Abstract
Pharmacological evidence shows that cytochrome P450 2D6 (CYP2D6) function is important in the conversion of tamoxifen to its active metabolites. Many retrospective analyses have assessed the role of both CYP2D6 genotype and concurrent administration of drug inhibitors of CYP2D6 on outcome of tamoxifen therapy. These studies have frequently been of small size and their data highly variable. A published data meta-analysis of trials evaluating outcomes of tamoxifen therapy in early breast cancer was undertaken. Hazard ratios (HRs) were extracted for disease-free survival (DFS) and overall survival (OS). Pooled estimates were computed using inverse-variance and random-effect modeling. Data from 10 studies assessing CYP2D6 genotype were included in a meta-analysis. There was significant heterogeneity in the definition of comparison groups between studies. When compared to reduced CYP2D6 function, normal function was associated with a trend toward improved DFS (HR 2.07, 95% CI 0.96–4.49, P = 0.06) but not OS (HR 1.36, 95% CI 0.73–2.52, P = 0.34). Pooling of data from two studies evaluating CYP2D6 drug inhibitors showed that concomitant administration of these with tamoxifen was associated with a non-significant association with DFS (HR 1.37, 95% CI 0.69–2.73, P = 0.37). Analysis of the effect of CYP2D6 drug inhibitors on OS was not possible. The effect of CYP2D6 genotype on breast cancer seems to be relatively small and may not warrant testing of CYP2D6 genotype in all women with hormone receptor positive breast cancer. The effect of CYP2D6 genotype on outcome in low-risk patients may not be clinically relevant, while the upfront use of aromatase inhibitors is a reasonable alternative to tamoxifen in high-risk post-menopausal women, irrespective of CYP2D6 genotype. There are limited data supporting the association of potent inhibitors of CYP2D6 and detrimental outcome, but avoidance of such drugs seems reasonable.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
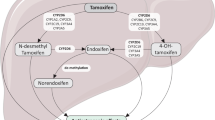
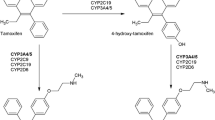
Tamoxifen is a selective estrogen receptor modulator, which is used in the treatment of hormone receptor positive breast cancer. Tamoxifen is a pro-drug, which is metabolized by hepatic cytochrome P450s (CYP) to produce its primary metabolites: N-desmethyltamoxifen and 4-hydroxy-tamoxifen (4-HT), which are formed by CYP3A and CYP2D6 [1, 2]. Oxidation of these metabolites results in the formation of the 4-hydroxy-N-desmethyltamoxifen (endoxifen) [2]. Endoxifen and 4-HT are the most potent metabolites of tamoxifen [3, 4]. In a steady state, plasma concentrations of endoxifen are 5–10 times higher than 4-HT and in contrast to 4-HT endoxifen also down-regulates expression of nuclear estrogen receptor-α (ER-α) in cancer cells [5, 6]. Therefore, endoxifen is considered to be the most important metabolite of tamoxifen.
CYP2D6 is an important enzyme in the conversion of tamoxifen into endoxifen, but its activity is not an absolute requirement. CYP2D6 activity is determined genetically but may also be affected by drugs (e.g., CYP2D6 inhibitors). To date, more than 75 allelic variants of CYP2D6 gene have been reported [7]. They have different activities: (a) functional or wild-type (wt) alleles (e.g., *1, *2), (b) alleles with reduced activity (e.g., *10, *17), (c) non-functional alleles (e.g., *3, *4, *5, *6), and (d) alleles with unknown activity. Whereas wt and non-functional alleles are the most frequent in Caucasian populations, alleles with reduced activity are more frequent in Asians, Africans, and Afro-Americans [8]. Activity of CYP2D6 enzyme in every individual is determined by two inherited alleles. Approximately 6–10% of Caucasians are homozygous for two non-functional alleles (i.e., poor metabolizers) while up to 30% of Asians are homozygous for two alleles with a reduced activity [8].
Pharmacokinetic studies in Caucasian and Asian women have shown that those who carry two non-functional alleles (e.g., *4) have an approximately fourfold decrease in plasma concentrations of endoxifen while those with two alleles with reduced activity (i.e., *10) have approximately twofold reduction in concentration compared to women who are homozygous for two wt alleles [9, 10]. In contrast to individuals who are homozygous or heterozygous for wt CYP2D6 alleles and have normal CYP2D6 function (i.e., normal or extensive metabolism), individuals homozygous or compound heterozygous for alleles with reduced and/or absent function are considered to have reduced CYP2D6 function (i.e., intermediate or poor metabolism) [8]. Drugs, which are strong CYP2D6 inhibitors (e.g., paroxetine) can further decrease plasma concentrations of endoxifen irrespective of genetic background [9].
Studies assessing the role of CYP2D6 genotype have been particularly prevalent with a large and diverse body of data having been published in this area. All studies published thus far have been retrospective cohort or case–control studies, many have been very small in size and results have been very variable. Furthermore, there has been heterogeneity in the comparison groups used in studies. Consequently, results from these studies have been conflicting [11–17]. Similarly, population-based studies did not consistently demonstrate that concurrent use of CYP2D6 inhibitors and tamoxifen is detrimental in women with early breast cancer [18–20]. The present study, therefore, aimed to pool data from differing sources and assess the impact of both CYP2D6 genotype and CYP2D6 drug inhibitors on both disease-free (DFS) and overall survival (OS) of tamoxifen-treated patients with early breast cancer.
Materials and methods
Study identification
Studies were identified using a computerized search of MEDLINE (host: OVID), January 1950 to February, week 6, 2010, American Society of Clinical Oncology Annual Meetings, 2006–2009, San Antonio Breast Cancer Symposium Annual Meetings 2006–2009. All review articles and references from manuscripts from retrieved articles were screened for pertinent studies.
Selection criteria
For inclusion in the meta-analysis, studies fulfilled the following criteria: patients received any duration of tamoxifen for early stage, invasive breast cancer (Stage I to III), and assessed DFS, event-free (EFS), relapse-free (RFS) or OS as outcomes in a proportional hazards analysis (univariable or multivariable). Pharmacodynamic studies were excluded as were studies in metastatic breast cancer.
Outcomes for analysis
Outcomes were dichotomized into two groups: DFS and OS. If data on DFS were not available, data on EFS or RFS were assessed. These data were expressed as hazard ratios (HRs) of genotypes associated with normal CYP2D6 function (i.e., extensive metabolism) of tamoxifen over reduced CYP2D6 function (i.e., intermediate or poor metabolism). 95% confidence intervals (CI) were calculated for each point estimate.
Data extraction
Both authors independently extracted information using pre-designed forms. The following details were recorded: study design, participants, setting, interventions, duration of treatment, and efficacy outcomes. Any discrepancies regarding the extraction of quantitative data or assessment of the quality were addressed by consensus. When a trial was presented in abstract form, further information was sought as necessary (from the internet, contacting authors or checking for other available resources/publications). For studies with more than one publication, data were extracted from all the publications; however, the final or updated version of each trial was considered the primary reference.
Assessment of study quality
Study quality was assessed using modified criteria for case–control studies developed by the US Preventive Services Task Force [21].
Data synthesis
Data were combined into a meta-analysis whenever possible. Categorical data were only included where it could be divided into dichotomous outcomes. If the data could not be combined in a meta-analysis it was summarized in the text and grouped by outcome as appropriate. All data were entered using RevMan 5 analysis software (The Cochrane Collaboration, Copenhagen, Denmark).
Pooled estimates of HR were computed using a random-effect model [22] according to the generic inverse-variance approach [23]. The random-effect model was utilized in view of clinical heterogeneity in the study population. The included studies assessed both Caucasian and Asian populations and tested for different polymorphisms in the CYP2D6 gene. There was also variability in the duration of tamoxifen therapy with some data including patients treated for 2 years and other data based on 5 years of treatment.
However, the most critical area of heterogeneity was the comparison groups used by the different studies. Consequently, for analysis purposes, studies were dichotomized into those that used a genotype-based approach and those comparing a function-based approach, as per Bradford criteria [8]. In studies using a genotype-based approach, patients homozygous for wt alleles (i.e., normal CYP2D6 function) were compared to those with one or no wt alleles (i.e., combination of normal and reduced CYP2D6 function); for example *wt/*wt vs. *4/*4 and *4/*wt [15, 17, 24]. In studies using a function-based approach patients homozygous or heterozygous for wt alleles (i.e., normal CYP2D6 function) were compared to those who were homozygous or compound heterozygous for reduced function alleles (i.e., reduced CYP2D6 function); for example, *wt/*wt vs. homozygous or compound heterozygous for *3, *4, *5, *10, *41, or *wt/*wt and *wt/*10 vs. *10/*10 [13, 14]. Data from all studies were pooled for descriptive purposes only. Sensitivity analyses were carried out to assess the interaction of ethnic origin, and duration of tamoxifen therapy. For all analyses, funnel plots were generated to assess for publication bias as evidenced by asymmetry.
Results
Included studies
Twenty-four studies were identified [11–20, 24–37]. Of these, six studies did not meet inclusion criteria: studies by Toyama et al. [33], Ramon y Cajal et al. [34], and Lash et al. [37] did not analyze data by proportional hazards analysis, Rae et al. [36] assessed the association of CYP2D6 and the proliferation marker Ki67 and not with survival outcomes, a study by Wegman et al. [32] compared a tamoxifen-treated group with a non-tamoxifen-treated group while Abraham et al. [31] did not use a standardized genotype definition. A further five studies were excluded due to repeated data: two studies from Goetz [11, 25] and one from Schroth [16] were combined and updated in a single, later publication [13] while Kiyotani published an expanded dataset [12], therefore, the original publication [30] was not used. Finally, a study by Goetz et al. [27] was excluded as its data were partly based on previously published reports of CYP2D6 and outcomes.
Three studies assessing the association between CYP2D6 drug inhibitors and breast cancer outcome met the inclusion criteria, and they were analyzed separately from studies assessing CYP2D6 genotype. Of these, studies by Aubert et al. [18] and Dezentje et al. [19] presented only DFS while Kelly et al. [20] only presented OS. Consequently, pooling of data was carried out for the two studies reporting DFS while Kelly et al. was assessed descriptively.
Therefore, 10 studies were included in the meta-analysis of CYP2D6 genotype. Of these nine studies [12–15, 17, 24, 26, 28, 29] were included in the analysis of DFS and four studies [13, 15, 26, 35] were included in the analysis of OS. These analyses included 3120 and 1570 patients, respectively. Table 1 presents these studies and their main outcomes. In the analysis of CYP2D6 drug inhibitors, two studies containing 3621 patients were pooled for DFS while the analysis for OS contained the 2430 patients included in the study by Kelly et al. [20].
Of all the above, nine studies were published as full papers [12–15, 17, 20, 26, 29, 35] and four studies were only available in abstract form [18, 19, 24, 28]. Sufficient data were available from publically available sources for all studies. Therefore, specific authors did not need to be contacted. There were no discrepancies in outcome data extracted by the two reviewers, therefore κ was not calculated.
Study quality
In total two studies were considered of good quality [13, 26], three studies were considered of fair quality [12, 14, 29] and eight studies were graded as poor quality [15, 17–20, 24, 28, 35] (see Table 1). The agreement between the two reviewers was moderate (κ = 0.650, 95% CI 0.109–0.901) and disagreement was resolved by consensus in all cases.
Association between CYP2D6 genotype and DFS
Single study HRs ranged from 0.33 to 9.52 and were statistically significant in four studies [12–14, 24]. Pooled analysis showed a non-significant trend toward an increase in risk of disease recurrence with a HR of 1.41 (95% CI 0.94–2.10, P = 0.08, see Fig. 1). Subgroup analysis by ethnic origin showed the pooled HR was 1.22 (95% CI 0.88–1.68, P = 0.24) and 2.94 (95% CI 0.52–16.55, P = 0.22) for Caucasians and Asians, respectively (see Table 2). Subgroup analysis by duration of tamoxifen therapy showed that the pooled HR for 5 years of tamoxifen was 1.78 (95% CI 0.57–5.54, P = 0.32), for less than 5 years of tamoxifen was 0.87 (95% CI 0.38–1.97, P = 0.74) and for an unspecified duration was 1.30 (95% CI 0.91–1.36, P = 0.14). Once again, none of these were statistically significant (see Table 3). Finally, as shown in Table 4, in those studies using a function-based comparison, the pooled HR was 2.07 (95% CI 0.96–4.49, P = 0.06) while in the studies using a genotype-based comparison, the pooled HR was 1.06 (95% CI 0.60–1.86, P = 0.85, see Fig. 1). The funnel plot (not shown) showed good symmetry suggesting a low-likelihood of publication bias.
Association between CYP2D6 genotype and OS
Single study HRs ranged from 0.77 to 2.5 and were not statistically significant in any of the five studies. Pooled analysis showed a small and non-significant association toward an increased risk of death with a HR of 1.24 (95% CI 0.93–1.67, P = 0.14, see Fig. 2). Subgroup analysis by ethnic origin could not be carried out as none of the Asian studies reported OS. Subgroup analysis by duration of tamoxifen therapy showed that the pooled HR for 5 years of tamoxifen was 2.50 (95% CI 0.76–8.20, P = 0.13), for less than 5 years of tamoxifen was 1.59 (95% CI 0.93–2.73, P = 0.09) and for an unspecified duration was 1.11 (95% CI 0.86–1.44, P = 0.43). None of these estimates were statistically significant (see Table 3). Finally, as shown in Table 4, in function-based comparison studies, the pooled HR was 1.36 (95% CI 0.73–2.52, P = 0.34) while in genotype-based comparison studies, the pooled HR was 1.20 (95% CI 0.60–2.40, P = 0.61, see Fig. 2). Once again, the funnel plot (not shown) showed good symmetry suggesting a low-likelihood of publication bias.
Association between CYP2D6 drug inhibitors and DFS
Two studies [18, 19] were pooled for this analysis. Single study HRs ranged from 0.95 to 1.92 and were statistically significant in one study [18]. Pooled analysis showed a small and non-significant association toward worse DFS with a HR of 1.37 (95% CI 0.69–2.73, P = 0.37).
Association between CYP2D6 drug inhibitors and OS
Only one study was identified as meeting inclusion criteria. Kelly et al. [20] assessed the impact of a number of selective serotonin reuptake inhibitors on breast cancer outcome of 2430 women treated with tamoxifen. While a multivariable analysis was carried out, it was weakened by the lack of inclusion of any breast cancer prognostic factors. Results showed that the strong CYP2D6 inhibitor paroxetine was associated with worse survival in a co-administration time-dependent manner. The HR for 25% co-treatment of tamoxifen and paroxetine was 1.24 and increased to 1.91 for 75% co-treatment. It should be noted, however, that the assessment of other potent inhibitors of CYP2D6 such as fluoxetine did not show such a significant adverse outcome and this impairs the face validity of these data.
Discussion
In contrast to men and to pre-menopausal women with early breast cancer, in post-menopausal women there are numerous therapeutic options for adjuvant endocrine therapy including both tamoxifen and aromatase inhibitors (AIs). In this setting, any tool which allows prediction of superiority of one treatment over the other has clinical utility. Assessment of CYP2D6 function has been suggested as one such tool [9] and a number of studies have been carried out to assess its role. Two separate approaches of assessing CYP2D6 function have been evaluated; the role of CYP2D6 genotype as well as the effect of concurrent administration of drug inhibitors of CYP2D6 on outcome of women with early breast cancer.
Pooled analysis of CYP2D6 genotype showed that there was a non-significant trend towards improved DFS in patients with normal (at least one wt allele) CYP2D6 genotype (HR 1.41, P = 0.10). However, as mentioned above, due to the heterogeneity in comparison group definition, such pooling is flawed. Studies were therefore dichotomized according to the Bradford criteria [8] into those that compared normal CYP2D6 function to reduced function (i.e., function-based approach) and those comparing normal to a combination of normal and reduced function groups (i.e., genotype-based approach). When this analysis was undertaken, there was a decrease in DFS trending towards statistical significance in the reduced function group compared to those with genotypes conferring normal function (HR 2.07, 95% CI 0.96–4.49, P = 0.06). This finding was not seen in the combined normal and reduced function group (HR 1.06, 95% CI 0.6–1.86, P = 0.85) likely due to the effect being diminished by the inclusion of patients who were heterozygous for wt alleles, a group considered to have normal CYP2D6 function. Results for OS, showed that genotypes associated with reduced CYP2D6 function were again associated with a non-significant detrimental outcome (HR 1.24, 95% CI 0.93–1.67, P = 0.14). These data are suggestive that CYP2D6 genotypes, which confer reduced enzyme function, may be associated with increased risk of breast cancer recurrence with adjuvant tamoxifen therapy, although the magnitude of this association appears both relatively small and highly variable between different patients. Clinical relevance of this association may be further diminished by the fact, that definition of breast cancer recurrences often includes loco-regional recurrences, and new primary breast tumors.
Pooled analysis of CYP2D6 drug inhibitors showed that there was a non-significant association between DFS and concomitant use of CYP2D6 inhibitors (HR 1.37, 95% CI 0.69–2.73, P = 0.37). Finally, only one poor quality study contributed to assessment of the effect of CYP2D6 drug inhibitors on OS [20]. This study showed significantly poorer OS among women taking paroxetine, but not other potent CYP2D6 inhibitors and interpretation of this study should be undertaken with caution.
Women who are poor metabolizers (i.e., homozygous for non-functional alleles) still produce some endoxifen and 4-HT [9]. However, it is unknown whether decreased plasma levels of metabolites of tamoxifen still have clinically relevant anti-cancer effect. The Oxford overview suggested no difference in outcome between trials using higher doses of tamoxifen (e.g., 30–40 mg/day) as compared to the more commonly used dose of 20 mg per day [38]. It is possible that lower doses than 20 mg/day of tamoxifen are equally effective, at least in patients considered to have normal CYP2D6 function (i.e., homozygous or heterozygous for wt alleles). It had been demonstrated in a small randomized clinical trial that lower doses of tamoxifen (e.g., 1 or 5 mg/day) show similar effects on the tumor proliferation marker Ki67 as higher doses (20 mg/day) [39]. It is, however, not known whether this effect has clinical relevance.
Our present analysis suggests there is a potential detrimental effect of impaired metabolism of tamoxifen on the basis of CYP2D6 genotype or concomitant administration of tamoxifen and CYP2D6 drug inhibitors. However, the magnitude of this effect seems relatively small and may not be clinically relevant in all scenarios, especially in women with a low-risk breast cancer. As compared to women with a low-risk disease, suboptimal or inefficient endocrine therapy can be associated with worse outcome in women with high-risk disease. In post-menopausal women with high-risk disease, upfront use of AIs is a reasonable alternative to tamoxifen, irrespective of CYP2D6 genotype [40, 41]. It remains unclear whether the advantage of upfront AIs over tamoxifen is mainly due to the minority of patients with CYP2D6 genotypes associated with reduced function as suggested by a model by Punglia [42]. Results of further analysis of adjuvant clinical trials with AIs, will hopefully answer this question and are eagerly awaited. In pre-menopausal women, tamoxifen is still the gold standard and clinical trials evaluating AIs in combination with ovarian ablation are ongoing.
The data described above do have limitations. All the individual studies from which data were extracted were retrospective and even though most studies used multivariable analysis to control for breast cancer prognostic factors, it is possible that other unknown confounders may bias the data. Importantly, none of the included studies assessed the impact of compliance with medication. Recent data show that patients who have normal CYP2D6 function are more likely to discontinue their tamoxifen possibly due to an increase in adverse effects [43]. It is possible that such reduced compliance in these patients may reduce the beneficial effect [19] of their metabolic status compared with those who have reduced CYP2D6 function and are more compliant. Furthermore, in this study we have pooled data from patients who have both intermediate and poor CYP2D6 function (i.e., reduced CYP2D6 function). It could be argued that such pooling is inappropriate. However, the aim of this study was to assess whether reduced CYP2D6 function was associated with detrimental breast cancer outcomes. We feel that the methodology addressed this aim sufficiently.
Of interest, similar findings were recently obtained from an analysis of published and unpublished studies by Goetz et al. [27]. This study, which evaluated the association between CYP2D6 genotype and breast cancer outcome is currently not publically available but data were obtained from the primary author. Results of this analysis, also showed a non-significant association between CYP2D6 genotype and DFS (HR = 1.07; 95% CI 0.88–1.31, P = 0.51), and OS (HR = 0.92; 95% CI 0.72–1.18, P = 0.50). It should be noted that similar to the current study, this analysis could also be substantially biased by retrospective data included in the analysis.
In conclusion, according to the results of pooled analysis of retrospective studies, the effect of CYP2D6 genotype on breast cancer seems to be relatively small and may not warrant testing of CYP2D6 genotype in all women with endocrine positive breast cancer. While in low-risk women effect of CYP2D6 genotype may not be clinically relevant, upfront use of AIs is a reasonable alternative to tamoxifen in high-risk post-menopausal women, irrespective of CYP2D6 genotype. Clear cut-offs for decisions of whether to perform CYP2D6 genetic testing or not cannot be recommended on the basis of the data presented. However, CYP2D6 genetic testing might be especially valuable in high risk post-menopausal women if they start tamoxifen as part of the sequencing/switching strategy, or in those already on an AI but having side effects for which switching to tamoxifen is considered. Tamoxifen remains the standard of care in pre-menopausal women and in men and therefore, there is currently no data to support CYP2D6 testing in these groups. Data evaluating the impact of CYP2D6 drug inhibitors remains insufficient for formal evaluation. However, in most settings, there are alternative drugs to potent inhibitors of CYP2D6 on the market and therefore, it would appear reasonable to avoid such drugs in combination with tamoxifen. Hopefully, prospective studies and retrospective analysis of adjuvant studies of AIs will provide us with definitive answers regarding the impact of CYP2D6 function in women with breast cancer on tamoxifen.
References
Dehal SS, Kupfer D (1997) CYP2D6 catalyzes tamoxifen 4-hydroxylation in human liver. Cancer Res 57:3402–3406
Desta Z, Ward BA, Soukhova NV, Flockhart DA (2004) Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro: prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther 310:1062–1075
Johnson MD, Zuo H, Lee KH, Trebley JP, Rae JM, Weatherman RV, Desta Z, Flockhart DA, Skaar TC (2004) Pharmacological characterization of 4-hydroxy-N-desmethyl tamoxifen, a novel active metabolite of tamoxifen. Breast Cancer Res Treat 85:151–159
Lim YC, Li L, Desta Z, Zhao Q, Rae JM, Flockhart DA, Skaar TC (2006) Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther 318:503–512
Wu XL, Hawse JR, Subramaniam M, Goetz MP, Ingle JN, Spelsberg TC (2009) The tamoxifen metabolite, endoxifen, is a potent antiestrogen that targets estrogen receptor alpha for degradation in breast cancer cells. Cancer Res 69:1722–1727
Stearns V, Johnson MD, Rae JM, Morocho A, Novielli A, Bhargava P, Hayes DF, Desta Z, Flockhart DA (2003) Active tamoxifen metabolite plasma concentrations after coadministration of tamoxifen and the selective serotonin reuptake inhibitor paroxetine. J Natl Cancer Inst 95:1758–1764
Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C (2007) Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther 116:496–526
Bradford LD (2002) CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics 3:229–243
Jin Y, Desta Z, Stearns V, Ward B, Ho H, Lee KH, Skaar T, Storniolo AM, Li L, Araba A, Blanchard R, Nguyen A, Ullmer L, Hayden J, Lemler S, Weinshilboum RM, Rae JM, Hayes DF, Flockhart DA (2005) CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst 97:30–39
Lim HS, Ju LH, Seok LK, Sook LE, Jang IJ, Ro J (2007) Clinical implications of CYP2D6 genotypes predictive of tamoxifen pharmacokinetics in metastatic breast cancer. J Clin Oncol 25:3837–3845
Goetz MP, Rae JM, Suman VJ, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Flockhart DA, Desta Z, Perez EA, Ingle JN (2005) Pharmacogenetics of tamoxifen biotransformation is associated with clinical outcomes of efficacy and hot flashes. J Clin Oncol 23:9312–9318
Kiyotani K, Mushiroda T, Imamura CK, Hosono N, Tsunoda T, Kubo M, Tanigawara Y, Flockhart DA, Desta Z, Skaar TC, Aki F, Hirata K, Takatsuka Y, Okazaki M, Ohsumi S, Yamakawa T, Sasa M, Nakamura Y, Zembutsu H (2010) Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol 28:1287–1293
Schroth W, Goetz MP, Hamann U, Fasching PA, Schmidt M, Winter S, Fritz P, Simon W, Suman VJ, Ames MM, Safgren SL, Kuffel MJ, Ulmer HU, Bolander J, Strick R, Beckmann MW, Koelbl H, Weinshilboum RM, Ingle JN, Eichelbaum M, Schwab M, Brauch H (2009) Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. JAMA 302:1429–1436
Xu Y, Sun Y, Yao L, Shi L, Wu Y, Ouyang T, Li J, Wang T, Fan Z, Fan T, Lin B, He L, Li P, Xie Y (2008) Association between CYP2D6*10 genotype and survival of breast cancer patients receiving tamoxifen treatment. Ann Oncol 19:1423–1429
Nowell SA, Ahn JY, Rae JM, Scheys JO, Trovato A, Sweeney C, MacLeod SL, Kadlubar FF, Ambrosone CB (2005) Association of genetic variation in tamoxifen-metabolizing enzymes with overall survival and recurrence of disease in breast cancer patients. Breast Cancer Res Treat 91:249–258
Schroth W, Antoniadou L, Fritz P, Schwab M, Muerdter T, Zanger UM, Simon W, Eichelbaum M, Brauch H (2007) Breast cancer treatment outcome with adjuvant tamoxifen relative to patient CYP2D6 and CYP2C19 genotypes. J Clin Oncol 25:5187–5193
Wegman P, Elingarami S, Carstensen J, Stal O, Nordenskjold B, Wingren S (2007) Genetic variants of CYP3A5, CYP2D6, SULT1A1, UGT2B15 and tamoxifen response in postmenopausal patients with breast cancer. Breast Cancer Res 9:R7
Aubert RE, Stanek EJ, Yao J, Teagarden JR, Subar M, Epstein RS, Skaar TC, Desta Z, Flockhart DA (2009) Risk of breast cancer recurrence in women initiating tamoxifen with CYP2D6 inhibitors. J Clin Oncol (Meeting Abstracts) 27:CRA508
Dezentje V, Van Blijderveen NJ, Gelderblom H, Putter H, Van Herk-Sukel MP, Casparie MK, Egberts AC, Nortier JW, Guchelaar HJ (2009) Concomitant CYP2D6 inhibitor use and tamoxifen adherence in early-stage breast cancer: a pharmacoepidemiologic study. J Clin Oncol (Meeting Abstracts) 27:CRA509
Kelly CM, Juurlink DN, Gomes T, Duong-Hua M, Pritchard KI, Austin PC, Paszat LF (2010) Selective serotonin reuptake inhibitors and breast cancer mortality in women receiving tamoxifen: a population based cohort study. BMJ 340:c693
Harris RP, Helfand M, Woolf SH, Lohr KN, Mulrow CD, Teutsch SM, Atkins D (2001) Current methods of the US Preventive Services Task Force: a review of the process. Am J Prev Med 20:21–35
Deeks JJ, Higgins, JPT, Altman DG (eds) (2006) Analysing and presenting results. Cochrane Handbook for Systematic Reviews of Interventions 4.2.5. In: The Cochrane Library, Chichester, UK, John Wiley & Sons, Ltd
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Gonzalez-Santiago S, Zarate R, Haba-Rodriguez J, Gomez A, Bandres E, Borrega P, Garcia-Foncillas J, Aranda E (2006) Genetic polymorphism CYP2D6(*4) interaction in clinical outcomes of tamoxifen-treated breast cancer patients. Ann Oncol 17:62–63
Goetz MP, Knox SK, Suman VJ, Rae JM, Safgren SL, Ames MM, Visscher DW, Reynolds C, Couch FJ, Lingle WL, Weinshilboum RM, Fritcher EG, Nibbe AM, Desta Z, Nguyen A, Flockhart DA, Perez EA, Ingle JN (2007) The impact of cytochrome P450 2D6 metabolism in women receiving adjuvant tamoxifen. Breast Cancer Res Treat 101:113–121
Newman WG, Hadfield KD, Latif A, Roberts SA, Shenton A, McHague C, Lalloo F, Howell S, Evans DG (2008) Impaired tamoxifen metabolism reduces survival in familial breast cancer patients. Clin Cancer Res 14:5913–5918
Goetz MP, Berry DA, Klein TE (2009) Adjuvant tamoxifen treatment outcome according to cytochrome P450 2D6 (CYP2D6) phenotype in early stage breast cancer: findings from the International Tamoxifen Pharmacogenomics Consortium. Cancer Res 69:492S–493S
Thompson A, Quinlan P, Bray S, Johnson A, Nikoloff M, Fontecha M, Ferraldeschi R, Howell A, Lawrence J, Newman W (2009) CYP2D6 genotype affects outcome in postmenopausal breast cancer patients treated with tamoxifen monotherapy. In: Proc Am Soc Clin Oncol Breast Cancer Symposium, Abstract 35.
Okishiro M, Taguchi T, Kim SJ, Shimazu K, Tamaki Y, Noguchi S (2009) Genetic polymorphisms of CYP2D6*10 and CYP2C19*2,*3 are not associated with prognosis, endometrial thickness, or bone mineral density in japanese breast cancer patients treated with adjuvant tamoxifen. Cancer 115:952–961
Kiyotani K, Mushiroda T, Sasa M, Bando Y, Sumitomo I, Hosono N, Kubo M, Nakamura Y, Zembutsu H (2008) Impact of CYP2D6*10 on recurrence-free survival in breast cancer patients receiving adjuvant tamoxifen therapy. Cancer Sci 99:995–999
Abraham JE, Maranian M, Driver K, Greenberg D, Shah MN, Earl HM, Dunning AM, Pharoah PPDP, Caldas C (2009) Cytochrome P450 2D6 variants and their association with breast cancer survival. Cancer Res 69:229S
Wegman P, Vainikka L, Stal O, Nordenskjold B, Skoog L, Rutqvist LE, Wingren S (2005) Genotype of metabolic enzymes and the benefit of tamoxifen in postmenopausal breast cancer patients. Breast Cancer Res 7:R284–R290
Toyama T, Yamashita H, Sugiura H, Kondo N, Iwase H, Fujii Y (2009) No association between CYP2D6*10 genotype and survival of node-negative Japanese breast cancer patients receiving adjuvant tamoxifen treatment. Jpn J Clin Oncol 39:651–656
Ramon y Cajal T, Altes A, Pare L, del Rio E, Alonso C, Barnadas A, Baiget M (2010) Impact of CYP2D6 polymorphisms in tamoxifen adjuvant breast cancer treatment. Breast Cancer Res Treat 119:33–38
Bijl MJ, van Schaik RHN, Lammers LA, Hofman A, Vulto AG, van Gelder T, Stricker BHC, Visser LE (2009) The CYP2D6*4 polymorphism affects breast cancer survival in tamoxifen users. Breast Cancer Res Treat 118:125–130
Rae JM, Thibert J, Sikora MJ, Johnson MD, Skene A, A’Hern R, Smith IE, Dowsett M (2009) Lack of correlation between genetic polymorphisms in tamoxifen metabolizing enzymes with primary endpoints in the IMPACT trial. Cancer Res 69:595S–596S
Lash TL, Cronin-Fenton D, Ahern TP, Rosenberg CL, Lunetta KL, Silliman RA, Hamilton-Dutoit S, Garne JP, Ewertz M, Sorensen HT, Pedersen L (2010) Breast cancer recurrence risk related to concurrent use of SSRI antidepressants and tamoxifen. Acta Oncol 49:305–312
(1998) Tamoxifen for early breast cancer: an overview of the randomised trials. Early Breast Cancer Trialists’ Collaborative Group. Lancet 351:1451–1467
Decensi A, Robertson C, Viale G, Pigatto F, Johansson H, Kisanga ER, Veronesi P, Torrisi R, Cazzaniga M, Mora S, Sandri MT, Pelosi G, Luini A, Goldhirsch A, Lien EA, Veronesi U (2003) A randomized trial of low-dose tamoxifen on breast cancer proliferation and blood estrogenic biomarkers. J Natl Cancer Inst 95:779–790
Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M (2008) Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol 9:45–53
Mouridsen H, Giobbie-Hurder A, Goldhirsch A, Thurlimann B, Paridaens R, Smith I, Mauriac L, Forbes JF, Price KN, Regan MM, Gelber RD, Coates AS (2009) Letrozole therapy alone or in sequence with tamoxifen in women with breast cancer. N Engl J Med 361:766–776
Punglia RS, Burstein HJ, Winer EP, Weeks JC (2008) Pharmacogenomic variation of CYP2D6 and the choice of optimal adjuvant endocrine therapy for postmenopausal breast cancer: a modeling analysis. J Natl Cancer Inst 100:642–648
Lynn HN, Rae JM, Li L, Azzouz F, Skaar TC, Desta Z, Sikora MJ, Philips S, Nguyen AT, Storniolo AM, Hayes DF, Flockhart DA, Stearns V (2009) Association between CYP2D6 genotype and tamoxifen-induced hot flashes in a prospective cohort. Breast Cancer Res Treat 117:571–575
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Seruga, B., Amir, E. Cytochrome P450 2D6 and outcomes of adjuvant tamoxifen therapy: results of a meta-analysis. Breast Cancer Res Treat 122, 609–617 (2010). https://doi.org/10.1007/s10549-010-0902-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-010-0902-3