Abstract
Mutations in RAD51 gene are believed to be associated with elevated breast cancer risk. However, several case–control studies focusing on the association between RAD51 135G>C and breast cancer risk failed to achieve consensus. To clarify the effect of RAD51 135G>C polymorphism on breast cancer, a meta-analysis was performed. By searching PubMed and EMBASE, a total of 14 case–control studies, containing 12,183 cases and 10,183 controls, were included. The strength of association between RAD51 135G>C polymorphism and breast cancer risk was assessed by odds ratio (OR) with the corresponding 95% confidence interval (95% CI). When all the eligible studies were pooled into the meta-analysis, an elevated cancer risk was revealed in additive model (OR, 1.34; 95% CI, 1.01–1.78; P = 0.044) and recessive model (OR, 1.37; 95% CI, 1.03–1.82; P = 0.032). In subgroup analyses by ethnicity, BRCA1/2 mutation status, and family history, a significant association was found only among BRCA2 mutation carriers (additive model: OR, 4.92; 95% CI, 1.11–21.83; P = 0.036; recessive model: OR, 4.88; 95% CI, 1.10–21.67; P = 0.037). Sensitivity analysis did not perturb the results. In conclusion, this meta-analysis suggests that RAD51 variant 135C homozygote is associated with elevated breast cancer risk among BRCA2 mutation carriers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Double-strand break (DSB) damage, causing cell death or loss of genetic material, is the most injurious lesion and responsible for cancer development. However, it can be repaired by several DSB repair genes such as BRCA1/2 in which mutations have been proven to contribute to high risk of cancer in women [1]. RAD51, a homolog of Escherichia coli RecA, is another important DSB repair gene and can interact with BRCA1 and BRCA2 proteins, functioning through homologous recombination and non-homologous end joining [2, 3]. Mutations in RAD51 gene have been believed to be associated with an elevated risk of cancer including breast cancer which is the most common cancer in women all over the world.
A functional single-nucleotide polymorphism (SNP) in the 5′ untranslated region (UTR) of RAD51, 135G>C (c.-98G>C, rs1801320), has been identified, which is involved in modifying promoter activity and the penetrance of BRCA1/2 mutations [4, 5], and therefore may be a good candidate for low-penetrance variants that contribute to breast cancer risk. Several case–control studies focusing on the association between RAD51 135G>C and breast cancer risk have been conducted and suggested that RAD51 135G>C modified the breast cancer risk of women only if they had a family history or carried BRCA2 mutations [5–10]. However, the results have been inconsistent and inconclusive [11–13] owing to the relatively small sample sizes and different patient populations. Meta-analysis is a powerful and rigorous method to resolve these problems. In this study, to clarify the association between RAD51 135G>C polymorphism and breast cancer susceptibility, a meta-analysis was performed.
Methods
Study identification and selection
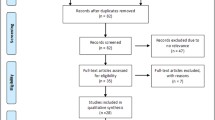
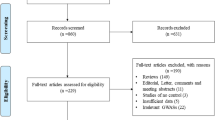
Before the study, inclusion criteria were defined as follows: (a) articles evaluating the association between RAD51 135G>C polymorphism and breast cancer risk; (b) study designed as case–control; (c) sufficient data available to estimate an odds ratio (OR) with its 95% confidence interval (95% CI). A literature search of PubMed and EMBASE (updated to 2010/04/10) was conducted using the following terms: ‘RAD51’, ‘polymorphism(s)’, ‘breast cancer’, or ‘breast carcinoma’, without restriction on language. The retrieved literatures were then read independently in their entirety to assess their appropriateness for the inclusion in this meta-analysis by the two authors (Zhou and Hu). The reference lists of reviews and retrieved articles were searched simultaneously to find additional eligible studies. If studies had partly overlapped subjects, only the study with a larger sample size was selected. Any disagreement was resolved by discussion between the two authors.
Data extraction
The following variables were extracted from each study if available: first author’s surname, publication year, patient ethnicity, matching criteria, sample size, and numbers of cases and controls in different RAD51 135G>C genotypes.
Statistical analysis
The strength of association between RAD51 135G>C polymorphism and breast cancer risk was assessed by OR with the corresponding 95% CI. The pooled OR was calculated by a fixed-effects model (the Mantel–Haenszel method) when between-study heterogeneity was absent [14]. Otherwise, a random-effects model (the DerSimonian and Laird method) [15] was selected. Statistical between-study heterogeneity was checked by the Q test [16] and was considered statistically significant with P < 0.10. The OR and its 95% CI in each comparison was assessed in dominant (GC/CC versus GG), additive (CC versus GG), and recessive (CC versus GG/GC) genetic models. In addition, subgroup analyses for ethnicity (Caucasian, Asian, and mixed population), BRCA1/2 mutation status (BRCA1 mutation carriers, BRCA2 mutation carriers, non-carriers, and unselected populations), and family history (familial and sporadic breast cancer) were conducted, and influence analysis was performed by omitting each study to find potential outliers [17]. In the control populations, Hardy–Weinberg equilibrium (HWE) was tested, but a deviation from HWE was allowed in a mixed control population. Sensitivity analysis was also conducted by excluding the HWE-violating studies. Potential publication bias was examined visually in a funnel plot of log [OR] against its standard error (SE), and the degree of asymmetry was tested by Egger’s test (P < 0.05 was considered a significant publication bias) [18]. This meta-analysis was performed using the software STATA version 10.0.
Results
Study characteristics
A total of 26 publications met the inclusion criteria. Of these studies, 12 [5, 11, 12, 19–27] were excluded because of their populations overlapped with another three included studies [8, 28, 29]. As a result, a total of 14 publications [6, 8–10, 13, 28–36] containing 12,183 cases and 10,183 controls were included in this meta-analysis. Table 1 lists the main characteristics of these studies. Among these publications, there were eight studies of Caucasian descent [6, 8–10, 28, 30, 32, 36], three of Asian descent [31, 33, 35], and three of mixed populations [13, 29, 34]. Two studies [6, 8] contained the subjects of BRCA1/2 mutation carriers, two [6, 29] contained non-carriers, and another ten [10, 13, 28, 30–36] contained unselected populations. In addition, five of these studies [10, 13, 28, 29, 36] presented RAD51 135G>C polymorphism genotype distributions according to family history (familial and sporadic breast cancer) (Table 2). All of the cases were histologically confirmed as breast cancer. Controls were mainly healthy populations, and matched with age or cancer-free. Genotype distributions in the controls of all studies were in agreement with HWE except one [36].
Meta-analysis results
As shown in Table 3, no between-study heterogeneity was found in overall comparisons in additive and recessive genetic models. When all the eligible studies were pooled into the meta-analysis, elevated breast cancer risks were revealed in additive model (OR, 1.34; 95% CI, 1.01–1.78; P = 0.044) and recessive model (OR, 1.37; 95% CI, 1.03–1.82; P = 0.032) (Fig. 1a). Next, the effect of RAD51 135G>C polymorphism was evaluated in subgroup analysis according to specific ethnicity, BRCA1/2 mutation status and family history. A significant association was found only among BRCA2 mutation carriers (additive model: OR, 4.92; 95% CI, 1.11-21.83; P = 0.036; recessive model: OR, 4.88; 95% CI, 1.10-21.67; P = 0.037) (Fig. 1b), but not in other subgroups (Table 3).
Sensitivity analysis
Influence analysis was performed to assess the influence of each individual study on the pooled OR by sequential removal of individual studies. The results suggested that no individual study significantly affected the pooled ORs (Fig. 2). Sensitivity analysis by excluding HWE-violating study did not perturb the overall results.
Influence analysis for GC/CC versus GG in the overall meta-analysis. This figure shows the influence of individual studies on the summary OR. The middle vertical axis indicates the overall OR and the two vertical axes indicate its 95% CI. Every hollow round indicates the pooled OR when the left study is omitted in this meta-analysis. The two ends of every broken line represent the 95% CI
Publication bias
Funnel plot and Egger’s test were performed to assess the publication bias. The shapes of the funnel plot did not indicate any evidence of obvious asymmetry in additive and recessive model (Fig. 3) and the Egger’s test suggested the absence of publication bias (additive model: P = 0.997; recessive model: P = 0.906).
Discussion
RAD51 plays an important role in maintenance of the genomic integrity through DSB repair mechanisms including homologous recombination and non-homologous end joining. However, accumulating evidence supports the hypothesis that genetic polymorphisms in RAD51 may result in reduced DNA repair capacity and be associated with increased susceptibility to breast cancer [4, 5]. In this meta-analysis, a total of 14 eligible case–control studies were pooled to explore the association between RAD51 135G>C polymorphism and breast cancer risk, and the results suggested that carriers with variant 135C allele homozygote in additive and recessive genetic model had an increased risk of breast cancer. Influence analysis by sequentially removing individual studies did not perturb the results. Neither did sensitivity analysis by excluding the HWE-violating study.
Ethnicity was significantly associated with breast cancer risk and genotype of RAD51 135G>C variants (P < 0.0001). Asian women had a lower risk of breast cancer than White women. Among Black women, 37.4% had at least one copy of the variant allele RAD51 135C, while among non-Jewish White, Jewish White, and other ethnic populations, the frequencies were 15.9, 9.6, and 17.3%, respectively [34]. This may lead to RAD51 135G>C polymorphism genotype distribution disequilibrium when all ethnic populations were pooled together. It was essential to conduct a subgroup analysis on ethnicities. In this meta-analysis, all subjects were classified into three ethnic groups (Caucasian (White), Asian, and mixed populations). The results revealed no association between RAD51 135G>C variants and breast cancer susceptibility in all ethnic groups.
As described above, the RAD51 gene product acts together with BRCA1 and BRCA2 proteins in homologous recombination and DSB repair. It is reasonable to assume that RAD51 and BRCA1/2 mutations may have interactive effects on breast cancer risk. Some previous studies presented an association of RAD51 variant allele 135C with an elevated breast cancer risk only in BRCA2 mutation carrier, but not in BRCA1 mutation carriers or non-carriers or unselected populations [5–8]. In contrast, Jakubowska et al. [11, 12] observed a significantly reduced risk of breast cancer among Polish female carriers of RAD51 135C allele and BRCA1 founder mutations (5382insC, 4153delA and 300T>G). Subgroup analysis on BRCA1/2 mutation status in this meta-analysis, however, confirmed the former result.
Furthermore, germline mutations in BRCA1/2 genes account for fewer than 2% of all breast cancer cases, while for about 20% of the familial breast cancer [37], suggesting that some differences in susceptibility genes may exist between familial and sporadic breast cancer. In order to define whether RAD51 135G>C polymorphism played different roles in familial and sporadic breast cancer risks or not, a subgroup analysis based on family history (familial and sporadic breast cancer) was performed. No association was found in both groups as the results suggested.
Although the results of this meta-analysis were powerful, some limitations still exist. First, this study is a study-level but not an individual patient-level meta-analysis. It is known that study-level analysis can lead to biased assessments and use of aggregated summary values has some limitations in explaining the heterogeneity [38]. Second, OR value was obtained without correction. More accurate OR should be corrected by age, menopause status, ethnicity and other exposure factors that are potentially associated with breast cancer risk. Third, of these 14 studies, most subjects were Caucasians and Asians, and no study presented RAD51 genotype distribution among Africans. Therefore, the conclusion about this association in African populations should be further investigated.
In conclusion, this study suggested that RAD51 variant 135C homozygote is associated with elevated breast cancer risk among BRCA2 mutation carriers, but not in BRCA1 mutation carriers or non-carriers or unselected populations.
References
Hughes DJ (2008) Use of association studies to define genetic modifiers of breast cancer risk in BRCA1 and BRCA2 mutation carriers. Fam Cancer 7:233–244. doi:10.1007/s10689-008-9181-0
Baumann P, West SC (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci 23:247–251. doi:10.1016/S0968-0004(98)01232-8
Thacker J (2005) The RAD51 gene family, genetic instability and cancer. Cancer Lett 219:125–135. doi:10.1016/j.canlet.2004.08.018
Hasselbach L, Haase S, Fischer D, Kolberg HC, Sturzbecher HW (2005) Characterisation of the promoter region of the human DBA-repair gene RAD51. Eur J Gynaecol Oncol 26:589–598
Wang WW, Spurdle AB, Kolachana P, Bove B, Modan B, Ebbers SM, Suthers G, Tucker MA, Kaufman DJ, Doody MM, Tarone RE, Daly M, Levavi H, Pierce H, Chetrit A, Yechezkel GH, Chenevix-Trench G, Offit K, Godwin AK, Struewing JP (2001) A single nucleotide polymorphism in the 5’ untranslated region of RAD51 and risk of cancer among BRCA1/2 mutation carriers. Cancer Epidemiol Biomarkers Prev 10:955–960
Kadouri L, Kote-Jarai Z, Hubert A, Durocher F, Abeliovich D, Glaser B, Hamburger T, Eeles RA, Peretz T (2004) A single-nucleotide polymorphism in the RAD51 gene modifies breast cancer risk in BRCA2 carriers, but not in BRCA1 carriers or noncarriers. Br J Cancer 90:2002–2005. doi:10.1038/sj.bjc.6601837
Levy-Lahad E, Lahad A, Eisenberg S, Dagan E, Paperna T, Kasinetz L, Catane R, Kaufman B, Beller U, Renbaum P, Gershoni-Baruch R (2001) A single nucleotide polymorphism in the RAD51 gene modifies cancer risk in BRCA2 but not BRCA1 carriers. Proc Natl Acad Sci USA 98:3232–3236. doi:10.1073/pnas.051624098
Antoniou AC, Sinilnikova OM, Simard J, Léoné M, Dumont M, Neuhausen SL, Struewing JP, Stoppa-Lyonnet D, Barjhoux L, Hughes DJ, Coupier I, Belotti M, Lasset C, Bonadona V, Bignon YJ; Genetic Modifiers of Cancer Risk in BRCA1/2 Mutation Carriers Study (GEMO), Rebbeck TR, Wagner T, Lynch HT, Domchek SM, Nathanson KL, Garber JE, Weitzel J, Narod SA, Tomlinson G, Olopade OI, Godwin A, Isaacs C, Jakubowska A, Lubinski J, Gronwald J, Górski B, Byrski T, Huzarski T, Peock S, Cook M, Baynes C, Murray A, Rogers M, Daly PA, Dorkins H; Epidemiological Study of BRCA1 and BRCA2 Mutation Carriers (EMBRACE), Schmutzler RK, Versmold B, Engel C, Meindl A, Arnold N, Niederacher D, Deissler H; German Consortium for Hereditary Breast and Ovarian Cancer (GCHBOC), Spurdle AB, Chen X, Waddell N, Cloonan N; Kathleen Cuningham Consortium for Research into Familial Breast Cancer (kConFab), Kirchhoff T, Offit K, Friedman E, Kaufmann B, Laitman Y, Galore G, Rennert G, Lejbkowicz F, Raskin L, Andrulis IL, Ilyushik E, Ozcelik H, Devilee P, Vreeswijk MP, Greene MH, Prindiville SA, Osorio A, Benitez J, Zikan M, Szabo CI, Kilpivaara O, Nevanlinna H, Hamann U, Durocher F, Arason A, Couch FJ, Easton DF, Chenevix-Trench G; Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA) (2007) RAD51 135G > C modifies breast cancer risk among BRCA2 mutation carriers: results from a combined analysis of 19 studies. Am J Hum Genet 81:1186–1200. doi:10.1086/522611
Jakubowska A, Jaworska K, Cybulski C, Janicka A, Szymańska-Pasternak J, Lener M, Narod SA, Lubiński J, IHCC-Breast Cancer Study Group (2009) Do BRCA1 modifiers also affect the risk of breast cancer in non-carriers? Eur J Cancer 45:837–842. doi:10.1016/j.ejca.2008.10.021
Costa S, Pinto D, Pereira D, Rodrigues H, Cameselle-Teijeiro J, Medeiros R, Schmitt F (2007) DNA repair polymorphisms might contribute differentially on familial and sporadic breast cancer susceptibility: a study on a Portuguese population. Breast Cancer Res Treat 103:209–217. doi:10.1007/s10549-006-9364-z
Jakubowska A, Narod SA, Goldgar DE, Mierzejewski M, Masojć B, Nej K, Huzarska J, Byrski T, Górski B, Lubiński J (2003) Breast cancer risk reduction associated with the RAD51 polymorphism among carriers of the BRCA1 5382insC mutation in Poland. Cancer Epidemiol Biomarkers Prev 12:457–459
Jakubowska A, Gronwald J, Menkiszak J, Górski B, Huzarski T, Byrski T, Edler L, Lubiñski J, Scott RJ, Hamann U (2007) The RAD51 135G > C polymorphism modifies breast cancer and ovarian cancer risk in Polish BRCA1 mutation carriers. Cancer Epidemiol Biomarkers Prev 16:270–275. doi:10.1158/1055-9965.EPI-06-0562
Dufloth RM, Costa S, Schmitt F, Zeferino LC (2005) DNA repair gene polymorphisms and susceptibility to familial breast cancer in a group of patients from Campinas, Brazil. Genet Mol Res 4:771–782
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Cochran WG (1954) The combination of estimates from different experiments. Biometrics 10:101–129
Tobias A (1999) Assessing the influence of a single study in the meta-analysis estimate. Stata Tecnol Bull 8:15–17
Egger M, Davey SG, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Gal I, Kimmel G, Gershoni-Baruch R, Papa MZ, Dagan E, Shamir R, Friedman E (2006) A specific RAD51 haplotype increases breast cancer risk in Jewish non-Ashkenazi high-risk women. Eur J Cancer 42:1129–1134. doi:10.1016/j.ejca.2005.09.035
Romanowicz-Makowska H, Smolarz B, Kulig A (2005) Germline BRCA1 mutations and G/C polymorphism in the 5’-untranslated region of the RAD51 gene in Polish women with breast cancer. Pol J Pathol 56:161–165
Romanowicz-Makowska H, Smolarz B (2006) Analysis of loss of heterozygosity and microsatellite instability RAD52, RAD54 and RAD54B gene and BRCA1 gene mutation in breast cancer. Pol Merkur Lekarski 21:548–550 Polish
Romanowicz-Makowska H, Smolarz B, Kulig A (2006) The G/C polymorphism of RAD51 gene in breast cancer. Pol Merkur Lekarski 21:55–58 Polish
Blasiak J, Przybyłowska K, Czechowska A, Zadrozny M, Pertyński T, Rykała J, Kołacińska A, Morawiec Z, Drzewoski J (2003) Analysis of the G/C polymorphism in the 5’-untranslated region of the RAD51 gene in breast cancer. Acta Biochim Pol 50:249–253
Romanowicz-Makowska H, Smolarz B, Zadrozny M, Kulig A (2006) Analysis of RAD51 polymorphism and BRCA1 mutations in Polish women with breast cancer. Exp Oncol 28:156–159
Synowiec E, Stefanska J, Morawiec Z, Blasiak J, Wozniak K (2008) Association between DNA damage, DNA repair genes variability and clinical characteristics in breast cancer patients. Mutat Res 648:65–72. doi:10.1016/j.mrfmmm.2008.09.014
Krupa R, Synowiec E, Pawlowska E, Morawiec Z, Sobczuk A, Zadrozny M, Wozniak K, Blasiak J (2009) Polymorphism of the homologous recombination repair genes RAD51 and XRCC3 in breast cancer. Exp Mol Pathol 87:32–35. doi:10.1016/jyexmp2009.04.005
Jara L, Acevedo ML, Blanco R, Castro VG, Bravo T, Gómez F, Waugh E, Peralta O, Cabrera E, Reyes JM, Ampuero S, González-Hormazábal P (2007) RAD51 135G > C polymorphism and risk of familial breast cancer in a South American population. Cancer Genet Cytogenet 178:65–69. doi:10.1016/j.cancergencyto.2007.05.024
Sliwinski T, Krupa R, Majsterek I, Rykala J, Kolacinska A, Morawiec Z, Drzewoski J, Zadrozny M, Blasiak J (2005) Polymorphisms of the BRCA2 and RAD51 genes in breast cancer. Breast Cancer Res Treat 94:105–109. doi:10.1007/s10549-005-0672-5
Jara L, Dubois K, Gaete D, de Mayo T, Ratkevicius N, Bravo T, Margarit S, Blanco R, Gómez F, Waugh E, Peralta O, Reyes JM, Ibáñez G, González-Hormazábal P (2010) Variants in DNA double-strand break repair genes and risk of familial breast cancer in a South American population. Breast Cancer Res Treat. doi:10.1007/s10549-009-0709-2
Kuschel B, Auranen A, McBride S, Novik KL, Antoniou A, Lipscombe JM, Day NE, Easton DF, Ponder BA, Pharoah PD, Dunning A (2002) Variants in DNA double-strand break repair genes and breast cancer susceptibility. Hum Mol Genet 11:1399–1407. doi:10.1093/hmg/11.12.1399
Lee KM, Choi JY, Kang C, Kang CP, Park SK, Cho H, Cho DY, Yoo KY, Noh DY, Ahn SH, Park CG, Wei Q, Kang D (2005) Genetic polymorphisms of selected DNA repair genes, estrogen and progesterone receptor status, and breast cancer risk. Clin Cancer Res 11:4620–4626. doi:10.1158/1078-0432.CCR-04-2534
Webb PM, Hopper JL, Newman B, Chen X, Kelemen L, Giles GG, Southey MC, Chenevix-Trench G, Spurdle AB (2005) Double-strand break repair gene polymorphisms and risk of breast or ovarian cancer. Cancer Epidemiol Biomarkers Prev 14:319–323. doi:10.1158/1055-9965.EPI-04-0335
Chang TW, Wang SM, Guo YL, Tsai PC, Huang CJ, Huang W (2006) Glutathione S-transferase polymorphisms associated with risk of breast cancer in southern Taiwan. Breast 15:754–761. doi:10.1016/j.breast.2006.03.008
Brooks J, Shore RE, Zeleniuch-Jacquotte A, Currie D, Afanasyeva Y, Koenig KL, Arslan AA, Toniolo P, Wirgin I (2008) Polymorphisms in RAD51, XRCC2, and XRCC3 are not related to breast cancer risk. Cancer Epidemiol Biomarkers Prev 17:1016–1019. doi:10.1158/1055-9965.EPI-08-0065
Hu R, Wei Y, Jiang WJ, Yao WX, Long QM, Zhang JH, Liang Y, Tang XL (2008) Association of polymorphisms of N372H in BRCA2 gene and 135G/C in RAD51 gene and breast cancers. Sichuan Da Xue Xue Bao Yi Xue Ban 39:973–975 Chinese
Akisik E, Yazici H, Dalay N (2010) ARLTS1, MDM2 and RAD51 gene variations are associated with familial breast cancer. Mol Biol Rep. doi:10.1007/s11033-010-0113-3
Wooster R, Weber BL (2003) Breast and ovarian cancer. N Engl J Med 348:2339–2347. doi:10.1056/NEJMra012284
Berlin JA, Santanna J, Schmid CH, Szczech LA, Feldman HI, Anti-Lymphocyte Antibody Induction Therapy Study Group (2002) Individual patient- versus group-level data meta-regressions for the investigation of treatment effect modifiers: ecological bias rears its ugly head. Stat Med 21:371–387
Author information
Authors and Affiliations
Corresponding author
Additional information
G.-W. Zhou and J. Hu equally contributed to this study.
Rights and permissions
About this article
Cite this article
Zhou, GW., Hu, J., Peng, XD. et al. RAD51 135G>C polymorphism and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 125, 529–535 (2011). https://doi.org/10.1007/s10549-010-1031-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-1031-8