Abstract
Epidemiological studies on the association between SULT1A1 codon 213 polymorphism and breast cancer risk are inconclusive. In order to derive a more precise estimation of the association, a meta-analysis was conducted in this article. Sixteen studies including 9,881 cases and 13,564 controls were collected for SULT1A1 codon 213 polymorphism by searching the databases of Medline, PubMed, Embase, and ISI Web of Knowledge. The strength of association between SULT1A1 codon 213 polymorphism and breast cancer susceptibility was assessed by calculating crude ORs with 95% CIs. When all the 21 studies were pooled into the meta-analysis, there was no evidence for significant association between SULT1A1 codon 213 polymorphism and breast cancer susceptibility (for Arg/Arg versus Arg/His: OR = 0.999, 95% CI = 0.941–1.061; for Arg/Arg versus His/His: OR = 1.121, 95% CI = 1.013–1.242; for dominant model: OR = 1.128, 95% CI = 1.01–1.26; for recessive model: OR = 1.151, 95% CI = 0.950–1.394). In the subgroup analysis by the source of controls, significant increased risk was found for hospital-based studies (for Arg/Arg versus Arg/His: OR = 1.173, 95% CI = 1.000–1.376; for Arg/Arg versus His/His: OR = 1.600, 95% CI = 1.134–2.256; for dominant model: OR = 1.269, 95% CI = 1.134–2.256; for recessive model: OR = 1.664, 95% CI = 1.070–2.588). In summary, the meta-analysis suggests that SULT1A1 codon 213 polymorphism may be associated with the hospital-based studies. However, large number of samples and representative hospital-based studies with homogeneous breast cancer patients and well-matched controls are warranted to confirm this finding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer is the most common cancer in women: about 1.15 million new cases with 41,000 deaths were reported in 2002 [1]. In the United States, an estimated 203,500 new cases will be diagnosed and approximately 39,600 women will die from breast cancer [2]. The SULT1A1 gene that encodes the SULT1A1 enzyme has been located on chromosome 16p12.1–p11.2 [3], which has a common functional polymorphism, located in the coding region (638 G to A), resulting in a substitution of histidine for arginine (Arg213His) [4–6]. SULT1A1 has also been shown to be highly expressed in breast cancer cell lines [7], which was associated with a decreased risk of recurrence or improved prognosis of breast cancer [8]. The sulfonation of estrogens was catalyzed by SULT1A1 to form water-soluble and biologically inactive estrogen sulfates, which reducing the level of estrogen exposure in their target tissues [9, 10]. Previous functional studies have demonstrated that the variant A allele is associated with significantly reduced sulfotransferase activity in platelets compared with the wild-type G allele [5, 6] The relevant SULT1A1 Arg213His single-nucleotide polymorphism studies have shown that 213His allele is linked to cancer susceptibility in several cancers [11–15]. The specific role of SULT1A1 codon 213 polymorphism in breast cancer susceptibility has been the research focus of numerous researches [11, 16–30]. However, the results of these studies have also been inconclusive, partially maybe the possible small effect of the polymorphism on breast cancer risk and the relatively small sample size in each of the published studies. Therefore, we performed this meta-analysis to derive a more precise estimation of the association between the SULT1A1 codon 213 polymorphism and the susceptibility for developing breast cancer.
Methods
Publication search
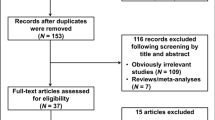
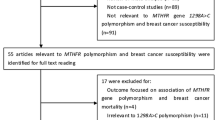
Medline, PubMed, Embase, and Web of Science (updated to April 20, 2010) were searched using the following search terms: “SULT1A1,” “Codon 213,” “polymorphism,” or “genotype” and “breast.” To search as many articles as possible, the relevant publications’ references were carefully evaluated. Only those published studies with full-text articles in English and Chinese were included in this meta-analysis. For overlapping and republished studies, only the first published or the study with the largest samples was included.
Selection criteria
The selection criteria were (1) evaluation the polymorphism of SULT1A1 Codon 213 and breast cancer risk, (2) case–control studies, (3) with sufficient published data to estimate an odds ratio (OR) with 95% confidence interval (CI), (4) written in English and Chinese language, (5) executing Hardy–Weinberg equilibrium in the control group (P < 0.01). The selected articles should meet all of the above criteria.
Data extraction
The relevant information was extracted from all selected articles independently by two of the authors (Yanlei Ma and Jianjun Yang) according to the above-mentioned inclusion criteria. The following data were extracted from each study: first author’s name, year of publication, countries which human samples came from, ethnicity, source of controls, total number of cases and controls, and numbers of cases and controls with the SULT1A1 Codon 213 polymorphism, respectively. Different ethnicities were categorized as Caucasian, Asian, and mixed. Study design was stratified to population-based studies, hospital-based studies. Any minimum number of patients to include was not defined in the current meta-analysis.
Statistical methods
Crude ORs with 95% CIs were used to assess the strength of association between the polymorphism of SULT1A1 Codon 213 and breast cancer risk. The pooled ORs were performed for co-dominant model (Arg/Arg versus Arg/His, Arg/Arg versus His/His), dominant model (His/Arg + His/His versus Arg/Arg), and recessive model (His/His versus His/Arg + Arg/Arg), respectively. Heterogeneity assumption was checked by the chi-square-based Q-test [31]. A P-value greater than 0.10 for the Q-test demonstrates a lack of heterogeneity among studies, so the pooled OR estimate of each study was calculated through the fixed-effects model (the Mantel–Haenszel method) [32]. Otherwise, the random-effects model (the DerSimonian and Laird method) was used [33]. Sensitivity analysis was performed to assess the stability of the results. Subgroup analysis was performed by ethnicity and study design. Each study involved in the meta-analysis was deleted each time to reflect the influence of the individual dataset to the pooled ORs [34]. An estimate of potential publication bias was executed by the funnel plot, where the standard error of log (OR) of each study was plotted against its log (OR). The significance of the intercept was determined by the t-test suggested by Egger (P < 0.05 was considered the representative of statistically significant publication bias) [35]. All the statistical tests were performed with STATA version 10.0 (Stata Corporation, College Station, TX, USA).
Results
Study characteristics
Our database search generated a total of 16 publications that met the inclusion criteria. In one of these publications, ORs were presented separately for each subgroup, so for all publications we considered each group separately for subgroup analysis. Hence, a total of 16 studies including 9,881 cases and 13,564 controls were involved in this meta-analysis. Table 1 lists the characteristics of each study. There were seven studies of Caucasians, seven of Asians, one study of mixed populations, and one study of not reported populations. Almost all cases were pathologically confirmed. Controls were primarily healthy individuals that were matched for age. Among these 16 studies, 8 were population-based, 7 were hospital-based, and 1 was mixed population.
Main results
When all the studies were pooled, no significant associations were found between the Arg213His polymorphism and breast cancer risk (His/Arg versus Arg/Arg: OR = 0.999, 95% CI = 0.941–1.061, P = 0.001 for heterogeneity; His/His versus Arg/Arg: OR = 1.121, 95% CI = 1.013–1.242, P = 0.012 for heterogeneity; His/Arg + His/His versus Arg/Arg: OR = 1.128, 95% CI = 1.01–1.26, P = 0.001 for heterogeneity; His/His versus His/Arg + Arg/Arg: OR = 1.151, 95% CI = 0.950–1.394, P = 0.012 for heterogeneity). However, in the stratified analysis by control sources, significant associations were observed in HB (His/Arg versus Arg/Arg: OR = 1.173, 95% CI = 1.000–1.376, P = 0.181 for heterogeneity; His/His versus Arg/Arg: OR = 1.600, 95% CI = 1.134–2.256, P = 0.125 for heterogeneity; His/Arg + His/His versus Arg/Arg: OR = 1.269, 95% CI = 1.036–1.555, P = 0.112 for heterogeneity; His/His versus His/Arg + Arg/Arg: OR = 1.664, 95% CI = 1.070–2.588, P = 0.194 for heterogeneity). To the contrary, no significant associations were detected in PB. Otherwise, in the stratified analysis by ethnicity, no significant associations were observed for all comparison models (Table 2).
Sensitivity analysis and publication bias
In overall studies, the results suggested that no significant influence of any of the individual dataset to the pooled OR values was observed. Begg’s funnel plot and Egger’s test similarly failed to reveal evidence of publication bias (data not shown).
Discussion
Although a number of recent studies [11, 16–31] have reported a significant association between the Arg213His polymorphism and breast cancer risk, others have found no such association. In order to resolve this conflict, we initiated this meta-analysis of 16 studies, involving 9,881 cases and 13,564 controls, so as to derive a more precise estimation of the exist or absence of this association.
We found that polymorphism in Arg213His of the SULT1A1 gene displayed no significant association with breast cancer risk, either when the included study populations were pooled or when they were subjected to a stratified analysis according to ethnicity. The latter result suggests that differences in genetic background and living environment do not influence any potential association between the Arg213His polymorphism and breast cancer risk. However, the current results indicated that significantly increased breast cancer risk in Arg213His of the SULT1A1 were found for the hospital-based studies but not in population-based studies. Therefore, using a proper and representative hospital-based control subjects is very important to reduce biases in such genetic association studies.
Two advantages of our study were the large number of samples included and our failure to find a significant association in any of the genetic models tested. However, some limitations were acknowledged. First of all, the controls in the included studies were not uniformly determinated, such that the control subjects in different studies might have had varying risks of developing breast cancer. Secondly, the results presented here are based on unadjusted estimates. A more precise analysis could be guided if more detailed individual data were available which allowed it to be adjusted according other covariates such as age, smoking and drinking status, menopausal status, lifestyle, basal metabolic index, obesity, and environmental factors.
In conclusion, this meta-analysis suggests that SULT1A1 Codon 213 polymorphism may be associated with the hospital-based studies. However, large number of samples and representative hospital-based studies with homogeneous breast cancer patients and well-matched controls are warranted to confirm this finding. Future studies should extend this investigation by incorporating other potential risk factors for breast cancer.
References
Parkin DM, Bray F, Ferlay J, Pisani P (2005) Global cancer statistics, 2002. CA Cancer J Clin 55:74–108
ACS (American Cancer Society): Cancer Facts and Figures. American Cancer Society, 2002
Carlini EJ, Raftogianis RB, Wood TC, Jin F, Zheng W, Rebbeck TR, Weinshilboum RM (2001) Sulfation pharmacogenetics: SULT1A1 and SULT1A2 allele frequencies in Caucasian, Chinese and African-American subjects. Pharmacogenetics 11:57–68
Nowell S, Ambrosone CB, Ozawa S, MacLeod SL, Mrackova G, Williams S, Plaxco J, Kadlubar FF, Lang NP (2000) Relationship of phenol sulfotransferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenetics 10:789–797
Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM (1997) Phenol sulfotransferase pharmacogenetics in humans: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun 239:298–304
Ozawa S, Tang YM, Yamazoe Y, Kato R, Lang NP, Kadlubar FF (1998) Genetic polymorphisms in human liver phenol sulfotransferases involved in the bioactivation of N-hydroxy derivatives of carcinogenic arylamines and heterocyclic amines. Chem Biol Interact 109:237–248
Falany JL, Falany CN (1996) Expression of cytosolic sulfotransferases in normal mammary epithelial cells and breast cancer cell lines. Cancer Res 56:1551–1555
Suzuki T, Nakata T, Miki Y, Kaneko C, Moriya T, Ishida T, Akinaga S, Hirakawa H, Kimura M, Sasano H (2003) Estrogen sulfotransferase and steroid sulfatase in human breast carcinoma. Cancer Res 63:2762–2770
Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB (1997) Sulfation and sulfotransferases 1: sulfotransferase molecular biology: cDNAs and genes. FASEB J 11:3–14
Falany CN (1997) Enzymology of human cytosolic sulfotransferases. FASEB J 11:206–216
Choi JY, Lee KM, Park SK, Noh DY, Ahn SH, Chung HW, Han W, Kim JS, Shin SG, Jang IJ, Yoo KY, Hirvonen A, Kang D (2005) Genetic polymorphisms of SULT1A1 and SULT1E1 and the risk and survival of breast cancer. Cancer Epidemiol Biomarkers Prev 14:1090–1095
Pachouri SS, Sobti RC, Kaur P, Singh J, Gupta SK (2006) Impact of polymorphism in sulfotransferase gene on the risk of lung cancer. Cancer Genet Cytogenet 171:39–43
Lilla C, Risch A, Verla-Tebit E, Hoffmeister M, Brenner H, Chang-Claude J (2007) SULT1A1 genotype and susceptibility to colorectal cancer. Int J Cancer 120:201–206
Boccia S, Cadoni G, La Torre G, Arzani D, Volante M, Cattel C, Gianfagna F, Paludetti G, Almadori G, Ricciardi G (2006) A case-control study investigating the role of sulfotransferase 1A1 polymorphism in head and neck cancer. J Cancer Res Clin Oncol 132:466–472
Boccia S, Persiani R, La Torre G, Rausei S, Arzani D, Gianfagna F, Romano-Spica V, D’Ugo D, Ricciardi G (2005) Sulfotransferase 1A1 polymorphism and gastric cancer risk: a pilot case-control study. Cancer Lett 229:235–243
Syamala VS, Syamala V, Sheeja VR, Kuttan R, Balakrishnan R, Ankathil R (2010) Possible risk modification by polymorphisms of estrogen metabolizing genes in familial breast cancer susceptibility in an Indian population. Cancer Invest 28:304–311
Chang-Claude J, Beckmann L, Corson C, Hein R, Kropp S, Parthimos M, Dünnebier T, Hamann U, Brors B, Eils R, Zapatka M, Brauch H, Justenhoven C, Flesch-Janys D, Braendle W, Brüning T, Pesch B, Spickenheuer A, Krankenhaus J, Ko YD, Baisch C, Dahmen N, Brauch H, Chang-Claude J, Corson C, Dünnebier T, Hein R, Justenhoven C, Parthimos M, Zapatka M (2010) Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Res Treat 119:463–474
Gulyaeva LF, Mikhailova ON, PustyInyak VO, Kim IV 4th, Gerasimov AV, Krasilnikov SE, Filipenko ML, Pechkovsky EV (2008) Comparative analysis of SNP in estrogen-metabolizing enzymes for ovarian, endometrial, and breast cancers in Novosibirsk, Russia. Adv Exp Med Biol 617:359–366
Hu MB, Xie W, Xiong B, Han DF, Li Y, Feng MH, Zhou YF (2006) Study on the relationship between polymorphisms of genes (CYP17, CYP19 and SULT1A1) and susceptibility to breast cancer in Chinese women. Zhonghua Liu Xing Bing Xue Za Zhi 27:351–355
Cheng TC, Chen ST, Huang CS, Fu YP, Yu JC, Cheng CW, Wu PE, Shen CY (2005) Breast cancer risk associated with genotype polymorphism of the catechol estrogen-metabolizing genes: a multigenic study on cancer susceptibility. Int J Cancer 113:345–353
Jerevall PL, Ahmadi A, Bergman M, Stål O, Wingren S (2005) Sulfotransferase1A1 and risk of postmenopausal breast cancer. Anticancer Res 25:2515–2517
Le Marchand L, Donlon T, Kolonel LN, Henderson BE, Wilkens LR (2005) Estrogen metabolism-related genes and breast cancer risk: the multiethnic cohort study. Cancer Epidemiol Biomarkers Prev 14:1998–2003
Lilla C, Risch A, Kropp S, Chang-Claude J (2005) SULT1A1 genotype, active and passive smoking, and breast cancer risk by age 50 years in a German case-control study. Breast Cancer Res 7:R229–R237
Yang G, Gao YT, Cai QY, Shu XO, Cheng JR, Zheng W (2005) Modifying effects of sulfotransferase 1A1 gene polymorphism on the association of breast cancer risk with body mass index or endogenous steroid hormones. Breast Cancer Res Treat 94:63–70
Chacko P, Rajan B, Mathew BS, Joseph T, Pillai MR (2004) CYP17 and SULT1A1 gene polymorphisms in Indian breast cancer. Breast Cancer 11:380–388
Han DF, Zhou X, Hu MB, Wang CH, Xie W, Tan XD, Zheng F, Liu F (2004) Sulfotransferase 1A1 (SULT1A1) polymorphism and breast cancer risk in Chinese women. Toxicol Lett 150:167–177
Langsenlehner U, Krippl P, Renner W, Yazdani-Biuki B, Eder T, Wolf G, Wascher TC, Paulweber B, Weitzer W, Samonigg H (2004) Genetic variants of the sulfotransferase 1A1 and breast cancer risk. Breast Cancer Res Treat 87:19–22
Tang D, Rundle A, Mooney L, Cho S, Schnabel F, Estabrook A, Kelly A, Levine R, Hibshoosh H, Perera F (2003) Sulfotransferase 1A1 (SULT1A1) polymorphism, PAH-DNA adduct levels in breast tissue and breast cancer risk in a case-control study. Breast Cancer Res Treat 78:217–222
Zheng W, Xie D, Cerhan JR, Sellers TA, Wen W, Folsom AR (2001) Sulfotransferase 1A1 polymorphism, endogenous estrogen exposure, well-done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 10:89–94
Seth P, Lunetta KL, Bell DW, Gray H, Nasser SM, Rhei E, Kaelin CM, Iglehart DJ, Marks JR, Garber JE, Haber DA, Polyak K (2000) Phenol sulfotransferases: hormonal regulation, polymorphism, and age of onset of breast cancer. Cancer Res 60:6859–6863
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Tobias A (1999) Assessing the influence of a single study in the meta-analysis estimate. Stata Tech Bull 8:15–17
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Qiu LX, Yao L, Mao C, Yu KD, Zhan P, Chen B, Yuan H, Zhang J, Xue K, Hu XC (2010) Lack of association of CYP1A2-164 A/C polymorphism with breast cancer susceptibility: a meta-analysis involving 17,600 subjects. Breast Cancer Res Treat. doi:10.1007/s10549-009-0731-4
Conflict of interest statement
None of the authors of this study has any applicable conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sun, Y., Zang, Z., Xu, X. et al. The association of SULT1A1 codon 213 polymorphism and breast cancer susceptibility: meta-analysis from 16 studies involving 23,445 subjects. Breast Cancer Res Treat 125, 215–219 (2011). https://doi.org/10.1007/s10549-010-0953-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-0953-5