Abstract
Male breast cancer (MBC) is a poorly characterized disease because of its rarity. Clinical management is based on results obtained from randomized trials conducted in women notwithstanding data in the literature suggesting relevant gender-associated differences in terms of biological and clinical behavior. However, a genome-wide characterization of MBC on a transcriptional level is lacking. In this study, gene expression profiles of 37 estrogen receptor positive (ER+) MBC specimens were compared to that of 53 ER+ Female Breast Cancer (FBC) samples similar for clinical and patho-biological features. Almost 1000 genes were found differentially expressed (FDR < 1%) between female and male patients and biological interpretation highlighted a gender-associated modulation of key biological processes ranging from energy metabolism to regulation of translation and matrix remodeling as well as immune system recruitment. Moreover, an analysis of genes correlated to steroid receptors and ERBB2 suggested a prominent role for the androgen receptor in MBC with a minor relevance for progesterone receptor and ERBB2, although, similarly to FBC, a genomic amplification could be observed. Our findings support the idea that breast cancer is a quite different disease in male and female patients and the underlying gender-related biological differences are likely to have clinical implications connected with different susceptibility to treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Male breast cancer (MBC) is a rare disease accounting for less than 1% of all cases of breast cancer diagnosed yearly, but its incidence rate continues to increase by 1.1% annually. Research on MBC has been limited by its rarity and most information on this disease has been extrapolated from results in female patients.
The etiology of MBC is unclear but, similarly to women, steroid hormones levels may play a role in the development of the disease while a family history confers a relative risk of 2.5. As in women, mutations in BRCA1 and BRCA2 genes increase the risk of developing breast cancer; BRCA1 mutations are relatively rare (occurring in up to 4% of the cases), while those in BRCA2 gene are more frequent accounting for 4–16% of men with breast cancer. The major genetic factors associated with an increased risk of breast cancer in men also include mutations within the DNA-binding domain of androgen receptor (AR), polymorphism in the CYP17 gene coding for an enzyme involved in steroid synthesis, mutation of CHEK2 and PTEN tumor suppressor genes; however, none of these genes has been demonstrated to have a causal association with MBC [1, 2].
The worse prognosis observed for male compared to female breast cancer (FBC) is mainly due to the delay in diagnosis and to a more advanced stage at presentation, since after adjustment for age at diagnosis and stage of disease, the gender-associated differences disappear [3]. The standard management of MBC derives from clinical data obtained in randomized trials conducted in women, and includes radical mastectomy plus axillary lymph node dissection, recently substituted by less invasive surgical procedures with no detectable decline in survival [2]. As for FBC, adjuvant chemotherapy is used to treat male patients who are at high risk of recurrence while the antiestrogen tamoxifen is considered the standard adjuvant hormonal therapy in male patients with steroid receptor-positive tumors [4].
Up to date, the characterization of MBC at molecular level has mainly focused on immunohistochemical analysis of a limited set of well know FBC biomarkers. As summarized in [1], a higher positivity rate of estrogen and progesterone receptors (ER and PgR) in MBC respect to FBC is well established. However, Munoz de Toro et al. [5] remarkably found that in MBC proliferation rate was higher in ER+/PgR+ compared to ER−/PgR− tumors, an opposite finding respect to FBC which strongly support the hypothesis that these receptors may play a different role in males compared to women. Controversial data were reported for AR positivity as well as for ERBB2 amplification rate, with positivity rates ranging from 0 to 95% among different studies [1, 2, 6, 7]. Furthermore, an immunohistochemical analysis of ER, AR, and functionally related proteins indicates a reduced functionality of ER but a significant role for AR in MBC [8]. Finally, a higher percentage of BCL2 positive tumors in male was also reported [8], suggesting a role for this antiapoptotic gene during carcinogenesis.
Globally, these data indicate many differences between the biology of MBC and FBC, an observation which might imply the need of distinct therapeutic approaches and management of the disease according to the gender.
During the last decade, the application of a number of high-throughput genomic technologies refined the taxonomy of breast cancer and identified genomic fingerprints likely to impact future clinical treatment approaches [9]. However, while FBC has been intensively investigated by comprehensive molecular analysis, at present no gene expression data are available for MBC. Recently, Fassan et al. reported a set of microRNA as differentially expressed by comparison of 23 MBC and 10 FBC samples [10].
Taking advantage of the availability in our Tumor Tissue Bank of a series of ER+ primary breast cancer specimens from male patients, we compared their gene expression profiles with the profile of breast cancer from female patients with similar clinical and patho-biological features, to investigate whether a gender-related biological difference does exist at transcriptomic level in breast cancer.
Our data suggest a profound biomolecular difference between MBC and FBC and highlight a set of differently regulated biological processes, ranging from energy metabolism to regulation of translation and matrix remodeling as well as immune system recruitment. Furthermore, correlation analysis supports a prominent role for AR in MBC with a minor relevance for PgR and ERBB2, although, similarly to FBC, a genomic amplification may be observed for the latter also in males.
Materials and methods
Patients and tumors features
We consecutively collected 37 snap-frozen ER+ primary tumor specimens from men submitted to radical mastectomy or conservative quadrantectomy plus axillary lymph nodes dissection for a primary breast cancer at the Istituto Nazionale dei Tumori of Milan during the period 1991–1997. All patients signed an informed consent to donate to the Istituto Nazionale dei Tumori the leftover tissue after completing diagnostic procedures. Fifty-three FBC with ER+ primaries for whom gene expression profiles were available were selected to obtain a distribution of the main clinical and patho-biological variables (age, tumor size, PgR status) comparable to that of MBC samples, limiting in this way confounding effects on comparison of gene expression data between the two genders. Clinical and patho-biological data are summarized in Table 1.
ER and PgR content was routinely evaluated at time of diagnosis according to the EORTC recommendations and within national and international quality control programs by a ligand binding assay as previously described [11] and results were expressed as fmol mg−1 of protein. Tumors with an ER concentration higher than 10 fmol mg−1 of protein or with a PgR concentration higher than 25 fmol mg−1 of protein were defined as ER-positive or PgR-positive, respectively.
RNA isolation and microarray hybridization
Total RNA extraction, probe labeling, and hybridization were performed as previously described [12]. Briefly, total RNA was extracted using Trizol reagent (Life Technologies, Inc. Grand Island, NY) following the manufacturer’s instructions and treated with DNaseI (Qiagen, Valencia, CA). Each sample was both directly and indirectly labeled with Cy-dCTPs (Amersham Biosciences; GE Healthcare, Piscataway, NJ) and with 3DNA Array900 detection kit (Genisphere, Montvale, NJ), respectively. Samples were co-hybridized together with a common RNA reference (Universal Human Reference RNA, Stratagene, La Jolla, CA) on custom made cDNA microarray slides containing 16457 sequence-verified I.M.A.G.E. clones (Research Genetics/Invitrogen) spotted in triplicate. cDNA microarrays were scanned using the GenePix 4000B microarray scanner at a resolution of 5 μm and the images were analyzed using GenePix Pro v.5.0 software (Axon Instruments, Union City, CA).
Data analysis and statistics
Array data
Microarray raw data were processed using marray package (version 1.22.0). After a quality control, a print-tip loess normalization was applied to each array, followed by an inter-array scale normalization. Intensities from flagged spots were removed, probes with more than 10% of missing values were filtered out and replicate probes were averaged. After this filtering, 13285 distinct clones remained corresponding to 7792 distinct genes according to annotation using the Stanford SOURCE Search website. The entire dataset is available at Array Express database (ID: E-TABM-810).
All class comparisons on gene expression data were performed applying the Significance Analysis of Microarrays (SAM) [13] to data at clone level, considering as threshold an FDR < 1% unless otherwise specified.
To aid the biological interpretation, Gene Ontology Analysis using the TopGO package (version 1.12.0) [14] using the Fisher’s exact test statistics was carried out.
Correlation analysis
Gene expression values of single clones of interest were correlated (Spearman correlation) to gene expression values of all the other clones, separately for MBC and FBC. Only probes with a P < 0.01 were considered significantly correlated. After array data filtering, no clones were found referring to the PgR gene, consequently in subsequent analysis PgR expression data were evaluated by qPCR.
Clustering analysis
Hierarchical clustering was performed using Euclidean distance and a complete linkage method. A Fisher’s exact test was used to evaluate difference in PgR status distribution in the two clusters identified by considering the first split in the hierarchical three.
All analyses were performed employing the R software (version 2.9.0).
Quantitative RT-PCR
The RNA expression of PgR was evaluated by quantitative RT-PCR on the same stored frozen tissue used for microarray analysis. Total RNA was reverse-transcribed using the High-Capacity cDNA Archive Kit and a TaqMan reaction was carried out in duplicate on an ABI PRISM 7700 machine, using Assays-on-Demand Gene Expression Product (Hs00172183_m1, Applied Biosystems, Foster City, CA). To normalize data, 18S ribosomal subunit was used as housekeeping (4319413E, Applied Biosystems, Foster City, CA, USA). Data analysis was performed using the Sequence Detector v1.9 software.
Results and discussion
Biological interpretation of differentially expressed genes
From the class comparison between MBC and FBC, we obtained 1273 clones differentially expressed at an FDR < 1%, corresponding to 920 distinct annotated genes.
Not surprisingly, the most significant gene was XIST, expressed exclusively from the X inactivation center of the inactive X chromosome.
Differentially expressed genes were subjected to Gene Ontology analysis using TopGO. Sixty-one terms of the Biological Process ontology, 20 of the Molecular Function and 16 of the Cellular Component ontology were found enriched at a significance level of P < 0.01 (Supplementary files 1–3).
Using the TopGo results as starting point, we identified a set of biological categories each involving a large number of differentially expressed genes, which were used to attempt an unbiased biological interpretation of the class comparison results. We report below a detailed description of the identified biological functions and of their role in cancer disease, highlighting key genes differentially expressed for each function. An overview of biological differences between genders is reported in Table 2. Instead, the complete lists of genes referred to a specific category are available as Supplementary file (Supplementary file 4). Genes mapping on the Y chromosome have not been considered.
Energy metabolism
A set of genes (e.g. CYCS, COX6C, NDUFs) coding for enzymes involved in different steps of mitochondrial oxidative phosphorylation (OXPHOS) were found down-regulated in FBC, suggesting a reduced OXPHOS compared to MBC. The observation of an impaired respiration and increased glycolysis in tumor cells dates to many decades ago [15] and is known as the Warburg’s hypothesis. An explanation on why tumor cells switch to a less efficient metabolism has been proposed [16], suggesting that malignant cells have important metabolic requirements that extend beyond ATP. The Warburg’s hypothesis was recently verified in breast cancer cells related to normal mammary cells [17], but interestingly our data suggest that this effect is at least reduced in MBC compared to FBC.
Strictly related to OXPHOS is ATP metabolism, a biological function that we also found modulated according to gender. In particular we noticed in MBC an increased expression of subunits of the mitochondrial ATP syntase complex and of the ATPIF1 gene, an inhibitor of the ATPase function of such a complex. Coherently with an increased availability of ATP, some ATPases were overexpressed, among them the vacuolar ATPase complex, which has important physiologic roles, but was also reported to be associated to tumor invasion as it is able to create a pH gradient across the membranes, mediating extracellular matrix acidification [18].
Furthermore, OXPHOS is the major source for ROS production, that is, therefore expected to be higher in MBC. It was not surprising to find 4 out of 6 human peroxiredoxins up-regulated in male tumors (PRDX1-4). Peroxiredoxins are antioxidant enzymes which reduce hydrogen peroxide and can indirectly affect cell proliferation: their role in cancer has been in fact extensively studied [19]. Similarly, the metabolism of glutathione, a potent reducing agent, seems to be affected. The GSH:GSSG ratio is critical for cell survival and influences S-glutathionylation, a chemical modification catalyzed by glutaredoxins, important for regulation of key cellular processes like the proteasomal protein degradation [20]. A rate limiting enzyme of glutathione synthesis (GCLC), a glutaredoxin (GLRX3), two glutathione peroxidase (GPX1, GPX2) and finally the glutathione reductase enzyme (GSR) were among differentially expressed genes in our data.
Translation
Protein synthesis is a fundamental process of eukaryotic cells and translation initiation represents a main regulatory step. Key molecules (e.g., EIF4 family members and poly(A) binding proteins) mediating this step were mostly found up-regulated in MBC. Particularly intriguing is EIF4E overexpression in MBC. This protein is frequently up-regulated in cancer and selectively enhances translation of key genes involved in tumorigenesis and cancer progression like MYC and BCL2 [21]. Consistently, these two genes are up-regulated in our data. EIF4E is a downstream effector of PI3K/AKT/mTOR mediated signals. Among signals acting on PI3K/AKT/mTOR pathway, there are many growth factors, but also the cell energy status, as a decreased AMP/ATP ratio promotes mTOR activity; mTOR itself, also overexpressed in male tumors, mediate phosphorylation of 4EBP, leading to EIF4E activation [22] (Fig. 1).
Regulation of translation initiation mediated by PI3K/AKT/mTOR pathway. The translation initiation factor EIF4E specifically promotes translation of cancer associated genes. Its activity is regulated by the PI3K/AKT/mTOR pathway, in turn regulated by AMP/ATP ratio. Genes in red were found up-regulated in MBC compared to FBC, and our data suggest a lower AMP/ATP ratio in MBC
Another result suggesting an increased protein synthesis in MBC is the upregulation of many ribosomal proteins of both the 40S (RPS8, RPS12, RPS16, RPS21) and 60S subunits (RPL18, RPL30, RPL35) and associated proteins.
Globally, an increased protein synthesis has been related to cell growth and proliferation, linking this process with malignancy [23]. As corollary, a group of DEAD-Box proteins (DDX3X, DDX5, DDX11, DDX12, DDX49, DDX55), involved in many aspects of RNA metabolism are also differentially expressed.
In eukaryotic cells, translation takes place also in mitochondria where 13 polypeptides that constitute the central core of the OXPHOS complex are translated [24]. Up-regulation of an elongation factor (GFM1) and 5 mitochondrial ribosomal proteins (MRPL14, MRPL23, MRPL30, MRPL34, MRPL42), indicates an enhanced translation of these mitochondrial genes in MBC, in agreement with what stated above.
Cytoskeleton and GTPase
Reorganization of the cytoskeleton is the primary mechanism of cell motility, is essential for most types of cell migration and is regulated by Rho family small GTPase. The tubulin/microtubule system is an important target for anticancer therapy with vinca alkaloids and taxanes, but also with newly developed drugs (e.g., epothilones [25] or eribulin [26]). Therefore, the observed gender divergence at level of cytoskeleton organization is particularly interesting. In fact, genes of both alfa (TUBA1A, TUBA3D, TUBA4A) and beta (TUBB, TUBB2C) tubulin families were up-regulated in MBC while two microtubule-associated proteins (MAP1B, MAP2), able to regulate the assembly–disassembly ratio, were down-regulated.
Actin reorganization is the underlying mechanism of cell migration both in physiological and malignant conditions. In MBC the actin polymerization process was altered by modulation of the ARP2/3 complex components and overexpression of the capping protein CAPZA1. This process is mainly regulated by Rho family small GTPase [27, 28]. RND2, a member of this family is more expressed in FBC. Two Rho GTPase activating proteins (SRGAP2, ARHGAP23) and a guanine nucleotide exchange factor, key regulators of Rho activity, are differentially expressed. Additionally, overexpression of TNK2 occurs in MBC. This gene is a tyrosine kinase that binds Cdc42 (a Rho family GTPase) and has been causally related to migration of cancer cells by enhancing cell surface localization of EGFR [29].
Rho GTPase are only one of 5 protein families composing the RAS GTPase superfamily: Rho, Ras, Rab, ARF, RAN family. Members belonging to all of these families showed a different expression pattern in MBC and FBC. In detail, we observed an increase in MBC of ARF (ARF6, ARL1, ARL8B) and RAB (RAB1A, RAB6A, RAB14, RAB22A, RAB31) GTPase as well as regulator molecules (respectively, ARFGEF1 and GDI2, RAB11FIP1). These GTPase are main factors in intracellular trafficking and their role in cancer is well documented as they are implicated in modulating growth factor receptor localization and supporting invadopodia and filopodia formation [30]. Specifically, ARF6, overexpressed in MBC, was shown to mediate MDA-MB-231 breast cancer cells invasive activities [31].
RAN and the regulatory RANBP1, also up-regulated in MBC, are instead involved in a wide range of cellular processes ranging from DNA synthesis and microtubules organization at mitosis to DNA and protein translocation through the nuclear pore [32]. Interestingly RAN is also an important AR coactivator as later discussed.
Finally also RAS family GTPase (RAP2C, RASL10A) and specific regulators of their activity were found transcriptionally altered comparing the two genders.
Tumor environment
Many recent studies are pointing out the role of the microenvironment during breast cancer development, growth and distant spreading. As a matter of fact, a prognostic role has been reported for some matrix metalloproteinases [33] and other ECM genes [34] as well as for stromal derived signatures [35].
Extracellular matrix is mainly constituted by two major components: the basement membrane and the interstitial matrix. Different types of collagen form a scaffold for both these components, laminins are the most abundant proteins of the basal membrane while among the proteins constituting the interstitial matrix it is worth to cite tenascin, proteoglycans and other glycoproteins. All these molecules strictly interact with a plethora of adhesion molecules expressed on cell surface, not only with a structural function but also to mediate signals regulating primary cell processes. Notably, genes belonging to all of cited extracellular matrix protein families and adhesion molecules as well were found as significantly both up- and down-regulated in our dataset. More in detail, various structural proteins like COL5A3, LAMC1, TNC, ECM2, FBN2, SPON1, THBS1 and the proteoglycans BGN, GPC3 as well as the UGDH, an enzyme which participate in glycosaminoglycan biosyntesis (e.g., hyaluronan) were up-regulated in MBC, although some were more expressed in FBC (FN1, MTN2, MGP, TMBS3).
Furthermore, extracellular matrix is not a static structure and is instead constantly remodeled by well regulated matrix protease [36]. MMPs (matrix metalloproteinases), ADAMTS (a disintegrin and metalloproteinase with trombospondin motifs), TIMPs (tissue inhibitors of metalloproteinases) along with the SERPIN superfamily of serine protease inhibitors constitute an important proteolitic axis. Many genes from the above families whose disregulation in breast cancer is well documented, were either up-regulated in MBC (MMP11, TIMP3, SERPINA5, SERPINA6, SERPINAB2, SPINT2) or in FBC (ADAMTS3, ADAMTS7, ADAMTSL4, MMP7).
Therefore, while it clearly appears that there are differences in the complex regulation of extracellular matrix remodeling, it is difficult to trace a gender-related model using only gene expression data, although it can be suggested that the gender differences at the level of the extracellular matrix may have a relevant impact on cell survival, growth and migration due to the central role of tissue remodeling in tumor biology.
The tumor microenvironment is also strongly affected by effectors of immune response. The myeloid and lymphoid tumor infiltrating cells has been reported for breast cancer with a frequency around 40% of tumors [37]. Moreover, chronic inflammation has recently been linked with an increased risk of distant metastases [38], while other studies suggest a protective role exerted by immune cells [39, 40].
Surprisingly, we found among differentially expressed genes many immune cells specific genes globally indicating a clear reduction of immune response in MBC. For instance, a numerous set of chemokines essentially produced during immune response (CCL25, CXCL1, CXCL9, CXCL10, IL23A, IL17B, IL32) were more expressed in female tumors. A similar pattern was observed for two members of the MHC II complex expressed by APC cells (HLA-DQB1, HLA-DRB1), a lymphocyte-specific protein tyrosine kinase (LCK), a member of the T-cell receptor complex (CD3D), a natural killer specific receptor (KLRK1) and a chitotriosidase secreted by activated macrophages (CHIT1). An exception to this is represented by the two B-cell specific genes found in our list (BCAP31, IGHG1) that are more expressed in male samples.
Membrane transport
Genes involved in membrane transport displayed differences between females and males. Two ATP-binding cassette transporters (ABCC2 and ABCC5) which have been linked to chemo-resistance in human cancer [41, 42] are more expressed in FBC. On the contrary, 21 members of the solute carrier family are either up- or down-regulated. This is, however, a highly heterogeneous family, including active transporters as well as proteins which facilitate transport, ion channels and aquaporins. Here a detailed analysis of single transporters would be useless, but it is worth to remember that they can regulate drug uptake as well as many cancer related processes [43]. For example changed expression of two glucose transporters (SLC2A1 and SLC2A3) could impact on glucose metabolism, consistently with what described about metabolic differences.
Growth factors response and apoptosis
Failure of inhibition of proliferation and overcome of apoptosis signals are crucial for cancer cell growth and survival. These two crucial aspects of breast cancer biology also appeared to be different according to the gender indirectly suggesting that male and female breast tumors adopt different survival strategies. For example, concerning growth signaling, ERBB2 and NRG1, a glycoprotein that enhances ERBB2 phosphorylation [44], GRB2, an EGFR interacting protein and MET, a growth factor receptor with a well established role in breast cancer, [45] are down-regulated in MBC. On the opposite, FGF receptor 2 is more expressed in MBC.
Focusing on apoptosis related genes, we found differentially expressed genes belonging to several apoptotic pathways: BCL2 with its interacting proteins BAG4 and BNIP3L, Fas-related proteins (FAIM3, FASTK), TNF mediated apoptosis genes (TNFRSF21, TNFRSF25, TNFSF13, TRADD). BCL2 was overexpressed in MBC, according to data reported elsewhere by IHC [8], supporting the role of this antiapoptotic gene in carcinogenesis.
Response to steroid stimulus
As expected, genes related to response to steroid hormones were quite different in MBC and FBC. Some genes coding for proteins functionally related to the AR as RAN, a GTPase able to enhance AR transactivation [46], RNF14, a coactivator interacting with AR signaling in prostate cancer [47] and PAK6, an AR and ER signaling inhibitor [48, 49] were overexpressed in MBC.
Correlation and clustering analysis
To further highlight similarities and differences between breast cancers in the two genders, we performed a correlation analysis as described. Given the well established role of steroid hormones in breast cancer, we separately searched in male and female breast tumors profiles for genes significantly correlated with AR expression values obtained from array data and PgR expression value obtained by qPCR. The same type of analysis was carried out for the ERBB2 gene, usually amplified in about 20% of FBC with a strong impact on prognosis and therapeutic strategy.
From the number of correlated genes and the overlap between genes correlated in the two genders, we could indirectly extrapolate some hints on how relevant and how similar the biological functions of these three genes were in the two genders. As shown in Table 3, huge differences were found in the numbers of receptor-correlated genes and few of them were common in the two genders.
2288 clones were significantly correlated with ERBB2 in the female samples dataset and only 243 were so in the male dataset with 45 common genes. This strongly suggests a reduced relevance of ERBB2 in MBC biology compared to FBC. Moreover, 12 out of the 45 common clones map near the ERBB2 locus (Table S1), with increasing correlation values for genes closer to ERBB2, in both genders (Fig. 2). Hence, similarly to what happen in females, also in male breast tumors ERBB2 overexpression is the consequence of a genomic amplification of its locus, according to literature results of FISH analysis in MBC samples [50, 51].
Correlation pattern of ERBB2 neighbor genes. Genes mapping near the ERBB2 locus on chromosome 17 are reported according to their correlation with ERBB2 in FBC (a) and MBC (b) samples. Genomic coordinates (Mb) are reported on x-axis. Correlation values increase for genes closer to ERBB2, in both MBC and FBC samples
An inverse behavior was observed for AR correlated genes, where only 86 clones were correlated to AR in FBC, against 441 clones in MBC. Interestingly, two genes up-regulated in MBC, BCL2 and GATA3, which are known to be associated to ER in FBC, were slightly inversely correlated with AR in FBC (S = −0.25, P = 0.1486 and S = −0.22, P = 0.1374, respectively), but significantly correlated to AR expression in MBC (S = 0.43, P = 0.0194 and S = 0.41, P = 0.0347).
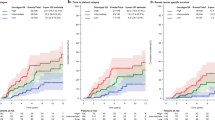
More PgR correlated genes were found in FBC (582) than in MBC (159) with a limited overlap (26 genes). Thus the PgR gene, more expressed in FBC (P = 0.0006), also seems to play a more important role in these tumors rather than in MBC. This hypothesis was further confirmed by a clustering analysis exploiting independent public data by Chanrion et al. [52]. From this dataset 145 ER+ FBC samples with clinico-pathological features comparable to our FBC samples were isolated. About 4000 clones (common to our data) were searched for genes differentially expressed between PgR+ and PgR− tumors. Fourteen clones were called as differentially expressed at FDR = 0. As expected, hierarchical clustering of Chanrion’s patients using this gene set revealed two main clusters significantly associated to PgR status (P = 0.00001, Fig. 3a). The same clustering in our FBC samples also identified two main clusters significantly associated to PgR status (P = 0.002, Fig. 3b). Whereas, clustering of MBC samples identified two clusters not at all associated to PgR status (P = 0.413, Fig. 3c). This finding further supports the concept that PgR, similarly to ER, AR, and ERBB2, may play a different role in MBC and FBC. The inverse approach searching for differentially expressed genes between PgR+ and PgR− in MBC was also performed. Hierarchical clustering with the 74 clones differentially expressed (FDR < 5%) identified a sub-cluster containing 7 out of 8 PgR− tumors, while a quite random distribution of PgR− samples was observed both in our FBC dataset and in the Chanrion FBC dataset (Supplementary Fig. S1).
Clustering analysis with PgR associated genes. Using a set of clones differentially expressed between PgR+ and PgR− FBC samples in the Chanrion dataset [52], a hierarchical clustering analysis was performed on the Chanrion dataset itself (a) as well as on our dataset, separately for FBC (b) and MBC (c) samples. Significant associations between the two main clusters (red and blue) and the distribution of PgR status were observed in FBC datasets but not in the MBC dataset
Conclusion
MBC is a rare disease poorly characterized and treated similarly to female counterpart. A few studies, reported a different expression in male samples of validated breast cancer biomarkers, but a comprehensive molecular characterization of these tumors is missing. At the best of our knowledge, our study is the first reporting gene expression data of MBC specimens compared to samples with similar clinical and patho-biological features of the female counterpart. The aim of the study was to give a genome wide description of these two tumors focusing on main biological relevant discrepancies in order to generate useful hypothesis to better understand MBC biology and to provide a guide for treatment strategies.
Class comparison uncovered profound differences between MBC and FBC transcriptomes, with around 1000 genes called as differentially expressed despite a stringent FDR. Biological interpretation of our results clearly pointed out the relevance of such a difference, as almost all processes we found to be differentially modulated, have a clear and well-known role in tumorigenesis and cancer progression. Furthermore in some cases, the observed differences might have strong implications on treatment choice and outcome. For example, inhibition of translation targeting either EIF4E or mTOR has been proposed as a promising new therapeutic strategy [21, 22] and an increased susceptibility to such treatment in MBC could be hypothesized based on our data. Our molecular data also suggest that drugs targeting tubulin/microtubules system like the vinca alkaloids and taxanes might also be more effective in MBC due to their overexpression of various tubulin subunits compared to FBC. It might also be hypothesized that MBC could be a preferred target for the newly developed epothilones [25].
An interesting difference appeared for ERBB2. In female patients amplification of ERBB2 characterizes a subgroup of tumors with a peculiar behavior in terms of aggressiveness and which is treated in different way from tumors without this amplification [53] The strong biological role played by ERBB2 in FBC is reflected by the high number of correlated genes. In MBC a tenfold lower number of genes was found to be correlated with ERBB2, suggesting a minor role for this gene in males and raising doubts about usefulness of treatment strategies targeting ERBB2 in males.
According to our data, the AR could instead be one of the driving genes in MBC biology as also suggested in previous studies [8]. Thus, a shift towards hormonal therapy targeting AR rather than ER could be envisioned.
The metabolic features of MBC, where the anaerobic glycolysis pathway does not seem to be the preferential glucose metabolic pathway as for most tumors, is also worth to be further investigated. It has biological implication which may be linked with tumorigenesis, as strong activation of oxidative phosphorylation is responsible for production of ROS, but also represent a warning when using diagnostic procedures as positron emission tomography which relay on 18F-deoxyglucose uptake.
Research recommendations on MBC collected in a specific multidisciplinary meeting were recently published [54] and strongly underline that MBC is a unique disease rather than an illness similar to postmenopausal FBC. As the majority of MBC are endocrine responsive the development of endocrine therapy options designed for MBC in specific clinical trials was considered as a priority. However, while waiting for results from large international multicenter studies to learn more on this disease, treatment cannot be simply derived from FBC, but should rely on single center studies like ours which highlight peculiar features of MBC. In this sense our results which suggest a different role of steroid receptors (and ERBB2) in males are a warning in the design of hormonal treatment in MBC.
In conclusion, our data highlighted the need to investigate the link between molecular characterization and outcome in MBC as done for FBC, in order to personalize the therapy and improve patient survival. These preliminary data may trace a path for future studies, however, due to the rarity of MBC, studies with a multicenter setting with a standardized biospecimens handling are essential.
References
Contractor KB, Kaur K, Rodrigues GS, Kulkarni DM, Singhal H (2008) Male breast cancer: is the scenario changing. World J Surg Oncol 6:58
Nahleh Z, Girnius S (2006) Male breast cancer: a gender issue. Nat Clin Pract Oncol 3:428–437
Willsher PC, Leach IH, Ellis IO, Bourke JB, Blamey RW, Robertson JF (1997) A comparison outcome of male breast cancer with female breast cancer. Am J Surg 173:185–188
Goss PE, Reid C, Pintilie M, Lim R, Miller N (1999) Male breast carcinoma: a review of 229 patients who presented to the Princess Margaret Hospital during 40 years: 1955–1996. Cancer 85:629–639
Munoz de Toro MM, Maffini MV, Kass L, Luque EH (1998) Proliferative activity and steroid hormone receptor status in male breast carcinoma. J Steroid Biochem Mol Biol 67:333–339
Curigliano G, Colleoni M, Renne G, Mazzarol G, Gennari R, Peruzzotti G, de Braud E, Robertson C, Maiorano E, Veronesi P, Nole F, Mandala M, Ferretti G, Viale G, Goldhirsch A (2002) Recognizing features that are dissimilar in male and female breast cancer: expression of p21Waf1 and p27Kip1 using an immunohistochemical assay. Ann Oncol 13:895–902
Bloom KJ, Govil H, Gattuso P, Reddy V, Francescatti D (2001) Status of HER-2 in male and female breast carcinoma. Am J Surg 182:389–392
Weber-Chappuis K, Bieri-Burger S, Hurlimann J (1996) Comparison of prognostic markers detected by immunohistochemistry in male and female breast carcinomas. Eur J Cancer 32A:1686–1692
Sotiriou C, Pusztai L (2009) Gene-expression signatures in breast cancer. N Engl J Med 360:790–800
Fassan M, Baffa R, Palazzo JP, Lloyd J, Crosariol M, Liu CG, Volinia S, Alder H, Rugge M, Croce CM, Rosenberg A (2009) MicroRNA expression profiling of male breast cancer. Breast Cancer Res 11:R58
Ronchi E, Granata G, Brivio M, Coradini D, Miodini P, Di Fronzo G (1986) A double-labeling assay for simultaneous estimation and characterization of estrogen and progesterone receptors using radioiodinated estradiol and tritiated Org 2058. Tumori 72:251–257
Cappelletti V, Gariboldi M, De Cecco L, Toffanin S, Reid JF, Lusa L, Bajetta E, Celio L, Greco M, Fabbri A, Pierotti MA, Daidone MG (2008) Patterns and changes in gene expression following neo-adjuvant anti-estrogen treatment in estrogen receptor-positive breast cancer. Endocr Relat Cancer 15:439–449
Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA 98:5116–5121
Alexa A, Rahnenfuhrer J, Lengauer T (2006) Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22:1600–1607
Warburg O (1956) On respiratory impairment in cancer cells. Science 124:269–270
Vander Heiden MG, Cantley LC, Thompson CB (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324:1029–1033
Putignani L, Raffa S, Pescosolido R, Aimati L, Signore F, Torrisi MR, Grammatico P (2008) Alteration of expression levels of the oxidative phosphorylation system (OXPHOS) in breast cancer cell mitochondria. Breast Cancer Res Treat 110:439–452
Sennoune SR, Luo D, Martinez-Zaguilan R (2004) Plasmalemmal vacuolar-type H+-ATPase in cancer biology. Cell Biochem Biophys 40:185–206
Neumann CA, Fang Q (2007) Are peroxiredoxins tumor suppressors? Curr Opin Pharmacol 7:375–380
Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD (2008) Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal 10:1941–1988
Graff JR, Konicek BW, Carter JH, Marcusson EG (2008) Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res 68:631–634
Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N (2006) mTOR, translation initiation and cancer. Oncogene 25:6416–6422
Caraglia M, Budillon A, Vitale G, Lupoli G, Tagliaferri P, Abbruzzese A (2000) Modulation of molecular mechanisms involved in protein synthesis machinery as a new tool for the control of cell proliferation. Eur J Biochem 267:3919–3936
Perez-Martinez X, Funes S, Camacho-Villasana Y, Marjavaara S, Tavares-Carreon F, Shingu-Vazquez M (2008) Protein synthesis and assembly in mitochondrial disorders. Curr Top Med Chem 8:1335–1350
Hunt JT (2009) Discovery of ixabepilone. Mol Cancer Ther 8:275–281
Cigler T, Vahdat LT (2010) Eribulin mesylate for the treatment of breast cancer. Expert Opin Pharmacother 11:1587–1593
Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420:629–635
Vega FM, Ridley AJ (2008) Rho GTPases in cancer cell biology. FEBS Lett 582:2093–2101
Howlin J, Rosenkvist J, Andersson T (2008) TNK2 preserves epidermal growth factor receptor expression on the cell surface and enhances migration and invasion of human breast cancer cells. Breast Cancer Res 10:R36
Chia WJ, Tang BL (2009) Emerging roles for Rab family GTPases in human cancer. Biochim Biophys Acta 1795:110–116
Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, Sabe H (2004) Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci USA 101:6647–6652
Rensen WM, Mangiacasale R, Ciciarello M, Lavia P (2008) The GTPase Ran: regulation of cell life and potential roles in cell transformation. Front Biosci 13:4097–4121
Kohrmann A, Kammerer U, Kapp M, Dietl J, Anacker J (2009) Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: new findings and review of the literature. BMC Cancer 9:188
Helleman J, Jansen MP, Ruigrok-Ritstier K, van Staveren IL, Look MP, Meijer-van Gelder ME, Sieuwerts AM, Klijn JG, Sleijfer S, Foekens JA, Berns EM (2008) Association of an extracellular matrix gene cluster with breast cancer prognosis and endocrine therapy response. Clin Cancer Res 14:5555–5564
Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M (2008) Stromal gene expression predicts clinical outcome in breast cancer. Nat Med 14:518–527
Bosman FT, Stamenkovic I (2003) Functional structure and composition of the extracellular matrix. J Pathol 200:423–428
Coronella-Wood JA, Hersh EM (2003) Naturally occurring B-cell responses to breast cancer. Cancer Immunol Immunother 52:715–738
de Visser KE, Eichten A, Coussens LM (2006) Paradoxical roles of the immune system during cancer development. Nat Rev Cancer 6:24–37
Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M (2008) The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res 68:5405–5413
Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D, Engels K, Karn T, Kaufmann M (2009) T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res 11:R15
Materna V, Stege A, Surowiak P, Priebsch A, Lage H (2006) RNA interference-triggered reversal of ABCC2-dependent cisplatin resistance in human cancer cells. Biochem Biophys Res Commun 348:153–157
Weaver DA, Crawford EL, Warner KA, Elkhairi F, Khuder SA, Willey JC (2005) ABCC5, ERCC2, XPA and XRCC1 transcript abundance levels correlate with cisplatin chemoresistance in non-small cell lung cancer cell lines. Mol Cancer 4:18
He L, Vasiliou K, Nebert DW (2009) Analysis and update of the human solute carrier (SLC) gene superfamily. Hum Genomics 3:195–206
Britsch S (2007) The neuregulin-I/ErbB signaling system in development and disease. Adv Anat Embryol Cell Biol 190:1–65
Lengyel E, Prechtel D, Resau JH, Gauger K, Welk A, Lindemann K, Salanti G, Richter T, Knudsen B, Vande Woude GF, Harbeck N (2005) C-Met overexpression in node-positive breast cancer identifies patients with poor clinical outcome independent of Her2/neu. Int J Cancer 113:678–682
Harada N, Ohmori Y, Yamaji R, Higashimura Y, Okamoto K, Isohashi F, Nakano Y, Inui H (2008) ARA24/Ran enhances the androgen-dependent NH2- and COOH-terminal interaction of the androgen receptor. Biochem Biophys Res Commun 373:373–377
Kang HY, Yeh S, Fujimoto N, Chang C (1999) Cloning and characterization of human prostate coactivator ARA54, a novel protein that associates with the androgen receptor. J Biol Chem 274:8570–8576
Lee SR, Ramos SM, Ko A, Masiello D, Swanson KD, Lu ML, Balk SP (2002) AR and ER interaction with a p21-activated kinase (PAK6). Mol Endocrinol 16:85–99
Schrantz N, da Silva Correia J, Fowler B, Ge Q, Sun Z, Bokoch GM (2004) Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J Biol Chem 279:1922–1931
Rudlowski C, Friedrichs N, Faridi A, Fuzesi L, Moll R, Bastert G, Rath W, Buttner R (2004) Her-2/neu gene amplification and protein expression in primary male breast cancer. Breast Cancer Res Treat 84:215–223
Fonseca RR, Tomas AR, Andre S, Soares J (2006) Evaluation of ERBB2 gene status and chromosome 17 anomalies in male breast cancer. Am J Surg Pathol 30:1292–1298
Chanrion M, Negre V, Fontaine H, Salvetat N, Bibeau F, Mac Grogan G, Mauriac L, Katsaros D, Molina F, Theillet C, Darbon JM (2008) A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clin Cancer Res 14:1744–1752
Ross JS, Slodkowska EA, Symmans WF, Pusztai L, Ravdin PM, Hortobagyi GN (2009) The HER-2 receptor and breast cancer: ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist 14:320–368
Korde LA, Zujewski JA, Kamin L, Giordano S, Domchek S, Anderson WF, Bartlett JMS, Gelmon K, Nahleh Z, Bergh J, Cutuli B, Pruneri G, McCaskill-Stevens W, Gralow J, Hortobagyi G, Cardoso F (2010) Multidisciplinary meeting on male breast cancer: summary and research recommendations. J Clin Oncol 28:2114–2122
Acknowledgments
Supported in part by Associazione Italiana per la Ricerca sul Cancro (PIs, MA Pierotti and MG Daidone), Progetto Integrato Oncologia from the Italian Health Ministry (PI, MG Daidone), Alleanza Contro il Cancro (PI, MG Daidone).
Conflict of interest statement
The authors declared that they have no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
10549_2010_1015_MOESM6_ESM.jpg
Clustering analysis with PgR associated genes. Using a set of clones differentially expressed between PgR+ and PgR− MBC samples, a hierarchical clustering analysis was performed on the MBC dataset itself (A) as well as on our FBC dataset (B) and on the Chanrion FBC dataset (C) samples. A sub-cluster containing 7/8 PgR− tumors was noticed in the clustering of MBC samples, while a quite random distribution of PgR− tumors was observed in FBC datasets (JPG 111 kb)
Rights and permissions
About this article
Cite this article
Callari, M., Cappelletti, V., De Cecco, L. et al. Gene expression analysis reveals a different transcriptomic landscape in female and male breast cancer. Breast Cancer Res Treat 127, 601–610 (2011). https://doi.org/10.1007/s10549-010-1015-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-1015-8