Abstract
Endocrine treatment of breast cancer is widely applied and effective. However, in advanced disease cases, the tumors will eventually progress into an estrogen-independent and therapy-resistant phenotype. To elucidate the molecular mechanisms underlying this endocrine therapy failure, we applied retroviral insertion mutagenesis to identify the main genes conferring estrogen independence to human breast cancer cells. Estrogen-dependent ZR-75-1 cells were infected with replication-defective retroviruses followed by selection with the anti-estrogen 4-hydroxy-tamoxifen. In the resulting panel of 79 tamoxifen-resistant cell lines, the viral integrations were mapped within the human genome. Genes located in the immediate proximity of the retroviral integration sites were characterized for altered expression and their capacity to confer anti-estrogen resistance when transfected into breast cancer cells. Out of 15 candidate BCAR (breast cancer anti-estrogen resistance) genes, seven (AKT1, AKT2, BCAR1, BCAR3, EGFR, GRB7, and TRERF1/BCAR2) were shown to directly underlie estrogen independence. Our results show that insertion mutagenesis is a powerful tool to identify BCAR loci, which may provide insights into the molecular and cellular mechanisms of breast tumor progression and therapy resistance thereby offering novel targets for the development of tailor-made therapeutical and prevention strategies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endocrine therapy of breast cancer has been applied widely and has proven to be effective. Breast tumor development and recurrence can be reduced by long-term anti-estrogen treatment with tamoxifen (or novel selective estrogen receptor modulators) and aromatase inhibitors [1]. In addition, tamoxifen and other endocrine treatment regimens have shown clinical benefit in advanced disease [2]. Tamoxifen competes with estrogen for the estrogen receptor (ER) α and interferes effectively with estrogen signaling. However, in many instances in advanced disease, endocrine treatments ultimately fail due to the development of therapy resistance. Notwithstanding the fact that estrogen antagonists and aromatase inhibitors exhibit alternative modes of action, in nearly all cases tumor cells eventually become estrogen-independent for growth. Despite the detailed insights in ER function, the mechanism of this general therapy failure is still poorly understood [3–7]. Gene expression profiling has become a powerful tool to identify gene sets associated with disease progression and clinical outcome [8–11]. However, only a minority of these genes hint at the underlying mechanisms, and most just represent markers. Here we have undertaken an extensive study into the mechanisms leading to estrogen independence using a genome-wide functional screen based on retrovirus-mediated insertion mutagenesis.
Insertion mutagenesis aims to introduce single genetic modifications in the target cells in a random manner leading to an altered and selectable phenotype. Retroviruses are well suited for this purpose since they integrate in a fairly random manner into the target cell genome as part of their life cycle. As a consequence, a particular target gene may become disrupted. More importantly, the presence of strong promoter and enhancer elements in the retroviral LTRs may increase the expression of genes in the immediate proximity [12]. Subsequently, retroviral sequences may be used as a tag to isolate the target loci. The identification of a common Virus Integration Site (cVIS, i.e. a small region on the genome with multiple integration events in independent cell clones) strongly indicates that the retrovirus inactivated or modulated expression of a target gene that underlies the selected phenotype. In recent years, large numbers of cVIS have been defined using high throughput technologies for various types of malignancies in mice [12–14].
The human ZR-75-1 breast cancer cell line is completely dependent on estrogen for growth in tissue culture and a representative in vitro model for ER-positive breast cancer. Under anti-estrogen-supplemented culture conditions, ZR-75-1 cell proliferation is completely blocked. In a proof of principle experiment, we have demonstrated that the introduction of a single gene (EGFR) can transform this phenotype [15]. Moreover, random modulation of gene expression by DNA demethylation also caused estrogen-independent cell proliferation [16]. These results provided the basis to initiate a genome-wide functional screen using replication-defective murine retrovirus for the identification of BCAR genes contributing to estrogen-independent breast cancer cell proliferation in vitro [17]. In this manuscript, we describe the comprehensive analysis of the integration events of the defective retrovirus and the target genes responsible for estrogen independence of breast cancer cells. Furthermore, the relevance of these BCAR genes in human breast cancer is established. Our results document the power of such an unbiased strategy and also provide insight into the multiple pathways contributing to evasion of anti-estrogen therapy in breast cancer patients.
Materials and methods
Cell culture
Culture of human ZR-75-1 breast cancer cells was performed as described previously [18]. Transfection experiments in ZR-75-1 cells with candidate BCAR genes were done with FuGENE 6 (Roche Diagnostics GmbH, Mannheim, Germany). Pools of transfectants were selected with G418 (Invitrogen) and subsequently assayed for growth in the presence of 4-hydroxy-tamoxifen (OH-Tam, Sigma–Aldrich Chemie BV, Zwijndrecht, The Netherlands) as described previously [18–20]. The panel of tamoxifen-resistant cell lines was previously generated by retrovirus infection and selection for OH-Tam resistance [17]. In brief, ZR-75-1 cells were infected with amphotropic, defective murine retrovirus and plated in medium containing 1 μM of OH-Tam. Within 5 weeks after start of selection, proliferating colonies were individually picked and expanded to stable cell lines (further details in the Supplementary M&M, Text S1).
General molecular procedures
To identify the retrovirus integration sites, we have performed inverse and Splinkerette PCR on high molecular weight genomic DNA from these cell lines. The inverse PCR procedure for circularized genomic DNA fragments has been detailed previously [17, 21]. For the Splinkerette PCR [22], the restriction enzyme-specific adaptor was ligated to digested genomic DNA. PCR was performed using primers selected for the retrovirus, and for the Splinkerette adaptor. Products were purified on agarose gel, sequenced and mapped on the human genome reference sequence assembly by BLAST or BLAT analyses (http://www.ncbi.nlm.nih.gov/blast/ and http://genome.ucsc.edu/cgi-bin/hgGateway). Genes located in the immediate proximity of the integration site were identified by manual inspection. Further details are provided in the Text S1.
For Northern blot analysis total RNA was isolated using RNAzol B (Campro Scientific, Veenendaal, NL). Ten microgram total RNA per lane was size fractioned on 1.2% agarose-formaldehyde gel and transferred onto nitrocellulose. Equivalent gel loading was judged from the Ethidium Bromide-stained 28S and 18S rRNA bands and from GAPD control hybridizations. Probes were derived from cDNA clones or generated by gene-specific PCR, and sequence verified. Sequence analysis was done on a LI-COR sequencer (LICOR, Inc., Lincoln, NE) using Thermo Sequenase DYEnamic direct cycle sequencing kits (Amersham plc, Buckinghamshire, United Kingdom) or on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA) using ABI Prism BigDye Terminators v3.0 cycle sequencing kits (Applied Biosystems) according to the protocols of the manufacturers. Probe labeling and hybridization was performed as previously described [16, 18]. Generation of expression constructs for candidate genes was achieved by inserting sequence verified cDNAs into the LZRS-IRES-Neo expression vector [19].
Protein analyses
BCAR2 antibodies against purified His-tagged polypeptides corresponding to a 25 kDa central and a 23.5 kDa C-terminal fragments of BCAR2α (Text S1) isolated from bacteria were raised in rabbits (Eurogentec Nederland BV, Maastricht, NL). Additional antibodies directed against EGFR and AKT2 (Upstate Biotechnology, Inc., Lake Placid NY), total AKT (Cell Signaling Technology, Inc. New England Biolabs, Hitchin, UK), and GRB7 (N-20, Santa Cruz Biotechnology, Santa Cruz, CA) were used according to the recommendations of the supplier. Western blot analyses using equivalent amounts of protein and ECL detection were performed as detailed in Text S1.
Results
Insertion mutagenesis of human breast cancer cells in vitro
To initiate this functional screen, over 800 million ZR-75-1 cells were subjected to infection with replication-defective retrovirus. From these infected cultures, proliferating colonies were recovered within 5 weeks after start of the selection with 4-hydroxy-tamoxifen (OH-Tam) and expanded to 79 cell lines exhibiting stable cell proliferation in the presence of this anti-estrogen. In contrast, no colonies were recovered from 200 million of mock-infected cells within this time interval [17]. These results strongly suggested that growth and colony formation in the presence of OH-Tam could only occur when a virus integration activated a growth control pathway independent of estrogen signaling.
Common Virus Integration Sites (cVIS), harboring the putative target gene [12], were identified using integration site specific probes and Southern blotting techniques, or Splinkerette PCR in combination with direct sequencing of the PCR products [17, 18, 22]. The majority of the integration sites of the defective virus were recovered and mapped onto the human genome reference sequence assembly and their most proximal genes were identified (Table S1). The distribution of the 160 integration sites over the human genome revealed 12 cVIS in independently derived cell lines, which were unlikely (P < 0.005) the consequence of random virus integration (Table 1). In each of these cVIS, the individual integration events were in near proximity (from 1 up to 107 kb) of a single target gene (Tables 1 and S1). All integration sites not belonging to the cVIS loci were considered as unique Virus Integration Sites (uVIS) in our cell panel. Since our selection system is nearly background free, it is strongly suggested that each cell line harbors a single VIS causative for estrogen independence (Table S2).
The hallmark of a target gene in a retroviral integration locus should be the alteration of its expression [12]. To identify targets, Northern blot analyses were performed for the adjacent gene(s) in the respective cell lines (Fig. 1, and summarized in Table 1). Genes found to exhibit altered expression as a consequence of the virus integration were subsequently introduced into estrogen-dependent ZR-75-1 cells and evaluated for their capacity to confer estrogen-independent cell proliferation. Genes meeting all these criteria represent bona fide BCAR genes causing tamoxifen resistance in these human breast cancer cells. Brief descriptions of the retroviral targets in cVIS and selected uVIS are presented below. Further details are provided in the Supplementary Results (Text S2).
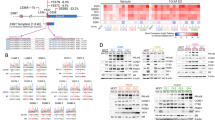
Retrovirus insertion modulates expression of flanking gene. Positions of ribosomal 28S RNA (◀) or 18S RNA (–) on the northern blots are given. Bands of interest are highlighted (◁). Abbreviations used: ZR = ZR-75-1 cells; IB indicates immuno blot probed with antibody for the indicated target. Hybridization with GAPD or ethidium bromide staining of rRNA was used for blot loading control. (a) Northern blots are shown for cVIS8 (EGFR ∼10 kb mRNA) and GAPD (∼1.3 kb). Western blot analysis of EGFR protein (∼170 kDa) was performed following immune precipitation of cell lysates with anti-EGFR. Positions of marker proteins (217 and 123 kDa) are indicated. (b) Northern blots for cVIS1 (BCAR1 ∼3.2 kb mRNA). (c) cVIS2 (BCAR2/TRERF1 ∼5 and 8 kb mRNAs) northern blots are shown. Protein lysates of virus-induced cVIS2 cell line (VIII-24) and control cVIS1 and cVIS3 cell lines were probed with BCAR2 antibody (αBCAR2/D). The position of the ∼130 kDa BCAR2 protein is indicated. (d) Northern blots for uVIS1 (AKT1 ∼2.7 kb) and cVIS12 (AKT2 ∼3.5 kb) and western blots for AKT proteins. (e) Northern blot analysis for uVIS2 (GRB7 ∼3–3.5 kb and ERBB2 ∼4.8 kb). Western blots were probed with antibodies directed against ERBB2 (markers positions 200 and 150 kDa) and the amino terminus of GRB7 (marker positions 75, 50 and 37 kDa), respectively
Targets in virus integration sites
Proof of concept for the insertion mutagenesis protocol was provided by the identification of the cVIS8 (Table 1). Expression levels of EGFR mRNA and protein were markedly enhanced in cell lines with a virus integration in cVIS8, while completely undetectable in the parental cells (Fig. 1a). Previously, the EGFR gene was used to demonstrate that single genetic alterations could transform the hormone dependency of ZR-75-1 cells [15] and was recently identified in transduction experiments of cDNA expression libraries [20]. From these observations it is concluded that EGFR was the target of the virus and caused the tamoxifen-resistant phenotype of these cell lines.
The power of the Splinkerette PCR and chromosome mapping protocol was illustrated by the finding of an additional integration event in the cVIS1 and two within the cVIS3 locus. These integrations occurred outside of the regions previously investigated with conventional Southern blotting [17, 18]. Northern analysis confirmed that the distant virus integration upstream of BCAR1 indeed increased its expression level (Fig. 1b).
The search for the target gene within the cVIS2/BCAR2 locus, identified by cell fusion-mediated gene transfer [21], revealed increased levels of BCAR2 mRNAs (∼5 and 8 kb, Fig. 1c). Sequence analysis of full-length cDNAs revealed two major splice variants, one (BCAR2β) encoding a 1,200 amino acid protein identical to TREP-132/TRERF1 and one unique variant containing 20 additional amino acids due to alternative splicing at the end of exon 8 (BCAR2α) [23, Text S1]. Rabbit polyclonal antibodies directed against BCAR2 revealed increased TRERF1 protein levels (approximately 130 kDa) in cell line VIII-24 (carrying the integration in the cVIS2 locus) compared with other ZR-75-1-derived cell lines (Fig. 1c, bottom).
In cVIS12, integrations in the proximity of AKT2 caused altered mRNA and protein levels (Table 1; Fig. 1d). As a close family member of AKT2, the integration near AKT1 (uVIS1) was further investigated (Table 1). In this particular cell line, the levels of the AKT1 mRNA and protein were clearly increased (Fig. 1d), strongly suggesting that this target was responsible for tamoxifen resistance. The integration event in uVIS2 was downstream of ERBB2 and positioned within intron 11 of GRB7. mRNA and protein levels of both genes were determined and showed that only GRB7 was modulated by the virus (Fig. 1e).
Northern blot analyses also identified NCOR2 (cVIS4), CITED2 (cVIS9), ZADH2 (cVIS6), TLE3 (uVIS3), SRC (uVIS5), RHOBTB3 (uVIS6), SETBP1 (uVIS7), and GLN3 (uVIS20) as putative BCAR genes with altered expression in the cell lines carrying a viral integration (Table 1 and Text S2). In contrast, the nearest targets within cVIS5 and cVIS10 appeared not modulated by the virus (Table 1). Another 10 genes in other uVIS were not regulated by the virus (Text S2). In approximately 30 tamoxifen-resistant cell lines candidate BCAR genes remain to be identified from the different integrations present (Table 1).
Retrovirus targets causing estrogen independence
Alteration of expression as a consequence of the virus integration does not provide proof for the causative role of the particular gene. To verify the involvement in estrogen-independent cell proliferation, several candidate genes were transfected into the parental cells and assayed. Cells with over-expression of the putative BCAR gene were tested for colony formation or proliferation in the presence of anti-estrogen. Transfection of EGFR, BCAR1 or BCAR3 into ZR-75-1 indeed caused estrogen-independent, and tamoxifen-resistant cell proliferation [15, 18–20]. Cell pools transfected with expression constructs carrying no insert or one of the BCAR2/TRERF1 variants, were tested in the presence of G418 plus estrogen or G418 plus OH-Tam in a colony assay. BCAR2/TRERF1 cells were able to form OH-Tam-resistant colonies within 18 days (BCAR2α and BCAR2β, respectively 360 and 110 colonies per 1,000 colonies formed with estrogen), while control vector-transfected cells failed to produce OH-Tam-resistant colonies (none out of nearly 40,000 colonies formed with estrogen). From these results we conclude that TRERF1 was the viral target responsible for estrogen independence in our cell model.
Pools of transfected cells carrying the AKT2 expression constructs supported estrogen-independent cell proliferation, while vector control cells failed (Fig. 2a). Similarly, transfected cells carrying the AKT1 expression construct also proliferated in the presence of anti-estrogen OH-Tam (Fig. 2b). A comparable estrogen-independent growth potential of AKT1-transfected cells was observed in the presence of the pure anti-estrogen ICI 182,780 (not shown). Following the same strategy, GRB7 transfected cells were also able to proliferate in the presence of anti-estrogen (Fig. 2c). These experiments demonstrate that both AKT family members and GRB7 were targets of retroviral integration events essential for the tamoxifen-resistant proliferation potential of these cell lines.
Estrogen-independent cell proliferation induced by AKT2, AKT1 and GRB7. Cell pools were generated by transfection of ZR-75-1 cells with empty LZRS-IRES-Neo vector or vector containing a cDNA of the complete coding region of AKT2, AKT1 or GRB7, respectively. Independent pools of transfected cells were expanded by G418 selection and subsequently plated in triplicate at a density of 0.7–1.0 million cells in medium containing 1 μM of OH-Tam. Cells were harvested at the indicated time points and re-plated at the initial density. Vector controls are shown with open symbol and dashed lines, transfectants are presented as solid symbols and lines. Cumulative cell numbers and SD are presented. The inserts show immuno blots of lysates of cell pools carrying the control vector (C) or the expression construct (T) probed with antibodies for AKT2 (panel A), total AKT (B) and GRB7 (C), respectively
Discussion
Insertion mutagenesis as a functional screen for estrogen independence
The primary goal was to identify genes causing estrogen-independent proliferation of estrogen-dependent human breast cancer cells. The strategy of mapping nearly all virus integration events in our tamoxifen-resistant cell panel has assigned 43 out of 79 cell lines to 12 cVIS. The target gene conferring the estrogen-independent phenotype has been conclusively identified for five of these cVIS and for two uVIS (Table 1). In several other VIS, a strong suggestion for the involvement of a particular gene (CITED2, NCOR2, RHOBTB3, SRC, TLE3, and ZADH2) is based on its altered transcription as a result of the nearby integration of a virus. But formal proof by gene transfer or knock down remains required in these cases. In some cVIS cases, clear candidate target genes have not yet been assigned. For cVIS7 this was caused by the absence of documented genes, while in other cases expression changes as a consequence of the virus integration events were not observed for the immediately adjacent genes (for example cVIS5 and cVIS10). This suggests the existence of yet undefined (non-) coding genes in the vicinity of the virus and/or the long-range action of the integrated virus on distantly located genes [13, 14]. In the tamoxifen-resistant cell lines not belonging to a cVIS, the responsible target genes remain to be determined. This implies that another 30 different BCAR genes hide out among the uVIS loci present in these cell lines (Tables 1 and S2).
Mechanisms of estrogen independence of breast cancer cells
This study demonstrates that AKT1, AKT2, BCAR1, BCAR3, EGFR, GRB7, and TRERF1 individually play a causative role in estrogen-independent and anti-estrogen-resistant proliferation of ZR-75-1 breast cancer cells. The proliferation capacity of the transfectant cells with over-expression of various genes (AKT1, BCAR1, BCAR3, and EGFR) was retained in the presence of a pure anti-estrogen as well, indicating that growth was not stimulated by tamoxifen through the ER [15, 19, 24]. The list of (candidate) target genes of retrovirus insertion mutagenesis causing estrogen independence comprises several functional categories of cellular signaling, i.e. cell surface receptor (EGFR), protein kinases (AKT1, AKT2, SRC), adaptor molecules (GRB7, BCAR3, BCAR1, RHOBTB3), and various transcription regulators (TRERF1, NCOR2, CITED2, TLE3). Analysis of literature and bioinformatics resources provides many functional links between these genes and some have been implicated in anti-estrogen-resistant cell proliferation.
Within the cytoplasmic compartment of the cell, signaling from the receptor tyrosine kinases along the RAS-ERK and/or the PI3K-AKT pathway involves several proteins (ERBB2, EGFR, PDGFRA and B, NRG1, FGF17, activated HRAS and RAF1, AKT1) contributing to estrogen independency [for review 4, 7, 20, 25–28]. Adaptor proteins like GRB7, BCAR1 and BCAR3 may modulate the outcome of these main cytoplasmic signaling routes through selective recruitment of other players [29–31]. In addition to its adaptor function, BCAR3 may also act as a GDP exchange factor for RAC1 [32], and in the cross talk between insulin and estrogen signaling [33]. RHOBTB3, a member of a subfamily of Rho-GTPases, may also contribute to small GTPase signaling and modulation of the cytoplasmic signaling events. Within the nuclear compartment, NCOR2 participates in a co-repressor complex resulting in chromatin condensation and may also modulate ligand dependency of hormone receptors and contribute to estrogen independency [34–36]. TRERF1 was reported to negatively modulate breast cancer cell proliferation by up-regulation of G1 cyclin-dependent kinase inhibitors and through co-activation of the progesterone receptor [23, 37]. Whether the transcription regulators CITED2 and TLE3 fit into the same pathways as the cytoplasmic targets or represent alternative signaling routes, remains an intriguing question.
BCAR genes and progression of breast cancer
In breast disease, RHOBTB3 belongs to a gene set capable of classifying tumors with evidence of hypoxia [38] and TRERF1 appears to be down-modulated in breast cancer in comparison to normal epithelium [23]. High expression of EGFR is associated with poor response to tamoxifen treatment [39]. Disruption of the AKT pathway by loss of PTEN, and mutation of PIK3CA and AKT1 is considered to contribute to the development and progression of breast cancer [40]. Furthermore, infrequent AKT2 amplification is observed in breast cancer [41]. High phospho-AKT and low AKT2 levels have been found associated with reduced survival of adjuvant tamoxifen-treated breast cancer patients [42]. We have demonstrated that BCAR1 protein levels are prognostic for disease recurrence and predictive for response to tamoxifen treatment [43–45], and BCAR1 also may be associated with HER2-Neu driven tumorigenesis [46, 47]. GRB7 belongs to the 21 gene set (Oncotype DX assay) for breast cancer prognosis prediction and is often co-amplified with ERBB2 in breast cancer [9, 29] and may contribute to the transformed cell phenotype [48]. We have applied quantitative RT-PCR to establish relations between the transcript levels of our (candidate) genes and the clinical parameters of breast cancer. These studies have shown that the majority of these BCAR genes are associated with tamoxifen resistance and/or tumor aggressiveness of breast cancer (Van Agthoven et al. manuscript in preparation).
In conclusions, this report documents that our non-biased functional screen provides a set of genes which contribute to estrogen independence in breast cancer cells and, at least in part, associate with clinical breast cancer progression. Future studies will provide detailed insights in the escape routes available to the tumor cells during endocrine therapy and may allow for the development of targeted treatments to block the progression of the disease.
References
Early Breast Cancer Trialists’ Collaborative Group (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365:1687–1717
Jaiyesimi IA, Buzdar AU, Decker DA et al (1995) Use of tamoxifen for breast cancer: twenty-eight years later. J Clin Oncol 13:513–529
Ali S, Coombes RC (2002) Endocrine-responsive breast cancer and strategies for combating resistance. Nat Rev Cancer 2:101–112
Clarke R, Liu MC, Bouker KB et al (2003) Antiestrogen resistance in breast cancer and the role of estrogen receptor signaling. Oncogene 22:7316–7339
Osborne CK, Shou J, Massarweh S et al (2005) Crosstalk between estrogen receptor and growth factor receptor pathways as a cause for endocrine therapy resistance in breast cancer. Clin Cancer Res 11:865s–870s
Lewis JS, Jordan VC (2005) Selective estrogen receptor modulators (SERMs): mechanisms of anticarcinogenesis and drug resistance. Mutat Res 591:247–263
Riggins RB, Schrecengost RS, Guerrero MS et al (2007) Pathways to tamoxifen resistance. Cancer Lett doi:10.106/j.canlet.2007.2003.2016
van de Vijver MJ, He YD, van’t Veer LJ et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009
Paik S, Shak S, Tang G et al (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351:2817–2826
Jansen MP, Foekens JA, van Staveren IL et al (2005) Molecular classification of tamoxifen-resistant breast carcinomas by gene expression profiling. J Clin Oncol 23:732–740
Foekens JA, Atkins D, Zhang Y et al (2006) Multicenter validation of a gene expression-based prognostic signature in lymph node-negative primary breast cancer. J Clin Oncol 24:1665–1671
Uren AG, Kool J, Berns A et al (2005) Retroviral insertional mutagenesis: past, present and future. Oncogene 24:7656–7672
Neil JC, Cameron ER (2002) Retroviral insertion sites and cancer: fountain of all knowledge? Cancer Cell 2:253–255
Theodorou V, Kimm MA, Boer M et al (2007) MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet 39:759–769
Van Agthoven T, Van Agthoven TLA, Portengen H et al (1992) Ectopic expression of epidermal growth factor receptors induces hormone independence in ZR-75-1 human breast cancer cells. Cancer Res 52:5082–5088
Van Agthoven T, Van Agthoven TLA, Dekker A et al (1994) Induction of estrogen independence of ZR-75-1 human breast cancer cells by epigenetic alterations. Mol Endocrinol 8:1474–1483
Dorssers LCJ, Van Agthoven T, Dekker A et al (1993) Induction of antiestrogen resistance in human breast cancer cells by random insertional mutagenesis using defective retroviruses: Identification of bcar-1, a common integration site. Mol Endocrinol 7:870–878
Van Agthoven T, Van Agthoven TLA, Dekker A et al (1998) Identification of BCAR3 by a random search for genes involved in antiestrogen resistance of human breast cancer cells. EMBO J 17:2799–2808
Brinkman A, Van der Flier S, Kok EM et al (2000) BCAR1, a human homologue of the adapter protein p130Cas and antiestrogen resistance in breast cancer cells. J Natl Cancer Inst 92:112–120
Meijer D, Van Agthoven T, Bosma PT et al (2006) Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res 4:379–386
Dorssers LCJ, Veldscholte J (1997) Identification of a novel breast-cancer-anti-estrogen-resistance (BCAR2) locus by cell-fusion-mediated gene transfer in human breast-cancer cells. Int J Cancer 72:700–705
Mikkers H, Allen J, Knipscheer P et al (2002) High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet 32:153–159
Gizard F, Robillard R, Barbier O et al (2005) TReP-132 controls cell proliferation by regulating the expression of the cyclin-dependent kinase inhibitors p21WAF1/Cip1 and p27Kip1. Mol Cell Biol 25:4335–4348
Dorssers LCJ, Van Agthoven T, Brinkman A et al (2005) Breast cancer oestrogen independence mediated by BCAR1 or BCAR3 genes is transmitted through mechanisms distinct from the oestrogen receptor signalling pathway or the epidermal growth factor receptor pathway. Breast Cancer Res 7:R82–R92. doi:10.1186/bcr1954
Dorssers LCJ, Van der Flier S, Brinkman A et al (2001) Tamoxifen resistance in breast cancer: elucidating mechanisms. Drugs 61:1721–1733
Stoica GE, Franke TF, Moroni M et al (2003) Effect of estradiol on estrogen receptor-alpha gene expression and activity can be modulated by the ErbB2/PI 3-K/Akt pathway. Oncogene 22:7998–8011
de Graffenried LA, Friedrichs WE, Russell DH et al (2004) Inhibition of mTOR activity restores tamoxifen response in breast cancer cells with aberrant Akt Activity. Clin Cancer Res 10:8059–8067
Glaros S, Atanaskova N, Zhao C et al (2006) Activation function-1 domain of estrogen receptor regulates the agonistic and antagonistic actions of tamoxifen. Mol Endocrinol 20:996–1008
Kairouz R, Parmar J, Lyons RJ et al (2005) Hormonal regulation of the Grb14 signal modulator and its role in cell cycle progression of MCF-7 human breast cancer cells. J Cell Physiol 203:85–93
O’Neill GM, Fashena SJ, Golemis EA (2000) Integrin signalling: a new cas(t) of characters enters the stage. Trends Cell Biol 10:111–119
Riggins RB, Thomas KS, Ta HQ et al (2006) Physical and functional interactions between Cas and c-Src induce tamoxifen resistance of breast cancer cells through pathways involving epidermal growth factor receptor and signal transducer and activator of transcription 5b. Cancer Res 66:7007–7015
Felekkis KN, Narsimhan RP, Near R et al (2005) AND-34 activates phosphatidylinositol 3-kinase and induces anti-estrogen resistance in a SH2 and GDP exchange factor-like domain-dependent manner. Mol Cancer Res 3:32–41
Yu Y, Hao Y, Feig LA (2006) The R-Ras GTPase mediates cross talk between estrogen and insulin signaling in breast cancer cells. Mol Cell Biol 26:6372–6380
Shou J, Massarweh S, Osborne CK et al (2004) Mechanisms of tamoxifen resistance: increased estrogen receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J Natl Cancer Inst 96:926–935
Perissi V, Rosenfeld MG (2005) Controlling nuclear receptors: the circular logic of cofactor cycles. Nat Rev Mol Cell Biol 6:542–554
Keeton EK, Brown M (2005) Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol Endocrinol 19:1543–1554
Gizard F, Robillard R, Gross B et al (2006) TReP-132 is a novel progesterone receptor coactivator required for the inhibition of breast cancer cell growth and enhancement of differentiation by progesterone. Mol Cell Biol 26:7632–7644
Dressman HK, Hans C, Bild A et al (2006) Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin Cancer Res 12:819–826
Nicholson S, Halcrow P, Farndon JR et al (1989) Expression of epidermal growth factor receptors associated with lack of response to endocrine therapy in recurrent breast cancer. Lancet i:182–185
Brugge J, Hung MC, Mills GB (2007) A new mutational AKTivation in the PI3K pathway. Cancer Cell 12:104–107
Bellacosa A, De Feo D, Godwin AK et al (1995) Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer 64:280–285
Kirkegaard T, Witton CJ, McGlynn LM et al (2005) AKT activation predicts outcome in breast cancer patients treated with tamoxifen. J Pathol 207:139–146
Van der Flier S, Brinkman A, Look MP et al (2000) Bcar1/p130Cas protein and primary breast cancer: prognosis and response to tamoxifen treatment. J Natl Cancer Inst 92:120–127
Dorssers LCJ, Grebenchtchikov N, Brinkman A et al (2004) The prognostic value of BCAR1 in patients with primary breast cancer. Clin Cancer Res 10:6194–6202
Dorssers LCJ, Grebenchtchikov N, Brinkman A et al (2004) Application of a newly developed ELISA for BCAR1 Protein for prediction of clinical benefit of tamoxifen therapy in patients with advanced breast cancer. Clin Chem 50:1445–1447
Defilippi P, Di Stefano P, Cabodi S (2006) p130Cas: a versatile scaffold in signaling networks. Trends Cell Biol 16:257–263
Cabodi S, Tinnirello A, Di Stefano P et al (2006) p130Cas as a new regulator of mammary epithelial cell proliferation, survival, and HER2-neu oncogene-dependent breast tumorigenesis. Cancer Res 66:4672–4680
Kao J, Pollack JR (2006) RNA interference-based functional dissection of the 17q12 amplicon in breast cancer reveals contribution of coamplified genes. Genes Chromosomes Cancer 45:761–769
Acknowledgements
We acknowledge Anton Berns (Amsterdam) for the splinkerette protocol, and Fred Sweep and Nicolai Grebenchtchikov (Nijmegen) for their support in generating antibodies. Excellent support of various current and former members of our research laboratory is gratefully acknowledged. For stimulating discussions and support, we are indebted to Ad Brinkman, Danielle Meijer, John Foekens, Maxime Look, Leendert Looijenga, Els Berns, John Martens, Maurice Jansen and Riccardo Fodde. Funding: This study was supported by grants of the Dutch Cancer Society (DDHK96-1245 & 99-1883), the Susan G. Komen Breast Cancer Foundation (BCTR0100675), the Association for International Cancer Research (04–148) and the Josephine Nefkens Stichting.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ton van Agthoven and Jos Veldscholte contributed equally to this research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
van Agthoven, T., Veldscholte, J., Smid, M. et al. Functional identification of genes causing estrogen independence of human breast cancer cells. Breast Cancer Res Treat 114, 23–30 (2009). https://doi.org/10.1007/s10549-008-9969-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-008-9969-5