Abstract
High levels of the cyclooxygenase-2 (COX-2) protein have been associated with invasion and metastasis of breast tumors. Both prostaglandin E2 (PGE2) and interleukin-8 (IL-8) have been shown to mediate the invasive activity of COX-2 in breast cancer cells. Here we expand these studies to determine how COX-2 uses PGE2 and IL-8 to induce breast cancer cell invasion. We demonstrated that PGE2 and IL-8 decreased the expression of the tumor suppressor protein Programmed Cell Death 4 (PDCD4). We hypothesized that suppression of PDCD4 expression is vital to the invasive activity of PGE2 and IL-8. In MCF-7 cells overexpressing PDCD4 (MCF-7/PDCD4), PGE2 and IL-8 failed to induce invasion, in contrast to the parental MCF-7 cells, thus indicating that PDCD4 blocks breast cancer cell invasion. MCF-7/PDCD4 cells produced higher levels of the Tissue Inhibitor of Metalloproteinases-2 (TIMP-2) than the parental cells. Silencing TIMP-2 mRNA in MCF-7/PDCD4 cells reversed the anti-invasive effects of PDCD4, allowing PGE2 and IL-8 to induce the invasion of these cells. Here we report the novel findings that suppression of PDCD4 expression is vital for the invasive activity of COX-2 mediated by PGE2 and IL-8, and that PDCD4 increases TIMP-2 expression to inhibit breast cancer cell invasion.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Metastasis is the primary cause of breast cancer mortality. The 5-year survival rate for women diagnosed with localized breast cancer is 98%, which contrasts dramatically with the 27% survival rate of women diagnosed with distant metastasis breast cancer [1]. Understanding the mechanisms of breast cancer metastasis is paramount to the development of novel therapeutic strategies to stop the spreading of the disease and to prolong patients’ lives.
Over 40% of human breast tumors have been found to have high expression of the cyclooxygenase-2 (COX-2) protein [2–4]. High COX-2 expression in breast tumors has been correlated with poor survival, distant metastasis and lower local-regional control of disease [2, 3]. Ectopic expression of COX-2 in breast cancer cell lines was found to increase the production of prostaglandin E2 (PGE2) and interleukin-8 (IL-8), which are essential to COX-2-induced breast cancer invasion [5–7]. However, which downstream factors are inhibited by the COX-2/ PGE2/IL-8-mediated pathway to induce breast cancer cell invasion are not known.
Zhang and DuBois [8] demonstrated that inhibition of COX-2 by the selective inhibitor NS398 increased the mRNA expression of Programmed Cell Death 4 (PDCD4) in colon cancer cells. PDCD4 suppresses the in vitro transformation of mouse keratinocytes induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) [9, 10] and the promotion and progression of skin carcinogenesis in response to 7,12-dimethylbenz(a)anthracene/TPA in animal models [11]. PDCD4 interacts with translation initiation factors eIF4A and eIF4G and inhibits AP-1 transactivation [9, 12]. PDCD4 has also been shown to induce expression of the cyclin-dependent kinase inhibitor p21 [13]. These findings indicate PDCD4 has tumor suppressor activity. Loss of PDCD4 expression has been found in several types of human cancer cell lines [14], primary lung cancer [15], primary pancreatic cancer [16], hepatocarcinoma [17], colon carcinoma [18], and gliomas [19]. Recently, Wen et al. [20] reported that compared to normal breast epithelial cells, PDCD4 expression is only slightly decreased in ductal carcinoma in situ samples, but is markedly decreased in invasive ductal carcinoma samples. This study implicates that PDCD4 may have an important role in regulating the invasive activity of breast tumors. Increased expression of PDCD4 has been shown to decrease invasion of colon cancer cells [21, 22], whereas downregulation of PDCD4 has been shown to promote invasion of colon cancer cells [23]. However, whether PDCD4 has a role in breast cancer cell invasion has not been reported.
Our unpublished cDNA microarray data revealed that overexpression of COX-2 led to decreased PDCD4 mRNA levels in breast cancer cells. Since COX-2 induces invasion and decreases PDCD4 expression, we hypothesize that COX-2 decreases PDCD4 expression as a mechanism to increase breast cancer cell invasion. Here we determine the effects and the mechanisms by which PDCD4 suppresses breast cancer cell invasion.
Materials and methods
Cells
MCF-7 cells were obtained from The American Type Culture Collection (Manassas, VA, USA). MCF-7/COX-2 cells were generated as previously described [24]. FUGENE 6 Transfection Reagent (Roche Diagnostics, Indianapolis, IN, USA) was used to transfect MCF-7 cells with pcDNA3.1 plasmids (Invitrogen Corporation, Grand Island, NY, USA) either empty (MCF-7/Vector) or encoding the human PDCD4 gene [9] (MCF-7/PDCD4). MCF-7/Vector and MCF-7/PDCD4 stable clones were selected based on their resistance to 500 μg/ml geneticin (Invitrogen Corporation). All cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM)/F12 (Invitrogen Corporation) supplemented with 5% heat-inactivated fetal bovine serum (FBS, Invitrogen Corporation). MCF-7/Vector, MCF-7/COX-2 and MCF-7/PDCD4 cells were routinely grown in media supplemented with 500 μg/ml geneticin, but this antibiotic was removed during experiments.
Western blots
Western blots were performed on protein lysates obtained from exponentially growing MCF-7 parental cells and MCF-7/COX-2 cells. Thirty and 50 μg of protein lysates were used for COX-2 and PDCD4 western blots, respectively. Levels of COX-2 and PDCD4 were normalized to that of β-actin. Western blot was used to determine the levels of PDCD4 levels in untreated MCF-7 cells, MCF-7 cells treated with PGE2 (Cayman Chemical) and MCF-7 cells treated with IL-8 (Sigma-Aldrich). MCF-7 parental cells (4 × 105) were cultured in 5% FBS-supplemented media for 24 h, and then changed to serum free media for 24 h. After serum starvation, cells were either left untreated or treated with 10 μM PGE2 or 100 ng/ml IL-8 for 24 h. The expression of PDCD4 was similarly detected in MCF-7/PDCD4, MCF-7/Vector and MCF-7 cells. Monoclonal antibodies specific for COX-2 and β-actin were purchased from Cayman Chemical (Ann Harbor, MI, USA) and Sigma-Aldrich (St. Louis, MO, USA), respectively. Polyclonal antibody specific for PDCD4 was obtained from Dr. Nancy Colburn [9]. Protein bands were visualized by enhanced chemiluminescence (GE Healthcare, Piscataway, NJ, USA). Images were scanned and quantified by an Alpha Innotech densitometer using the Alpha Imager application program (San Leandro, CA, USA).
Matrigel invasion assays
The invasiveness of the cells was assessed by Matrigel invasion assays as previously described [6]. Briefly, MCF-7, MCF-7/Vector and MCF-7PDCD4 cells (4 × 105) were cultured and treated with 10 μM PGE2 or 100 ng/ml IL-8 for 24 h prior to the invasion assay. All cells were then collected and washed. Cells (4 × 105) were resuspended in 1 ml of serum free DMEM/F12 medium and added into transwell inserts (8-μm pore-size, Fisher Scientific, Middleton, VA, USA) coated with a Matrigel basement membrane (0.7 mg/ml; BD Bioscience, Bedford, MA, USA). The lower chambers were filled with 2 ml of DMEM/F12 medium supplemented with 10% FBS. Cells in the transwell inserts were then treated with either PGE2 or IL-8, at the same concentrations used before. Seventy-two hours later, non-invading cells on the upper surface of the filter were removed with cotton swabs. Cells that had invaded through the Matrigel onto the lower side of the filters were fixed, stained with Hema-3 Stain System (Fisher Scientific), and photographed. For each filter, the number of invaded cells was counted in five fields. The invasiveness of the cells is expressed as the mean number of cells of triplicate measurements that invaded to the lower side of the filter. Then, two-sample t-tests were used for comparison of the number of invading cells. The overall significance level of 0.05 was used on comparative analyses.
Tissue Inhibitor of Metalloproteinase-2 (TIMP-2) ELISA assay
MCF-7 or MCF-7/PDCD4 cells (4 × 105) were cultured in 5% FBS-supplemented media for 24 h before changed to serum free media. After 24 h of serum starvation, cells were treated with PGE2 or IL-8 as described above. The next day, conditioned media was collected, clarified by centrifugation and concentrated by using Centricon Ultracel YM-10 spin columns (Millipore, Bedford, MA, USA). An ELISA assay (EMD Biosciences, La Jolla, CA, USA) was used to determine the levels of TIMP-2 secreted into the supernatants. Samples were run in triplicates and TIMP-2 levels were normalized to cell number and expressed as means ± standard deviations (s.d.).
Reverse transcriptase polymerase chain reaction (RT-PCR)
MCF-7 and MCF-7/PDCD4 cells were cultured and treated with PGE2 or IL-8 as described above. Total RNA was extracted using the RNeasy Mini Kit (Qiagen Inc, Valencia, CA, USA) and 10 ng were used in the RT-PCR using SuperScript III One-Step RT-PCR System (Invitrogen Corporation). The sequences of the urokinase plasminogen activator receptor (uPAR) primers were: forward, 5′-GAG CTG GTG GAG AAA AGC TG-3′ and reverse, 5′-TGT TGC AGC ATT TCAGGA AG-3′. The sequences of the TIMP-2 primers were: forward, 5′-GGT CTC GCT GGA CGT TGG AG-3′ and reverse, 5′-GGA GCC GTC ACT TCT CTT G-3′. The sequences of the GAPDH primers were: forward, 5′-GCC AAG GTC ATC CAT GAC AAC-3′ and reverse, 5′-GTC CAC CAC CCT GTT GCT GTA-3′. The PCR conditions for all reactions were: 55°C for 30 min, 94°C for 2 min, 30 cycles of 94°C for 15 s, 57°C for 30 s, 68°C for 1 min, and 68°C for 5 min.
TIMP-2 knockdown with siRNA
MCF-7/PDCD4 cells (2 × 105 cells/well) were plated in 6-well plates. The next day, cells were transfected with 200 nM human TIMP-2 or non-silencing control siRNA (Dharmacon Inc., Lafayette, CO, USA) using Fugene 6 Transfection Reagent. Forty-eight hours later, cells were collected and RT-PCR as described above was used to confirm that the level of TIMP2 mRNA was reduced in cells transfected with TIMP-2 siRNA. Similarly after cells were transfected with TIMP-2 or non-silencing control siRNA, cells were collected and invasion assays as described above were performed. After invasion assays were set up, cells were re-transfected with TIMP-2 or non-silencing control siRNA, treated with either PGE2 or IL-8 as described above, and incubated for 72 h.
Results and discussion
Activation of the COX-2/PGE2/IL-8 pathway decreases PDCD4 expression in breast cancer cells
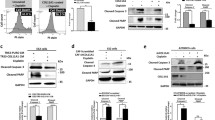
Western blot analysis showed elevated COX-2 expression in the three MCF-7/COX-2 clones tested (clones 8, 12, and 13) (Fig. 1a). Western blot analysis also showed that the level of PDCD4 protein expression in the MCF-7/COX-2 clones was 55 to 68% lower than that in the MCF-7 parental cells (Fig. 1a). Similarly, when MCF-7 cells were treated with PGE2 or IL-8, PDCD4 protein levels were decreased by 67 and 35%, respectively (Fig. 1b). These results indicate that activation of the COX-2/PGE2/IL-8 pathway suppresses PDCD4 expression.
COX-2, PGE2, and IL-8 decreased PDCD4 protein levels. (a) The expression of COX-2 and PDCD4 proteins in three independent clones of MCF-7/COX-2 cells and MCF-7 parental cells was determined by immunoblotting equal amounts of total cell lysates with COX-2 monoclonal and PDCD4 polyclonal antibodies, respectively. COX-2 and PDCD4 levels were analyzed by densitometry, normalized to β-actin level, and expressed as percentages of MCF-7 parental cells. (b) MCF-7 cells were untreated or treated with 10 μM PGE2 or 100 ng/ml IL-8 for 24 h. Protein lysates were immunoblotted with PDCD4 and β-actin antibodies. PDCD4 levels were analyzed by densitometry, normalized to the corresponding β-actin levels, and the ratios were expressed as percentage of untreated cells
PDCD4 blocks PGE2- and IL-8-induced breast cancer cell invasion
To determine whether PDCD4 has a role in COX-2-mediated breast cancer invasion, MCF-7 cells were stably transfected with plasmids encoding the human PDCD4 cDNA (MCF-7/PDCD4). MCF-7 cells were also transfected with empty vector (MCF-7/Vector) as a control. Western blot was performed to confirm that MCF-7/PDCD4 cells had higher PDCD4 levels than the parental and the vector control cells (Fig. 2). Densitometric analysis showed that PDCD4 expression in MCF-7/PDCD4 cells was approximately 9-fold higher than that in the MCF-7 parental or the MCF-7/Vector cells. PGE2 and IL-8 increased the invasion of MCF-7 and MCF-7/Vector cells across a Matrigel basement membrane (Fig. 3a, b). However under identical conditions, PGE2 and IL-8 did not increase the invasion of MCF-7/PDCD4 cells (Fig. 3a, b). Our results indicate that PDCD4 blocks PGE2- and IL-8-induced invasion of breast cancer cells. Flow cytometric analysis did not show any significant difference in cell cycling between transfected and parental MCF-7 cells (data not shown). Thus, difference in the number of invaded cells is attributed to the cell’s invasive activity and not its proliferation activity.
PDCD4 expression was higher in MCF-7/PDCD4 cells than in MCF-7 and MCF-7/Vector cells. Western blot was used to compare the expression of PDCD4 in MCF-7 parental cells, MCF-7 cells stably transfected with plasmid encoding the PDCD4 gene (MCF-7/PDCD4), and MCF-7 cells stably transfected with empty vector (MCF-7/Vector). β-actin was used as a loading control. PDCD4 levels were analyzed by densitometry, normalized to the corresponding β-actin levels, and expressed as percentage of MCF-7 parental cells
PDCD4 inhibited PGE2- and IL-8-induced breast cancer cells invasion. (a) A matrigel invasion assay was performed using MCF-7, MCF-7/Vector and MCF-7/PDCD4 cells. All cells were treated with the indicated concentrations of PGE2 or IL-8, and incubated for 72 h. Cells that had invaded through the Matrigel onto the lower side of the filters were fixed, stained, and photographed at 150× magnification. (b) The number of invaded cells for each filter was counted in five fields. The invasiveness of the cells is expressed as the number of cells that invaded to the lower side of the filter. Columns indicate the mean of triplicate measurements ± standard deviation. An asterisk (*) indicates a significant increase in the number of invaded cells relative to the untreated cells, P < 0.05, determined by Student t-test
PDCD4 increases TIMP-2 expression to inhibit breast cancer invasion
Leupold et al. [21] demonstrated that PDCD4 inhibited the invasion of colon cancer cells by a mechanism that involved decreased transcription of uPAR. We determined the mRNA levels of uPAR in MCF-7 and MCF-7/PDCD4 cells. The basal level of the uPAR mRNA was higher in MCF-7/PDCD4 cells than in MCF-7 cells (Fig. 4). Similarly upon PGE2 or IL-8 treatment, the uPAR mRNA level was higher in MCF-7/PDCD4 cells than in MCF-7 cells (Fig. 4). These results indicate that, in contrast to colon cancer cells, PDCD4 does not suppress uPAR transcription in breast cancer cells.
uPAR transcription levels remained unchanged upon IL-8 or PGE2 treatment. MCF-7 and MCF-7/PDCD4 cells were treated with 10 μM PGE2 or 100 ng/ml IL-8 for 24 h. RNA was isolated and RT-PCR was used to determine the levels of uPAR transcript. GAPDH levels were included as a loading control. uPAR levels were analyzed by densitometry, normalized to the corresponding GAPDH levels and expressed as uPAR/GAPDH ratio
To investigate the mechanisms by which PDCD4 inhibits breast cancer invasion, a protein array was used to analyze the levels of metalloproteinases (MMPs) and tissue inhibitor of metalloproteinases (TIMPs) in the conditioned medium of MCF-7 and MCF-7/PDCD4 cells. The array results revealed that the level of TIMP-2 was higher in MCF-7/PDCD4 than MCF-7 cells (data not shown). The balance between MMPs/TIMPs is an important determinant in the invasion process. TIMPs act by binding to MMPs and reversibly inhibiting their extracellular matrix degradation activity. An ELISA was performed to confirm that MCF-7/PDCD4 cells secreted higher amount of TIMP-2 than MCF-7 cells. TIMP-2 was not detected in the media of untreated MCF-7 cells, nor in the media of PGE2- or IL-8-treated MCF-7 cells (Fig. 5a). However, TIMP-2 was detected at a concentration of 4–6 ng/ml in the media of MCF/PDCD4 cells (Fig. 5a), indicating that PDCD4 increases TIMP-2 secretion.
PDCD4 increased TIMP-2 expression to inhibit breast cancer invasion. (a) PDCD4 significantly increased the secretion of TIMP-2. The levels TIMP-2 secreted into the supernatants of MCF-7 or MCF-7/PDCD4 cultures were analyzed by ELISA. Samples were run in triplicates and TIMP-2 levels were normalized to cell number and expressed as means ± s.d. ND = not detected. (b) TIMP-2 siRNA reduced the expression of TIMP2 mRNA in MCF-7/PDCD4 cells. MCF-7/PDCD4 cells were transfected with non-silencing control siRNA or siRNA targeted to human TIMP2. RT-PCR was used to determine the levels of TIMP2 mRNA in MCF-7 and MCF-7/PDCD4 cells transfected with siRNA. GAPDH levels were included as a loading control. The level of TIMP2 mRNA was measured densitometrically and normalized to that of GAPDH. The ratio of TIMP-2/GAPDH obtained with the TIMP-2 siRNA was compared to that of the non-silencing control siRNA. (c) TIMP-2 downregulation reversed the anti-invasive effects of PDCD4. Matrigel invasion assays were performed using MCF-7/PDCD4 cells transfected with either non-silencing control siRNA or TIMP-2 targeted siRNA. PGE2 (10 μM) and IL-8 (100 ng/ml) were used to induce cell invasion. The number of invaded cells was counted in five fields of each filter. Columns indicate the mean of triplicate measurements ±standard deviation. Asterisks (*) indicate a significant increase in the number of invaded cells relative to untreated or control siRNA-treated cells, P < 0.05, determined by Student t-test
To determine whether TIMP-2 is essential to the PDCD4-mediated inhibition of breast cancer invasion, siRNA was used to knock down TIMP2 expression in MCF-7/PDCD4 cells (Fig. 5b). MCF-7/PDCD4 cells were transfected with TIMP-2 siRNA or non-silencing control siRNA and the levels of TIMP2 mRNA in these cells as well as in MCF-7 parental cells were determined by RT-PCR. The TIMP2 mRNA levels were significantly higher in the MCF-7/PDCD4 cells than in the MCF-7 parental cells (Fig. 5b), thus supporting our TIMP-2 ELISA data and further indicating that PDCD4 increases TIMP-2 expression transcriptionally. TIMP-2 siRNA significantly decreased TIMP2 mRNA, approximately 62%, in MCF-7/PDCD4 cells (Fig. 5b). MCF-7/PDCD4 cells were transfected with TIMP-2 siRNA or non-silencing control siRNA and then used for a Matrigel invasion assay. In the absence of PGE2 or IL-8, there was no difference in the number of invaded MCF-7/PDCD4 cells transfected with TIMP-2 siRNA and the number of invaded MCF-7/PDCD4 cells transfected with control siRNA (Fig. 5c). However in the presence of PGE2 or IL-8, the number of invaded MCF-7/PDCD4 cells transfected with TIMP-2 siRNA was 2.5-fold higher than the number of invaded MCF-7/PDCD4 cells tranfected with control siRNA (Fig. 5c). TIMP-2 knock down reversed the suppressive effects of PDCD4 on PGE2- and IL-8-induced invasion, indicating that TIMP-2 is essential to the anti-invasive effects of PDCD4 in breast cancer cells. TIMP-2 has been shown to inhibit the invasiveness of breast cancer cells in vitro and in vivo. Overexpression of TIMP-2 decreased the in vitro invasion of ras-transformed breast epithelial cells [25]. Mice injected with TIMP-2-transfected MDA-MB-231 breast cancer cells had a lower number of osteolytic bone metastases and a higher survival rate than mice injected with nontransfected cells [26]. Liposome-complexed TIMP2 DNA constructs administered to MMTVneu transgenic mice reduced tumor growth and effectively inhibited the occurrence of lung metastases [27]. Our present findings are consistent with these of TIMP-2 acting as a suppressor of cell invasion. On the other hand, high levels of TIMP-2 have also been correlated with distant metastasis of breast tumors [28, 29]. TIMPs have been shown to be multifunctional proteins regulating many different cells processes, some seemly paradoxical for tumorigenesis, through MMP-dependent or independent pathways [30]. In either case, TIMP-2 can be pivotal to tumor progression. Additional studies are warranted to discover the determinants of TIMP-2 functions.
Relatively little is known about the mechanisms of suppressing PDCD4 expression in cancer. The phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) signaling pathway has been shown to induce the proteolysis of PDCD4 [31] and reduce the transcription of PDCD4 [32]. MicroRNA-21 has been reported to block PDCD4 expression transcriptionally [33] and post-transcriptionally [34]. COX-2 has been found to suppress PDCD4 transcription [8]. Here we demonstrated that the mechanisms by which COX-2 suppresses PDCD4 expression is via PGE2 and IL-8.
Microarray analysis of the NCI60 cells revealed low levels of PDCD4 mRNA in human breast cancer cell lines [14], and immunohistochemical analysis revealed low levels of PDCD4 protein in invasive breast carcinoma samples [20]. Low levels of PDCD4 protein were inversely correlated with the anti-tumor activity of the anti-estrogen tamoxifen. PDCD4 antisense decreased tamoxifen sensitivity, whereas overexpression of PDCD4 increased breast cancer cell sensitivity to tamoxifen [14]. Here we report for the first time that suppression of PDCD4 expression is vital for the invasive activity of COX-2 mediated by PGE2 and IL-8, and that PDCD4 increases TIMP-2 expression to inhibit breast cancer cell invasion. These data strongly suggest that rescuing PDCD4 expression may have therapeutic effects against breast carcinoma.
References
Ries L, Melbert D, Krapcho M et al (2007) SEER Cancer Statistic Review, 1975–2004, National Cancer Institute. Bethesda, MD. Available via INTERNET http://seer.cancer.gov/csr/1975_2004/, based on November 2006 SEER data submission, posted to the SEER web site, 2007. Cited 2 Feb 2007
Denkert C, Winzer KJ, Muller BM et al (2003) Elevated expression of cyclooxygenase-2 is a negative prognostic factor for disease free survival and overall survival in patients with breast carcinoma. Cancer 97(12):2978–2987
Ristimaki A, Sivula A, Lundin J et al (2002) Prognostic significance of elevated cyclooxygenase-2 expression in breast cancer. Cancer Res 62(3):632–635
Soslow RA, Dannenberg AJ, Rush D et al (2000) COX-2 is expressed in human pulmonary, colonic, and mammary tumors. Cancer 89(12):2637–2645
Larkins TL, Nowell M, Singh S et al (2006) Inhibition of cyclooxygenase-2 decreases breast cancer cell motility, invasion and matrix metalloproteinase expression. BMC Cancer 6:181–193
Simeone AM, Nieves-Alicea R, McMurtry VC et al (2007) Cyclooxygenase-2 uses the protein kinase C/interleukin-8/urokinase-type plasminogen activator pathway to increase the invasiveness of breast cancer cells. Int J Oncol 30(4):785–792
Singh B, Berry JA, Shoher A et al (2005) COX-2 overexpression increases motility and invasion of breast cancer cells. Int J Oncol 26(5):1393–1399
Zhang Z, DuBois RN (2001) Detection of differentially expressed genes in human colon carcinoma cells treated with a selective COX-2 inhibitor. Oncogene 20(33):4450–4456
Yang HS, Jansen AP, Nair R et al (2001) A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene 20(6):669–676
Cmarik JL, Min H, Hegamyer G et al (1999) Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc Natl Acad Sci USA 96(24):14037–14042
Jansen AP, Camalier CE, Colburn NH (2005) Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res 65(14):6034–6041
Yang HS, Jansen AP, Komar AA et al (2003) The transformation suppressor Pdcd4 is a novel eukaryotic translation initiation factor 4A binding protein that inhibits translation. Mol Cell Biol 23(1):26–37
Goke R, Barth P, Schmidt A et al (2004) Programmed cell death protein 4 suppresses CDK1/cdc2 via induction of p21(Waf1/Cip1). Am J Physiol Cell Physiol 287(6):C1541–C1546
Jansen AP, Camalier CE, Stark C et al (2004) Characterization of programmed cell death 4 in multiple human cancers reveals a novel enhancer of drug sensitivity. Mol Cancer Ther 3(2):103–110
Chen Y, Knosel T, Kristiansen G et al (2003) Loss of PDCD4 expression in human lung cancer correlates with tumour progression and prognosis. J Pathol 200(5):640–646
Ma G, Guo KJ, Zhang H et al (2005) Expression of programmed cell death 4 and its clinicopathological significance in human pancreatic cancer. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 27(5):597–600
Zhang H, Ozaki I, Mizuta T et al (2006) Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene 25(45):6101–6112
Mudduluru G, Medved F, Grobholz R et al (2007) Loss of programmed cell death 4 expression marks adenoma-carcinoma transition, correlates inversely with phosphorylated protein kinase B, and is an independent prognostic factor in resected colorectal cancer. Cancer 110(8):1697–1707
Gao F, Zhang P, Zhou C et al (2007) Frequent loss of PDCD4 expression in human glioma: possible role in the tumorigenesis of glioma. Oncol Rep 17(1):123–128
Wen YH, Shi X, Chiriboga L et al (2007) Alterations in the expression of PDCD4 in ductal carcinoma of the breast. Oncol Rep 18(6):1387–1393
Leupold JH, Yang HS, Colburn NH et al (2007) Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene 26(31):4550–4562
Yang HS, Matthews CP, Clair T et al (2006) Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol Cell Biol 26(4):1297–1306
Wang Q, Sun Z, Yang HS (2008) Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta-catenin/Tcf and AP-1-dependent transcription in colon carcinoma cells. Oncogene 27(11):1527–1535
Tari AM, Simeone AM, Li YJ et al (2005) Cyclooxygenase-2 protein reduces tamoxifen and N-(4-hydroxyphenyl)retinamide inhibitory effects in breast cancer cells. Lab Invest 85(11):1357–1367
Ahn SM, Jeong SJ, Kim YS et al (2004) Retroviral delivery of TIMP-2 inhibits H-ras-induced migration and invasion in MCF10A human breast epithelial cells. Cancer Lett 207(1):49–57
Yoneda T, Sasaki A, Dunstan C et al (1997) Inhibition of osteolytic bone metastasis of breast cancer by combined treatment with the bisphosphonate ibandronate and tissue inhibitor of the matrix metalloproteinase-2. J Clin Invest 99(10):2509–2517
Sacco MG, Cato EM, Ceruti R et al (2001) Systemic gene therapy with anti-angiogenic factors inhibits spontaneous breast tumor growth and metastasis in MMTVneu transgenic mice. Gene Ther 8(1):67–70
Ree AH, Florenes VA, Berg JP et al (1997) High levels of messenger RNAs for tissue inhibitors of metalloproteinases (TIMP-1 and TIMP-2) in primary breast carcinomas are associated with development of distant metastases. Clin Cancer Res 3(9):1623–1628
Vizoso FJ, Gonzalez LO, Corte MD et al (2007) Study of matrix metalloproteinases and their inhibitors in breast cancer. Br J Cancer 96(6):903–911
Jiang Y, Goldberg ID, Shi YE (2002) Complex roles of tissue inhibitors of metalloproteinases in cancer. Oncogene 21(14):2245–2252
Dorrello NV, Peschiaroli A, Guardavaccaro D et al (2006) S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science 314(5798):467–471
Ozpolat B, Akar U, Steiner M et al (2007) Programmed cell death-4 tumor suppressor protein contributes to retinoic acid-induced terminal granulocytic differentiation of human myeloid leukemia cells. Mol Cancer Res 5(1):95–108
Frankel LB, Christoffersen NR, Jacobsen A et al (2008) Programmed Cell Death 4 (PDCD4) is an important functional target of the MicroRNA miR-21 in breast cancer cells. J Biol Chem 283(2):1026–1033
Asangani IA, Rasheed SA, Nikolova DA, et al (2007) MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. Advance online publication October 29
Acknowledgements
We thank Vanity McMurtry and Wendy Schober for their technical assistance. This work was supported in part by the Susan G. Komen Breast Cancer Foundation (to AMT) and the Cancer Center Core Grant CA16672.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nieves-Alicea, R., Colburn, N.H., Simeone, AM. et al. Programmed Cell Death 4 inhibits breast cancer cell invasion by increasing Tissue Inhibitor of Metalloproteinases-2 expression. Breast Cancer Res Treat 114, 203–209 (2009). https://doi.org/10.1007/s10549-008-9993-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-008-9993-5