, smaller studies [28, 35, 36]. However, modification of the cut-off level for HER2 positivity may be necessary in the case of therapeutic decision-making.
AdnaTest BreastCancer was the other approach used in our current trial. Here, CTCs are isolated by immunomagnetic beads labeled with antibodies against MUC1 and EpCAM. After isolation of the mRNA, transcripts encoding epithelial-specific markers (GA 73.3, MUC1 and HER2) were amplified by a multiplex PCR and detected by microfluid gel electrophoresis. In contrast to RT-PCR approaches mRNA expression levels cannot be quantified, which would be desirable for therapy monitoring. In the present study, the overall detection rate for CTCs was 39%, which is within in the range of other published studies [33, 37]. However, using the CellSearch assay, 50% of patients were CTC positive with ≥5 CTCs per 7.5 ml blood despite the fact that only EpCAM was used for CTC enrichment and RT-PCR approaches are supposed to be more sensitive [38].
Overall agreement between AdnaTest BreastCancer and the CellSearch assay was 64%, which is low given that both assays should detect HER2-positive CTCs. Only one other study has so far evaluated both techniques in a head-to-head comparison [33]. The concordance rate was nearly 80% but another cut-off level (≥2 CTCs) was used for the CellSearch assay. In our study, we analyzed the concordance rate between the assays using three different cut-off levels (1, 2, and 5 cells). The concordance rate was highest using the cut-off of five cells, which is mostly used for patients with metastatic breast cancer. Nevertheless, concordance rates of only 52–64% could be obtained (Table 3).
Patients with HER2-positive CTCs were observed in 41 and 47% of cases using the CellSearch assay and AdnaTest BreastCancer, respectively. However, no correlation was observed between these assays regarding HER2 evaluation of CTCs. Several explanations may account for this observation. The CellSearch assay evaluates the HER2 status of individual cells by immunofluorescence, which is important due to heterogeneity of CTCs. AdnaTest BreastCancer determines the average HER2 expression of all tumor cells. The test is not able to detect the heterogeneity in HER2 expression between different CTCs and to determine the percentage of HER2-positive CTCs. To define a gold standard in an ideal study setting, the HER2 status has to be re-assessed by each method and compared to the HER2 status of the metastatic tissue. In our study, biopsy of the metastatic tissue was optional and all patients were encouraged to participate. One potential drawback of our study is that only 30 of 252 patients finally accepted a tissue biopsy. Since most of these patients were CTC negative, no meaningful comparison could be performed.
Should we use CTCs for HER2 assessment despite these caveats? Our findings and case reports from other studies suggest that (1) initially HER2-negative patients can have HER2-positive CTCs and (2) HER2-positive CTCs are eliminated by HER2-targeted therapy, resulting in an objective clinical response in initially HER2-negative patients [27, 28].
Our study is the largest to compare HER2 CTC testing with different methods. Advantages are the blinded data entry, technical performance of the assays, and the multicenter setting. To implement the HER2 status of CTCs as a stratification parameter in future clinical trials it will be necessary to define a gold standard for HER2 assessment. Since repeated tissue biopsies are not feasible, an alternative seems to investigate the clinical response to HER2-targeted therapy based on the changes in HER2-positive CTCs in cancer patients [25]. Our present study is a step toward the implementation of HER2 CTC status determination as a novel biomarker for the use of HER2-targeted therapies.
Abbreviations
- CK:
-
Cytokeratin
- CTC:
-
Circulating tumor cell
- DAPI:
-
4′,6-diamidino-2-phenylindole.
- FDA:
-
Food and drug administration
- HER2:
-
Human epidermal growth factor receptor 2
- EpCam:
-
Epithelial cell adhesion molecule
- ER:
-
Estrogen receptor
- FISH:
-
Fluorescence in situ hybridization
- FITC:
-
Fluorescein isothiocyanate
- IHC:
-
Immunohistochemistry
- MUC1:
-
Mucin 1
- N/A:
-
Not available
- PR:
-
Progesterone receptor
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
References
Coussens L, Yang-Feng TL, Lioa YC, Chen E, Gray A, Mc Grath J, Seeburg PH, Libermann TA, Schlessinger J, Francke U, Levison A, Ulrich A (1985) Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science 230:1132–1139
Schechter AL, Stern DF, Vaidyanathan L, Decker SJ, Drebin JA, Greene MI, Weinberg AR (1984) The neu oncogene: an erbB-related gene encoding a 185,000-Mr tumour antigen. Nature 312:513–516
Slamon DJ, Goldolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, Press MF (1989) Studies of the HER-2/neu proto-oncogene in human breast cancer and ovarian cancer. Science 244:707–712
Konecny G, Pauletti G, Pegram M, Untch M, Dandekar S, Aguilar Z, Wilson C, Rong HM, Bauerfeind I, Felber M, Wang HJ, Beryt M, Seshadri R, Hepp H, Slamon DJ (2003) Quantitative association between HER-2/neu and steroid hormone receptors in hormone receptor-positive primary breast cancer. J Natl Cancer Inst 95:142–153
Moliterni A, Ménard S, Valagussa P, Biganzoli E, Boracchi P, Balsari A, Casalini P, Tomasic G, Marubini E, Pilotti S, Bonadonna G (2003) HER2 overexpression and doxorubicin in adjuvant chemotherapy for resectable breast cancer. J Clin Oncol 21:458–462
Burstein HJ, Sun Y, Dirix LY, Jiang Z, Paridaens R, Tan A, Awada A, Ranade A, Jiao S, Schwartz G, Abbas R, Powell C, Turnbull K, Vermette J, Zacharchuk C, Badwe R (2010) Neratinib, an irreversible ErbB receptor tyrosine kinase inhibitor, in patients with advanced ErbB2-positive breast cancer. J Clin Oncol 28:1301–1307
Gianni L, Lladó A, Bianchi G, Cortes J, Kellokumpu-Lehtinen PL, Cameron DA, Miles D, Salvagni S, Wardley A, Goeminne JC, Hersberger V, Baselga J (2010) Open-label, phase II, multicenter, randomized study of the efficacy and safety of two dose levels of pertuzumab, a human epidermal growth factor receptor 2 dimerization inhibitor, in patients with human epidermal growth factor receptor 2-negative metastatic breast cancer. J Clin Oncol 28:1131–1137
Baselga J, Gelmon KA, Verma S, Wardley A, Conte P, Miles D, Bianchi G, Cortes J, McNally VA, Ross GA, Fumoleau P, Gianni L (2010) Phase II trial of pertuzumab and trastuzumab in patients with human epidermal growth factor receptor 2-positive metastatic breast cancer that progressed during prior trastuzumab therapy. J Clin Oncol 28:1138–1144
Wolff AC, Hammond ME, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, Dowsett M, Fitzgibbons PL, Hanna WM, Langer A, McShane LM, Paik S, Pegram MD, Perez EA, Press MF, Rhodes A, Sturgeon C, Taube SE, Tubbs R, Vance GH, van de Vijver M, Wheeler TM, Hayes DF (2007) American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 25:118–145
Vincent-Salomon A, Pierga JY, Couturier J, d’Enghien CD, Nos C, Sigal-Zafrani B, Lae M, Fréneaux P, Diéras V, Thiéry JP, Sastre-Garau X (2007) HER2 status of bone marrow micrometastasis and their corresponding primary tumours in a pilot study of 27 cases: a possible tool for anti-HER2 therapy management? Br J Cancer 96:654–659
Zidan J, Dashkovsky I, Stayerman C, Basher W, Cozacov C, Hadary A (2005) Comparison of HER-2 overexpression in primary breast cancer and metastatic sites and its effect on biological targeting therapy of metastatic disease. Br J Cancer 93:552–556
Gancberg D, Di Leo A, Cardoso F, Rouas G, Pedrocchi M, Paesmans M, Verhest A, Bernard-Marty C, Piccart MJ, Larsimont D (2002) Comparison of HER-2 status between primary breast cancer and corresponding distant metastatic sites. Ann Oncol 13:1036–1043
Regitnig P, Schippinger W, Lindbauer M, Samonigg H, Lax SF (2004) Change of HER-2/neu status in a subset of distant metastases from breast carcinomas. J Pathol 203:918–926
Edgerton SM, Moore D 2nd, Merkel D, Thor AD (2003) erbB-2 (HER-2) and breast cancer progression. Appl Immunohistochem Mol Morphol 2003(11):214–221
Carlsson J, Nordgren H, Sjöström J, Wester K, Villman K, Bengtsson NO, Ostenstad B, Lundqvist H, Blomqvist C (2004) HER2 expression in breast cancer primary tumours and corresponding metastases. Original data and literature review. Br J Cancer 90:2344–2348
Aitken SJ, Thomas JS, Langdon SP, Harrison DJ, Faratian D (2009) Quantitative analysis of changes in ER, PR and HER2 expression in primary breast cancer and paired nodal metastases. Ann Oncol. doi:10.1093/annonc/mdp427
Tanner M, Järvinen P, Isola J (2001) Amplification of HER-2/neu and topoisomerase II alpha in primary and metastatic breast cancer. Cancer Res 61:5345–5348
Simmons C, Miller N, Geddie W, Gianfelice D, Oldfield M, Dranitsaris G, Clemons MJ (2009) Does confirmatory tumor biopsy alter the management of breast cancer patients with distant metastases? Ann Oncol 20:1499–1504
Liedtke C, Broglio K, Moulder S, Hsu L, Kau SW, Symmans WF, Albarracin C, Meric-Bernstam F, Woodward W, Theriault RL, Kiesel L, Hortobagyi GN, Pusztai L, Gonzalez-Angulo AM (2009) Prognostic impact of discordance between triple-receptor measurements in primary and recurrent breast cancer. Ann Oncol 20:1953–1958
Riethdorf S, Fritsche H, Müller V, Rau T, Schindlbeck C, Rack B, Janni W, Coith C, Beck K, Jänicke F, Jackson S, Gornet T, Cristofanilli M, Pantel K (2007) Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch System. Clin Cancer Res 13:920–928
Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF (2004) Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 351:781–791
Budd GT, Cristofanilli M, Ellis MJ, Stopeck A, Borden E, Miller MC, Matera J, Repollet M, Doyle GV, Terstappen LW, Hayes DF (2006) Circulating tumor cells versus imaging: predicting overall survival in metastatic breast cancer. Clin Cancer Res 12:6403–6409
Fehm T, Hoffmann O, Aktas B, Becker S, Solomayer EF, Wallwiener D, Kimmig R, Kasimir-Bauer S (2009) Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. doi:10.1186/bcr2349
Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S (2009) Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. doi: 10.1186/bcr2333
Riethdorf S, Müller V, Zhang L, Rau T, Loibl S, Komor M, Roller M, Huober J, Fehm T, Schrader I, Hilfrich J, Holms F, Tesch H, Eidtmann H, Untch M, von Minckwitz G, Pantel K (2010) Detection and HER2 expression of circulating tumor cells: prospective monitoring in breast cancer patients treated in the neoadjuvant Geparquattro trial. Clin Cancer Res 16:1634–2645
Hauch S, Zimmermann S, Lankiewicz S, Zieglschmid V, Böcher O, Albert WH (2007) The clinical significance of circulating tumour cells in breast cancer and colorectal cancer patients. Anticancer Res 27:1337–1341
Hayes DF, Walker TM, Singh B, Vitetta ES, Uhr JW, Gross S, Rao C, Doyle GV, Terstappen LW (2002) Monitoring expression of HER-2 on circulating epithelial cells in patients with advanced breast cancer. Int J Oncol 21:1111–1117
Meng S, Tripathy D, Shete S, Ashfaq R, Haley B, Perkins S, Beitsch P, Khan A, Euhus D, Osborne C, Frenkel E, Hoover S, Leitch M, Clifford E, Vitetta E, Morrison L, Herlyn D, Terstappen LW, Fleming T, Fehm T, Tucker T, Lane N, Wang J, Uhr J (2004) HER-2 gene amplification can be acquired as breast cancer progresses. Proc Natl Acad Sci USA 101:9393–9398
Hayes DF, Ethier S, Lippman ME (2006) New guidelines for reporting of tumor marker studies in breast cancer research and treatment: remark. Breast Cancer Res Treat 100:237–238
McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM (2005) Statistics Subcommittee of the NCI-EORTC Working Group on Cancer Diagnostics: Reporting recommendations for tumor marker prognostic studies (remark). J Natl Cancer Inst 97:1180–1184
Thurm H, Ebel S, Kentenich C, Hemsen A, Riethdorf S, Coith C, Wallwiener D, Braun S, Oberhoff C, Jänicke F, Pantel K (2003) Rare expression of epithelial cell adhesion molecule on residual micrometastatic breast cancer cells after adjuvant chemotherapy. Clin Cancer Res 9:2598–2604
Sieuwerts AM, Kraan J, Bolt J, van der Spoel P, Elstrodt F, Schutte M, Martens JMW, Gratama JW, Sleijfer S, Foekens JA (2009) Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst 101:61–66
Van der Auwera I, Peeters D, Benoy IH, Benoy IH, Elst HJ, Van Laere SJ, Prové A, Maes H, Huget P, van Dam P, Vermeulen PB, Dirix LY (2010) Circulating tumour cell detection: a direct comparison between the CellSearch System, the AdnaTest and CK-19/mammaglobin RT-PCR in patients with metastatic breast cancer. Br J Cancer 102:276–284
Hayes DF, Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Miller MC, Matera J, Allard WJ, Doyle GV, Terstappen LW (2006) Circulating tumor cells at each follow-up time point during therapy of metastatic breast cancer patients predict progression-free and overall survival. Clin Cancer Res 12:4218–4224
Pestrin M, Bessi S, Galardi F, Truglia M, Biggeri A, Biagioni C, Cappadona S, Biganzoli L, Giannini A, Di Leo A (2009) Correlation of HER2 status between primary tumors and corresponding circulating tumor cells in advanced breast cancer patients. Breast Cancer Res Treat 118:523–530
Fehm T, Becker S, Duerr-Stoerzer S, Sotlar K, Mueller V, Wallwiener D, Lane N, Solomayer E, Uhr J (2007) Determination of HER2 status using both serum HER2 levels and circulating tumor cells in patients with recurrent breast cancer whose primary tumor was HER2 negative or of unknown HER2 status. Breast Cancer Res. doi:10.1186/bcr1783
Tewes M, Aktas B, Hauch S, Kimmig R, Kasimir-Bauer S (2009) Molecular profiling and predictive value of circulating tumor cells in patients with metastatic breast cancer: an option for monitoring response to breast cancer related therapies. Breast Cancer Res Treat 115:581–590
Reuben JM, Lee BN, Li C, Broglio KR, Valero V, Jackson S, Ueno NT, Krishnamurthy S, Hortobagyi GN, Cristofanilli M (2007) Genomic of circulating tumor cells in metastatic breast cancer [abstract 1002]. J Clin Oncol 25(Suppl)
Acknowledgements
This work was supported by an unrestricted research grant of Roche Pharma AG, Germany. Furthermore, this study was partly supported by Adnagen (Langenhagen, Germany). We thank Cornelia Coith, Susanne Hoppe, Sylke Krenkel, Oliver Mauermann, Malgorzata Stoupiec, and Silke Dürr-Störzer for excellent technical assistance. Our special thanks to Ute Hilcher for coordinating the study. In addition, we would like to thank all doctors (Michaela Becker, Julia Jüstock, Dina Mury, Mitras Tewes, and Julia Zeitz) for help with patients and data collection, Dr. Elke Heidrich-Lorsbach (Alcedis GmbH) for statistical analysis and data management, and Lee Miller from Miller Medical Communications for editing and reviewing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
This study is conducted on behalf of the DETECT Study Group.
T. Fehm and V. Müller contributed equally to this work.
Rights and permissions
About this article
Cite this article
Fehm, T., Müller, V., Aktas, B. et al. HER2 status of circulating tumor cells in patients with metastatic breast cancer: a prospective, multicenter trial. Breast Cancer Res Treat 124, 403–412 (2010). https://doi.org/10.1007/s10549-010-1163-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-1163-x