Abstract
Three known non-synonymous polymorphisms (Ala394Thr, Ser471Leu and Pro690Ala) in the largest circadian gene, Neuronal PAS domain protein 2 (NPAS2), were genotyped in a breast cancer case-control study conducted in Connecticut, USA (431 cases and 476 controls). We found that women with the heterozygous Ala394Thr genotype were significantly associated with breast cancer risk compared to those with the common homozygous Ala394Ala (OR = 0.61, 0.46–0.81, P = 0.001). This is the first evidence demonstrating a role of the circadian gene NPAS2 in human breast cancer, suggesting that genetic variations in circadian genes might be a novel panel of biomarkers for breast cancer risk.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Disruption of the circadian rhythm has been reported to increase the risk of breast cancer in several epidemiologic studies that involved women working night shifts [1–3]. These findings suggest a role of circadian rhythms in breast tumorigenesis and lead to the circadian gene hypothesis, which states that genetic variants in genes responsible for maintaining circadian rhythms might act as a panel of biomarkers associated with an individual’s risk of breast cancer [4]. The hypothesis is supported by emerging data from animal models that demonstrate a substantial impact of circadian genes on several tumor-related biological pathways such as cell proliferation, cell cycle control, and apoptosis [5]. However, very few studies have tested this hypothesis in human populations.
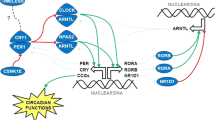
NPAS2, the largest circadian gene, maps on chromosome 2 at 2q11.2 and covers 176.68 kb according to the NCBI Build 35 (www.ncbi.nlm.nih.gov/IEB/Research/Acembly). It encodes for a member of the basic helix-loop-helix-PAS class of transcription factors and is expressed primarily in the mammalian forebrain. NPAS2 forms heterodimers with BMAL1 and then transcriptionally activates expression of the circadian genes per and cry, which are required for maintaining biological rhythms in many organisms [6, 7]. In animal studies, altered circadian patterns of sleep and behavioral adaptability have been observed in NPAS2-deficient mice [8].
The public NCBI dbSNP database (www.ncbi.nlm.nih.gov/SNP) contains 560 single nucleotide polymorphisms (SNPs) identified in the NPAS2 gene, of which the SNP rs2305160 (A/G) is the only non-synonymous mutation altering an amino acid at codon 394, Ala394Thr. Two more non-synonymous mutations, Ser471Leu (hCV2153849) and Pro690Ala (hCV25757546), were subsequently found in the Celera SNP database. The current case-control study aimed to screen these three NPAS2 polymorphisms for 431 breast cancer patients and 476 healthy controls in order to test whether these non-synonymous mutations could serve as circadian biomarkers associated with breast cancer risk.
Materials and methods
Study population
Study subjects came from a recently completed breast cancer case-control study conducted in Connecticut. All breast cancer cases, aged 30–80 years old, had no previous diagnosis of cancer except for nonmelanoma skin cancer. Detailed information regarding the study population was described previously [9]. Briefly, the cases were histologically confirmed breast cancer patients from New Haven County and Tolland County, Connecticut, USA. For New Haven County, eligible cases were identified from the major hospital of the county, the Yale-New Haven Hospital (YNHH), through the computer database system at the Department of Surgical Pathology. Controls were also randomly selected from the computer database from women who were histologically confirmed to be without breast cancer. The participation rates were 77% for cases and 71% for controls in New Haven County. For Tolland County, since there was no major hospital in this county, newly diagnosed breast cancer cases were identified from area hospital records by the Rapid Case Ascertainment system at the Yale Comprehensive Cancer Center. Controls from Tolland County were recruited through random digit dialing methods for those under age 65 and randomly selected from Health Care Financing Administration (HCFA) files for those aged 65 and over. The participation rates were 74% for cases and 64% for controls in Tolland County.
Data collection
Informed consent was obtained from all study participants prior to the collection of epidemiological data by personal interview. In addition to demographic characteristics of cases and controls, data on menstrual and reproductive factors (such as age at menarche, age at menopause, and lifetime lactation history) and family history of breast cancer were obtained. Menopausal status was assessed at the time of diagnosis. At the completion of the interview, blood was drawn into sodium-heparinized tubes for immediate DNA isolation and subsequent molecular analysis.
TaqMan genotyping
The TaqMan Assay was used to determine genotypes using archived genomic DNA isolated from peripheral blood lymphocytes. Assays-on-Demand primers and probes were C_15976652_10, C_2153849_10 and C_25757546_10 for SNPs rs2305160, hCV2153849 and hCV25757546 respectively (Applied Biosystems, Inc., Foster City, CA). The TaqMan assays were performed following manufacture’s instructions (Applied Biosystems, Inc.,) in MX3000P instrument (Stratagene, La Jolla, CA). Each genotyping plate contained positive and negative controls. Approximately 10% of the samples were duplicated to assure quality control in genotyping and two reviewers separately performed genotype scoring to confirm results.
Statistical analysis
All statistical analyses were performed using the STATA statistical software (StataCorp.; College Station, TX). Genotype frequencies at SNP loci in the control population were first checked for compliance with Hardy–Weinberg equilibrium. Genotype data were analyzed with the homozygotes of the common allele as the reference group. Adjusted Odds ratios (ORs) with 95% confidence intervals (CI) were calculated by logistic regression to estimate the relative risk associated with different genotypes. The analyses were adjusted by age, family history of breast cancer in first-degree relatives, study site, and reproductive factors including lifetime months of breast feeding, age at menarche, menopausal status, age at first full-term birth and number of live births.
Results
Table 1 presents the distribution of selected baseline characteristics for cases and controls. There were significantly more postmenopausal women in the case population (76.6%) than that in controls (65.5%), indicating an increased risk of breast cancer associated with menopausal status. No other baseline factors showed a material difference between the cases and controls.
Genotyping data was available for 431 breast cancer cases and 476 controls in this study. We achieved 99% of genotype concordance for our quality control samples. Statistical analyses were performed for two SNPs, rs2305160 and hCV2153849, which changed the amino acid at codon 394 and 471 respectively. The third SNP hCV25757546 had a very low minor allele frequency (<0.5%) and was therefore not included in our analyses. The Hardy–Weinberg equilibrium of genotype frequencies was met in control groups at both loci rs2305160 (chi2 = 2.23, P = 0.135) and hCV2153849 (chi2 = 1.46, P = 0.293).
Among all of these participants, logistic regression analysis showed that individuals with the heterozygous Ala394Thr genotype were significantly associated with breast cancer risk compared to those having the common Ala394Ala genotype after adjustment for breast cancer confounding factors (OR = 0.61; 95% CI: 0.46–0.81, P = 0.001) (Table 2). However, no association was detected between the homozygous Thr394Thr genotype and breast cancer risk (OR = 1.00; 95% CI: 0.63–1.57, P = 0.988). After stratifying by menopausal status, the significant relationship of the heterozygous Ala394Thr genotype with breast cancer was observed in both premenopausal and postmenopausal women. Interestingly, this heterozygous effect was more evident in premenopausal women (OR = 0.44, 95% CI 0.25–0.77, P = 0.004) than in postmenopausal women (OR = 0.65, 95% CI 0.46–0.91, P = 0.014).
Analysis of the SNP hCV2153849 (Ser471Leu) did not yield any significant results. Here we only reported the results for all subjects, with adjustments for race, but a data analysis of only Caucasians provided very similar findings. Moreover, analyses for two study sites separately yielded similar results as well.
Discussion
Our findings demonstrate for the first time an association of the NPAS2 Ala394Thr variation with risk for breast cancer, supporting the hypothesis that genetic variations in circadian genes may confer inherited susceptibility to breast cancer. This is in accordance with our previous observations that associate a structural genetic variation in another circadian gene, PERIOD3 (PER3), with breast cancer risk [10].
The possible involvement of NPAS2 in tumorigenesis has also been supported by an animal model study that found that the BMAL1/NPAS2 heterodimers might mediate the promoter of the oncogene c-myc and suppress its transcription [11]. Moreover, a heterozygous missense mutation in the NPAS2 Leu471Ser has been recently linked to the reduced risk of human seasonal affective disorder (SAD) [12], in which circadian rhythms such as diurnal preference have been suggested to play a role [13]. Recent findings of ours have also demonstrated a robust association of the NPAS2 variant Thr genotypes (Ala/Thr & Thr/Thr) with reduced risk of non-Hodgkin’s lymphoma [14]. Therefore, NPAS2 might serve as biomarkers for an individual’s risk of human cancers, although explicit mechanisms are currently far from clear.
The greater impact of the Ala394Thr genotype on breast cancer risk among young people corresponded with the suggested theory that breast cancer occurrence among young women is more likely related to inherited genetic factors. Moreover, the many environmental factors that can accrue with age may attenuate any estimate of the genetic role in the disease. For example, there is evidence that shows circadian variation of endothelial function is present in pre-menopausal women but disappears after menopause [15]. It is also possible that circadian genes may play a role in hormone regulation associated with menopausal status, which could account for its different impact between pre- and post-menopausal women. For example, VEGF is a growth factor and modulator of endothelial growth and function, and VEGF levels predict breast cancer prognosis. Recent findings suggest that expression of VEGF is modulated in cancer cells both by the circadian and menstrual/estrous cycles [16, 17]. This circadian and menstrual clock controlled VEGF gene and protein expression may be responsible not only for the menstrual cycle modulation of breast cancer outcome in premenopausal women [18] but also for the early post operative breast cancer recurrence discovered by screening mammography in young women with breast cancer [19].
An interesting finding in our study is the heterozygote effect of the NPAS2 genotypes in risk estimate of breast cancer. A previous study reported a similar observation that the NPAS2 heterozygous Leu471Ser genotype was associated with the reduced risk for human seasonal affective disorder (SAD) [12]. This heterozygote effect, however, may be due to the small number of individuals with the homozygous variant that limits the statistical power for detecting any associations in our current study.
The non-synonymous mutation (rs2305160, Ala394Thr) may have several potential impacts on NPAS2 functionalities. First, an amino acid change (Ala → Thr) caused by this polymorphism may alter the NPAS2 protein structure. Second, our bioinformatics search (ProDom; http://prodes.toulouse.inra.fr/prodom/current/html/form.php) predicts that the NPAS2 Ala394Thr is located in the PAS sensory domain. Therefore, it may affect protein sensory capacity directly or interfere with NPAS2/BMAL1 heterodimerization, which is essential for normal sensory function. Third, based on our search using the ESEfinder [20], this polymorphism (A/G) affects the DNA sequences of a putative exonic splicing enhancer (ESE) responsive to the human SR protein SRp55, which may influence the alternative splicing of the NPAS2.
In summary, our findings provide the first evidence linking the circadian gene NPAS2 and breast cancer risk. As a group of genes regulating many fundamental biological processes, circadian genes might be a novel panel of promising biomarkers for breast cancer risk and prognosis that warrants further investigation. A potential limitation of our study is that we included benign breast disease patients as the control group in New Haven County and a population-based control group in Tolland County. However, as described earlier, the multivariate analysis by study sites reached the same conclusions as the combined analysis. Further work is needed to confirm our findings, including replication in independent large sample sets, preferably using methods to control for the risk of population stratification as well as studies of the possible biological effects of NPAS2 genotypes.
References
Davis S, Mirick DK, Stevens RG (2001) Night shift work, light at night, and risk of breast cancer. J Natl Cancer Inst 93:1557–1562
Schernhammer ES, Laden F, Speizer FE et al (2001) Rotating night shifts and risk of breast cancer in women participating in the nurses’ health study. J Natl Cancer Inst 93:1563–1568
Lie JA, Roessink J, Kjaerheim K (2006) Breast cancer and night work among Norwegian nurses. Cancer Causes Control 17:39–44
Stevens RG (2005) Circadian disruption and breast cancer: from melatonin to clock genes. Epidemiology 16:254–258
Fu L, Lee CC (2003) The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 3:350–361
Vitaterna MH, King DP, Chang AM et al (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264:719–725
Bunger MK, Wilsbacher LD, Moran SM et al (2000) Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103:1009–1017
Dudley CA, Erbel-Sieler C, Estill SJ et al (2003) Altered patterns of sleep and behavioral adaptability in NPAS2-deficient mice. Science 301:379–383
Zheng T, Holford TR, Mayne ST et al (2000) Risk of female breast cancer associated with serum polychlorinated biphenyls and 1,1-dichloro-2,2’-bis(p-chlorophenyl)ethylene. Cancer Epidemiol Biomarkers Prev 9:167–174
Zhu Y, Brown HN, Zhang Y, Stevens RG, Zheng T (2005) Period3 Structural Variation: A circadian biomarker associated with breast cancer in young women. Cancer Epidemiol Biomarkers Prev 14:268–270
Fu L, Pelicano H, Liu J, Huang P, Lee C (2002) The circadian gene Period2 plays an important role in tumor suppression and DNA damage response in vivo. Cell 111:41–50
Johansson C, Willeit M, Smedh C et al (2003) Circadian clock-related polymorphisms in seasonal affective disorder and their relevance to diurnal preference. Neuropsychopharmacology 28:734–739
Lam RW, Levitan RD (2000) Pathophysiology of seasonal affective disorder: a review. J Psychiatry Neurosci 25:469–480
Zhu Y, Leaderer D, Guss C et al (2007) Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer 120:432–435
Walters JF, Hampton SM, Deanfield JE et al (2006) Circadian variation in endothelial function is attenuated in postmenopausal women. Maturitas 54:294–303
Koyanagi S, Kuramoto Y, Nakagawa H et al (2003) A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res 63:7277–7283
Wood PA, Du-Quiton J, You S, Hrushesky WJ (2006) Circadian clock coordinates cancer cell cycle progression, thymidylate synthase, and 5-fluorouracil therapeutic index. Mol Cancer Ther 5:2023–2033
Hrushesky WJ, Bluming AZ, Gruber SA, Sothern RB (1989) Menstrual influence on surgical cure of breast cancer. Lancet 2:949–952
Retsky M, Demicheli R, Hrushesky W (2001) Breast cancer screening for women aged 40–49 years: screening may not be the benign process usually thought. J Natl Cancer Inst 93:1572
Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3:285–298
Acknowledgments
This study was supported by funds from Yale University. The work was also supported by the NIH grants CA62986, CA110937, CA108369, ES11659 and CA122676.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, Y., Stevens, R.G., Leaderer, D. et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk . Breast Cancer Res Treat 107, 421–425 (2008). https://doi.org/10.1007/s10549-007-9565-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10549-007-9565-0