Abstract
Numerous independent groups from a range of countries have reported a high prevalence of Mouse Mammary Tumour Virus (MMTV)-like env sequences in human breast cancer specimens, including a prevalence of almost 40% in Australia. MMTV-like sag sequences and a completely integrated provirus have also been described. Recently, it was reported that MMTV is capable of productive infection of human breast cells in vitro. Conclusive demonstration of an association between MMTV and human breast cancer has remained elusive, and negative findings from a number of independent studies have questioned the role of MMTV as an aetiological agent. We used breast cancer specimens from women in the Australian Breast Cancer Family Study (ABCFS) who were diagnosed with first primary invasive breast cancer before the age of 40 years. Specimens were selected for higher grade cancers and for diagnosis relatively soon after childbirth. We searched for MMTV-like env sequences in tumour-enriched DNA using a nested PCR designed to detect all MMTV variants represented in GenBank, including those reportedly detected in human breast cancers. Forty-two specimens were deemed adequate for testing based on strong β-globin PCR. Despite the MMTV nested PCR regimen consistently detecting five copies of control plasmid against a background of MMTV-negative human genomic DNA, no MMTV env sequence was detected in any of the breast cancer specimens. Our findings appear inconsistent with previous reports on Australian breast cancer specimens but consistent with a growing number of independent negative reports internationally. We recommend caution in inferring a role for MMTV or a closely related virus in human breast cancer and suggest that universally regarded alternative lines of evidence such as highly specific serology data will be required to support such an association.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Worldwide, approximately 800,000 incident cases of breast cancer are diagnosed each year [1]. For several decades, the retrovirus Mouse Mammary Tumour Virus (MMTV) or a close relative (collectively termed ‘MMTV’) has been suggested as a possible aetiological agent for human breast cancer. Aspects of the biology of MMTV and some recent reports suggest a strong case. MMTV was the first virus shown to cause cancer in mammals [2]. In the mouse, MMTV can be transmitted through the germline or via milk to suckling pups. Tumorigenesis occurs due to disruptive non-site-specific proviral integration in mammary epithelia during puberty and pregnancy [3]. MMTV env sequences have been reported at relatively high frequencies in unselected breast cancer specimens but at dramatically lower frequencies in healthy breast tissues from affected or unaffected women or in other cancer tissues [4–12]. Frequencies in breast cancer tissues of 20–40% have been reported for women in the USA, Italy, Mexico, Brazil, Argentina and Australia, and frequencies of 12, 10–17 and 0.8% have been reported for women in Japan, China and Vietnam, respectively. The geographical variation in the incidence of female breast cancer has been reported to correlate with the regional dominance of the mouse species Mus domesticus, suggesting a possible zoonotic connection [13]. MMTV env sequences have been reported to be especially common in higher grade breast cancers [9], breast cancers diagnosed during gestation (62%) [14] or lactation [6], and cases from multiple-case families [6, 15], as well as in male breast cancers (62%) [9]. MMTV transcripts and the MMTV sag gene have also been reported as common in breast cancer specimens [16, 17], and a complete 9.9 kb provirus has been reported [18]. It was recently reported that MMTV is capable of active infection of the human aneuploid mammary epithelial cell line Hs578T [19].
However, MMTV’s candidature as an aetiological agent for human breast cancer is controversial [20–23]. Null findings have been reported by studies of UK, Austrian, Swedish, Japanese and Italian breast cancer specimens [24–28]. In apparent discord are reports of 13.9 or 74% env-positivity in Tunisian breast cancer specimens [29, 30] and 4.2 or 24% in Mexican cases [31, 32].
The implications for public health, should MMTV eventuate as a causative agent for human breast cancer, are great. We sought to validate previous reports by attempting to detect MMTV env sequences in a population-based sample of breast cancer specimens from the Australian Breast Cancer Family Study (ABCFS) using a specifically engineered nested PCR. Studied specimens were enriched for higher grade breast cancers and cases for which diagnosis occurred relatively soon after childbirth.
Materials and methods
ABCFS specimens
Thirty-eight 5 μm and twelve 3 μm Formalin Fixed Paraffin Embedded (FFPE) breast cancer tissue sections representing breast cancers from ABCFS participants were selected to enrich for breast cancers of relatively high histological grade and cancers from cases in which there was a relatively short time between the woman’s last childbirth and her date of breast cancer diagnosis. H&E stained neighbouring sections were used to confirm the presence of breast cancer tissue [33, 34].
p203 and human genomic DNA controls
The plasmid p203 in E. coli was purchased from the American Type Culture Collection (ATCC). p203 harbours the relevant portion of MMTV targeted by the nested PCR described here. A small-scale plasmid preparation was performed according to ATCC instructions and using the QIAprep Spin Miniprep Kit (Qiagen, Doncaster, VIC, Australia). p203 concentration was determined using the Quant-It system (Invitrogen, Mulgrave, VIC, Australia) and Qubit instrument (Invitrogen, Mulgrave, VIC, Australia) according to the manufacturer’s instructions. QIAamp Kit (Qiagen, Doncaster, VIC, Australia)-purified human genomic DNA from a healthy control individual whole blood sample was sourced and the concentration determined using the Quant-IT system with the Qubit instrument.
DNA extractions
Formalin Fixed Paraffin Embedded tissues (full face sections) were scraped from slides onto aluminium foil using scalpel blades and transferred to microfuge tubes using syringe needles. Separate foil, blades and needles were used for each specimen. DNA was extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen, Doncaster, VIC, Australia) according to the manufacturer’s instructions but with the following exceptions: histolene was used instead of xylene, two ethanol spin/wash steps were employed instead of one to facilitate thorough washing without losing tissue during aspiration of supernatant, no RNaseA was used, and elution was performed using 50 μl ATE buffer.
β-Globin PCR specimen adequacy check
GH20 and PC04 β-globin PCR primers [35] were ordered from Integrated DNA Technologies (IDT) at standard desalting grade. PCRs were performed in 30 μl reaction volumes containing 1.5 U AmpliTaq Gold (Applied Biosystems, Mulgrave, VIC, Australia), 200 μM dNTPs (Bioline, Alexandria, NSW, Australia), 100 nM PC04 and GH20 primers and 5 μl template DNA and overlaid with 25 μl molecular biology grade mineral oil (Sigma-Aldrich, Sydney, NSW, Australia) added from a fresh aliquot by Gilson pipette. PCR was performed using the following thermal parameters: 95°C for 10 min, followed by 40 cycles of 94°C for 45 s, 56°C for 45 s and 72°C for 45 s, followed by 72°C for 5 min. PCR products (268 bp) were analysed using agarose gel electrophoresis.
Nested PCR primers and primer design
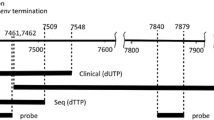
ClustalW2 multiple sequence alignments of MMTV env were performed using GenBank-deposited sequences to include a range of MMTV strains and sequences reported to be derived from human tissue specimens (see Fig. 1). Primer sites were selected with a view to yielding small amplicons (less than 200 bp), limiting the degree of mismatch across the aligned sequences, especially near the 3-prime terminus, targeting a GC content approaching 50% without excessively long GC runs, and avoiding unintended ‘off target’ priming from HERV-K10 (the reported most similar human endogenous retrovirus) or the human genome, excessive primer secondary structure formation and primer-dimer formation. Primer-BLAST, OligoAnalyzer 3.1 and ClustalW2 were employed to facilitate these design requirements. Remaining points of mismatch with respect to the alignment group were addressed by introducing degeneracy at corresponding primer positions. 5-prime clamp sequence was included to facilitate primer participation by all members of the degenerate primer ‘pool’. Resulting first round PCR primers were (5′ to 3′, lower case indicates 5′ clamp sequence): DPenvF2: ctatagggGGYTACTTTGGGATTTCTCCCTTC and DPenvR2: ctatagggACCAWGTAGGTTCTACCCATC. Resulting second round (nested) PCR primers were (5′ to 3′): DPenvF3: ctatagggTCGCCTARTRTAGATCAGTCAGA and DPenvR3: ctatagggCATCYTGCYTCATACCATCGATGAAC. Nested PCR primers were ordered from IDT at standard desalting grade. Anticipated product sizes from PCRs employing DPenvF2 and DPenvR2 or DPenvF3 and DPenvR3 are 166 and 123 bp, respectively. MMTV env sequences spanned by these PCRs are 150 and 107 bp, respectively.
ClustalW2 multiple sequence alignment of MMTV env within the human breast cancer-associated region [4]. The range of MMTV strains includes strains reported to be derived from human breast cancer tissue (prefixed ‘H’). Database accession codes for the respective sequences are NC_001503, D16249, AF228551, AF228550, AF071010, X01811, AF239172, AF346816 and AF243039. Boxed arrows indicate nested PCR primer binding sites used in this study. These are annotated with the degenerate bases included in the primers (Y, R or W). The vertical arrow indicates degeneracy designed into primer DPenvR3 based on the sequences AY600606, AY600599, AY600611, AY600614, AY600604, AY600605, AY600597 and AY600608 which, together with AY600615, AY600613, AY600610 and AY600598 [9], were also aligned to aid primer design (not shown)
Nested PCR
For assay sensitivity assessment, nested PCRs (PCR1 containing DPenvF2 and DPenvR2, followed by PCR2 containing DPenvF3 and DPenvR3) were performed using different cycle numbers (30 or 35 cycles for each PCR) or annealing temperatures (50–56°C) using decreasing copies of p203 (tested down to five copies) against a backdrop of 10 ng of MMTV-negative human genomic DNA, followed by gel electrophoretic analysis. All conditions clearly detected five copies of p203 against a backdrop of 10 ng MMTV-negative human genomic DNA. In all cases 10 ng MMTV-negative human genomic templates and water controls yielded negative results. Conditions selected to screen the breast cancer specimens were: PCR1: 30 μl reaction volumes containing 1.5 U AmpliTaq Gold (Applied Biosystems, Mulgrave, VIC, Australia), 200 μM dNTPs (Bioline, Alexandria, NSW, Australia), 100 nM DPenvF2 and DPenvR2 primers and 5 μl template DNA overlaid with 25 μl molecular biology grade mineral oil (Sigma-Aldrich, Sydney, NSW, Australia) added from a fresh aliquot by Gilson pipette. PCR1 was performed using the following thermal parameters: 95°C for 10 min, followed by 35 cycles of 94°C for 45 s, 50°C for 45 s and 72°C for 45 s, followed by 72°C for 5 min. PCR2: 30 μl reaction volumes containing 1.5 U AmpliTaq Gold (Applied Biosystems, Mulgrave, VIC, Australia), 200 μM dNTPs (Bioline, Alexandria, NSW, Australia), 100 nM DPenvF3 and DPenvR3 primers and 2 μl PCR1 product template overlaid with 25 μl molecular biology grade mineral oil (Sigma-Aldrich, Sydney, NSW, Australia) added from a fresh aliquot by Gilson pipette. PCR2 was performed using the following thermal parameters: 95°C for 10 min, followed by 35 cycles of 94°C for 45 s, 55°C for 45 s and 72°C for 45 s, followed by 72°C for 5 min. These conditions were selected as sufficiently relaxed to enable efficient specific amplification in cases of unforeseen primer-template mismatch whilst yielding ‘clean’ product profiles. PCR2 product was sequenced using DPenvF3 as the sequencing primer and BigDye3.1chemistry (Applied Biosystems, Mulgrave, VIC, Australia).
Agarose gel electrophoresis
2.4% Agarose TBE gels containing ethidium bromide were loaded with 5 μl PCR products alongside 250 ng of 100 bp ladder (New England Biolabs distributed by Genesearch, Arundel, QLD, Australia). Following electrophoresis, gels were photographed under UV light transillumination.
Results and discussion
Fifty ABCFS specimens were tested for ‘sample adequacy’ using PCR to amplify β-globin sequence. Strong bands of 268 bp following PCR and agarose gel electrophoresis were interpreted as being derived from adequate specimens, and the corresponding specimens were subsequently tested for MMTV env sequences by nested PCR. A relatively conservative approach was applied, faint products of 268 bp or no apparent products of 268 bp were interpreted as being derived from inadequate specimens and the corresponding specimens were not subsequently tested for MMTV env sequences. Of the 50 original specimens, 42 were deemed adequate (Fig. 2a), derived from thirty-four 5 μm and eight 3 μm sections.
2.4% Agarose TBE gels depicting the assessment of ABCFS specimen adequacy following β-globin PCR (a) and MMTV env sequences following nested PCR (b). ‘M’ indicates 250 ng 100 bp ladder marker; ‘−’ indicates a no template control; numbers 1–50 indicate the 50 ABCFS specimens screened for specimen adequacy; ‘F’ indicates a specimen interpreted as inadequate; A1–A42 indicate the 42 adequate specimens tested for MMTV env sequence by nested PCR; ‘+’ indicates a ‘five copies of p203 with 10 ng MMTV-negative human genomic DNA’ control
All 42 adequate specimens represented in this study were collected from women living in the Melbourne (22/42) or Sydney (20/42) metropolitan areas that were diagnosed with invasive breast cancer before the age of 40 years. The mean age at time of diagnosis was 35.1 years (28–39 years), and 83.3% (35/42) of specimens exhibited infiltrating ductal profiles. 76.2% of the specimens (32/42) presented as histological grade 3 cancers, the remainder being grade 2 cancers. Overall, 78.6% of women (33/42) had given birth and 90.9% of specimens (30/33) were from women who had breastfed. Of the women who had given birth, the median time from the last birth to date of diagnosis was 54 months (0–244 months). Based on previously reported prevalences of MMTV env detection for Australian breast cancers (30–40%) [8, 9], reported increased prevalence for higher grade breast cancers [9] and reported increased prevalence for cases diagnosed during gestation (62%) [14] or lactation [6], approximately 30–50% of the 42 breast cancer specimens included in this study might have been expected to harbour MMTV env sequences, i.e. approximately 12–21 specimens. From the Poisson distribution, the probability of observing zero prevalence is 0.000006 when the true prevalence is 30%, and 0.05 when the true prevalence is 7%. That is, the true prevalence for Australian women is highly unlikely to be as high as previous reports, though we cannot exclude a low prevalence.
The nested PCR regimen used in this study to look for MMTV env sequences proved to be robust and extremely sensitive. In all experiments conducted, each using fresh dilutions of p203 to yield five plasmid copies against 10 ng MMTV-negative human genomic DNA as a parallel sensitivity control for separate PCR runs, MMTV env was detected as a strong 123 bp band following agarose gel electrophoresis. ‘No template’ controls always yielded profiles free of visible PCR product. Sequencing and BLAST analysis of the assay sensitivity control products confirmed the detection of MMTV env-derived sequences. None of the 42 adequate breast cancer specimens tested yielded visible MMTV env-derived products (Fig. 2b). Given the nature of the sample set tested and the demonstrated high sensitivity of the nested PCR regimen employed, including design features and PCR conditions to account for unforseen discrepancies between potential sample set MMTV env sequences and sequences included in the primer design alignment group, it seems unlikely that any of the 42 adequate ABCFS specimens tested contained MMTV env sequences. These findings appear inconsistent with previous reports of associations between MMTV and human breast cancer and, in particular, reports of 30–40% prevalence for unselected Australian breast cancer specimens [8, 9]. Given the risk of PCR contamination and the potential to contribute to false findings [22], we recommend caution when interpreting findings derived from highly sensitive PCR regimens which are not consistently supported by other studies.
Mouse Mammary Tumour Virus has not been found in human breast milk and is not detected in healthy tissue adjacent to cancerous tissue in humans, unlike in the mouse, and the histopathology of murine MMTV tumours is dissimilar to human breast cancers (hyperplasias vs. typically invasive ductal carcinomas) [36]. In a large study, post-menopausal women who were breastfed as infants did not have a higher risk of breast cancer compared with those who were not [37], and a separate study has reported that human breast cancer rates have increased at the same time that breastfeeding rates have decreased [38]. Pregnancy in humans appears to have a protective effect with respect to breast cancer risk, unlike MMTV-associated breast cancer in the mouse [39, 40], and serological conversion to yield antibodies cross-reactive with MMTV in human breast cancer patients remains to be demonstrated at a population level [41] despite the fact that mice produce strong antibody responses to MMTV. Further, Transferrin Receptor 1 acts as an MMTV entry receptor in the mouse but not in humans [42, 43].
The above observations together with the findings presented in this study and a growing number of null findings from independent groups internationally suggest that MMTV is unlikely to be a causative agent for female breast cancer. Alternative sources of evidence, for example, derived from universally accepted specific serological analysis will probably be required to definitively demonstrate an association between MMTV and female breast cancer. Consistent demonstration by independent groups of such an association and the detection of MMTV infection prior to the development of disease will be required to demonstrate causality [44, 45].
References
Parkin D, Pisani M, Ferlay J (1999) Global cancer statistics. CA Cancer J Clin 49:33–64
Bittner JJ (1936) Some possible effects of nursing on the mammary gland tumor incidence in mice. Science 84:162
Ross SR (2008) MMTV infectious cycle and the contribution of virus-encoded proteins to transformation of mammary tissue. J Mammary Gland Biol Neoplasia 13:299–307
Wang Y, Holland JF, Bleiweiss IJ, Melana S, Liu X, Pelisson I, Cantarella A, Stellrecht K, Mani S, Pogo BG (1995) Detection of mammary tumor virus ENV gene-like sequences in human breast cancer. Cancer Res 55:5173–5179
Etkind P, Du J, Khan A, Pillitteri J, Wiernik PH (2000) Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin Cancer Res 6:1273–1278
Pogo BGT, Melana SM, Holland JF, Mandeli JF, Polotti S, Casalini P, Menard S (1999) Sequences homologous to the mouse mammary tumor virus env gene in human breast cancer correlate with overexpression of laminin receptor. Clin Cancer Res 5:2108–2111
Melana S, Holland JF, Pogo BGT (2001) Search for mouse mammary tumor-like env sequences in cancer and normal breast from the same individuals. Clin Cancer Res 7:283–284
Ford CE, Tran D, Deng Y, Ta VT, Rawlinson WD, Lawson JS (2003) Mouse mammary tumor virus-like gene sequences in breast tumors of Australian and Vietnamese women. Clin Cancer Res 9:1118–1120
Ford CE, Faedo M, Crouch R, Lawson JS, Rawlinson WD (2004) Progression from normal breast pathology to breast cancer is associated with increasing prevalence of mouse mammary tumor virus-like sequences in men and women. Cancer Res 64:4755–4759
Holland JF, Pogo BGT (2004) Mouse mammary tumor virus-like viral infection and human breast cancer. Clin Cancer Res 10:5647–5649
Holland JF, Melana S, Wang Y, Fernandez-Cobo M, Jiang JD, Pogo BGT (2003) Human mammary tumor virus (HMTV) is horizontally, not vertically transmitted. Proc Am Soc Clin Oncol 22:2003 (abstr 3511)
Luo T, Wu XT, Zhang MM, Qian K (2006) Study of mouse mammary tumor virus-like gene sequences expressing in breast tumors of Chinese women. Sichuan Da Xue Xue Bao Yi Xue Ban 37:844–846
Stewart TH, Sage RD, Stewart AF, Cameron DW (2000) Breast cancer incidence highest in the ranges of one species of house mouse, Mus domesticus. Br J Cancer 82:446–451
Wang Y, Melana SM, Baker B, Bleiweiss I, Fernandez-Cobo M, Mandeli SM, Holland JF, Pogo BG (2003) High prevalence of MMTV-like env gene sequences in gestational breast cancer. Med Oncol 20:233–236
Etkind PR, Stewart AFR, Wiernik PH (2008) Mouse mammary tumor virus (MMTV)-like DNA sequences in the breast tumors of father, mother, and daughter. Infect Agents Cancer 3:2
Wang Y, Go V, Holland JF, Melana SM, Pogo BG (1998) Expression of mouse mammary tumor virus-like env gene sequences in human breast cancer. Clin Cancer Res 4:2565–2568
Wang Y, Jiang JD, Xu D, Li Y, Qu C, Holland JF, Pogo BGT (2004) A mouse mammary tumor virus-like long terminal repeat superantigen in human breast cancer. Cancer Res 64:4105–4111
Liu B, Wang Y, Melana SM, Pelisson I, Najfeld V, Holland JF, Pogo BG (2001) Identification of a proviral structure in human breast cancer. Cancer Res 61:1754–1759
Indik S, Gunzburg WH, Kulich P, Salmons B, Rouault F (2007) Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology 4:73
Stewart A (2003) MMTV-related env sequences in human breast tumor. Int J Cancer 106:138
Zangen R, Sidransky D (2003) Response to ‘MMTV-related env sequences in human breast tumors’. Int J Cancer 106:139
Mant C, Cason J (2004) A human murine mammary tumour virus-like agent is an unconvincing aetiological agent for human breast cancer. Rev Med Virol 14:169–177
Mant C, Cason J (2005) Mouse mammary tumor virus and human breast cancer. Cancer Res 65:1112–1113
Mant C, Gillett C, D’Arrigo C, Cason J (2004) Human murine mammary tumour virus-like agents are genetically distinct from endogenous retroviruses and are not detectable in breast cancer cell lines or biopsies. Virology 318:393–404
Witt A, Hartmann B, Marton E, Zeillinger R, Schreiber M, Kubista E (2003) The mouse mammary tumor virus-like env gene sequence is not detectable in breast cancer tissue of Austrian patients. Oncol Rep 10:1025–1029
Bindra A, Muradrasoli S, Kisekka R, Nordgren H, Warnberg F, Blomberg J (2007) Search for DNA of exogenous mouse mammary tumor virus-related virus in human breast cancer samples. J Gen Virol 88:1806–1809
Fukuoka H, Moriuchi M, Yano H, Nagayasu T, Moriuchi H (2008) No association of mouse mammary tumor virus-related retrovirus with Japanese cases of breast cancer. J Med Virol 80:1447–1451
Zangen R, Harden S, Cohen D, Parrella P, Sidransky D (2002) Mouse mammary tumor-like env gene as a molecular marker for breast cancer? Int J Cancer 102:304–307
Hachana M, Trimeche M, Ziadi S, Amara K, Gaddas N, Mokni M, Korbi S (2008) Prevalence and characteristics of the MMTV-like associated breast carcinomas in Tunisia. Cancer Lett 271:222–230
Levine PH, Pogo BGT, Klouj A, Coronel S, Woodson K, Melana SM, Mourali N, Holland JF (2004) Increasing evidence for a human breast carcinoma virus with geographic differences. Cancer 101:721–726
Zapata-Benavides P, Saavedra-Alonso S, Zamora-Avila D, Vargas-Rodrate C, Barrera-Rodriguez R, Salinas-Silva J, Rodriguez-Padilla C, Tamez-Guerra R, Trejo-Avila L (2007) Mouse mammary tumor virus-like gene sequences in breast cancer samples of Mexican women. Intervirology 50:402–407
Holland JF, Melana SM, Wang Y, Bleiweiss I, Levine P, Gombe Mbalawa C, Kalengayi M, Ndom P, Ramirez M, Cervantes G, Pogo BGT (1998) Geographic variation in proportion of breast cancers with sequences homologous to MMTV env gene. Proc Am Assoc Cancer Res 39:55
Dite GS, Jenkins MA, Southey MC, Hocking JS, Giles GG, McCredie MRE, Venter DJ, Hopper JL (2003) Familial risks, early-onset breast cancer, and BRCA1 and BRCA2 germline mutations. J Natl Cancer Inst 95:448–457
John EM, Hopper JL, Beck JC, Knight JA, Neuhausen SL, Senie RT, Ziogas A, Andrulis IL, Anton-Culver H, Boyd N, Buys SS, Daly MB, O’Malley FP, Santella RM, Southey MC, Venne VL, Venter DJ, West DW, Whittemore AS, Seminara D (2004) The Breast Cancer Family Registry: an infrastructure for cooperative multinational, interdisciplinary and translational studies of the genetic epidemiology of breast cancer. Breast Cancer Res 6:R375–R389
Ip SM, Wong LC, Xu CM, Cheung ANY, Tsang PCK, Ngan HYS (2002) Detection of human papillomavirus DNA in malignant lesions from Chinese women with carcinomas of the upper genital tract. Gynecol Oncol 87:104–111
Callahan R, Smith GH (2000) MMTV-induced mammary tumorigenesis: gene discovery, progression to malignancy and cellular pathways. Oncogene 19:992–1001
Titus-Ernstoff L, Egan KM, Newcomb PA, Baron JA, Stampfer M, Greenberg ER, Cole BF, Ding J, Willett W, Trichopoulos D (1998) Exposure to breast milk in infancy and adult breast cancer risk. J Natl Cancer Inst 90:921–924
MacMahon B, Cole P, Brown J (1973) Etiology of human breast cancer: a review. J Natl Cancer Inst 50:21–42
Kampert JB, Whittemore AS, Paffenbarger RS Jr (1988) Combined effect of childbearing, menstrual events, and body size on age-specific breast cancer risk. Am J Epidemiol 128:962–979
Nicholl CS, Tucker HA (1965) Estimates of parenchymal, stromal, and lymph node deoxyribonucleic acid in mammary glands of C3H/Crgl-2 mice. Life Sci 4:993–1001
Goedert JJ, Rabkin CS, Ross SR (2006) Prevalence of serologic reactivity against four strains of mouse mammary tumor virus among US women with breast cancer. Br J Cancer 94:548–551
Ross SR, Schofield JJ, Farr CJ, Bucan M (2002) Mouse transferrin receptor 1 is the cell entry receptor for mouse mammary tumor virus. Proc Natl Acad Sci USA 99:12386–12390
Wang E, Albritton L, Ross SR (2006) Identification of the segments of the mouse transferrin receptor 1 required for mouse mammary tumor virus infection. J Biol Chem 281:10243–10249
Evans AS, Mueller NE (1990) Viruses and cancer. Causal associations. Ann Epidemiol 1:71–92
Vonka V (2000) Causality in medicine: the case of tumours and viruses. Philos Trans R Soc Lond 355:1831–1841
Acknowledgements
We would like to thank the women who participate in this study and gave consent for access to their tumour samples. This study was supported by the NHMRC Australia Fellowship number 466668 awarded to JLH, who is also a Victorian Breast Cancer Research Consortium Group Leader. MCS is a National Health and Medical Research Council Senior Research Fellow and a Victorian Breast Cancer Research Consortium Group Leader. The authors would like to thank Ms Veronika Gazdik for histological assistance. The ABCFS has been funded by NHMRC, the Victorian Health Promotion Foundation, and the NSW Cancer Council.
Conflict of interest statement
The authors declare no conflicting interests pertaining to this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, D.J., Southey, M.C., Giles, G.G. et al. No evidence of MMTV-like env sequences in specimens from the Australian Breast Cancer Family Study. Breast Cancer Res Treat 125, 229–235 (2011). https://doi.org/10.1007/s10549-010-0946-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-0946-4